Abstract
Dicer is an RNase III enzyme responsible for cleaving double stranded RNAs into small interfering RNAs and microRNAs, which either target mRNA transcripts for degradation, or inhibit translation. Dicer protein levels have been examined in breast cancer with contradictory results. Our goal was to resolve whether Dicer levels differ in breast cancer versus normal breast epithelium, and between estrogen receptor alpha positive (ER+) or negative (ER−) primary breast cancers.
We compared three different Dicer antibodies: Abcam 4A6, Abcam ab5818, and Sigma HPA000694, using immunohistochemistry and western blot analyses. All three Dicer antibodies detected higher levels of Dicer in ER+ breast cancer cell lines versus ER−, and all three recognized exogenous overexpressed Dicer. In clinical specimens, all three antibodies detected higher Dicer in ER+ breast cancers versus triple-negative breast cancer (TNBC), but had very different staining patterns by immunohistochemistry on the same tumor samples.
Using the optimal antibody, ab5818, selected for its sensitivity and specificity, Dicer protein expression was significantly higher in ER+ versus TNBC clinical specimens of primary tumor (p <0.0001, unpaired t-test). Dicer was also significantly higher in adjacent normal breast epithelium versus TNBC (p<0.0001, paired t-test, n=18 pairs).
Differences in antibody performance may explain contrasting results observed in the literature regarding Dicer protein in breast cancer. If Dicer becomes more clinically relevant as a prognostic indicator, further antibody optimization and standardization will be critical.
Keywords: Dicer, breast cancer, estrogen receptor, TNBC, Immunohistochemistry
1. Introduction
Human Dicer is an RNase III enzyme responsible for cleaving double stranded RNAs (dsRNAs) into small interfering RNAs (siRNA) or microRNAs (miRNA). These small RNAs are then incorporated into a multiprotein RNA-induced silencing complex (RISC) which uses them as templates for targeting specific mRNAs leading to their degradation or inhibiting their translation. siRNAs and miRNAs play major roles in both development and disease [1].
Dicer is a haploinsufficient tumor suppressor since dysfunctional miRNA processing leads to enhanced transformation and tumorigenesis [2–4]. However, studies of Dicer levels in relation to tumor progression and outcome in various cancers have demonstrated conflicting results [5–8]. For example, in breast cancer, a gradual loss of Dicer protein with disease progression was detected by immunohistochemistry (IHC) in clinical breast cancer specimens (normal > ductal carcinoma in situ > invasive carcinoma > nodal metastases) [9], while other studies have reported higher Dicer expression in nodal metastases compared to primary tumors by quantitative real-time polymerase chain reaction (qRT-PCR) [10], and IHC [11].
Overall, estrogen receptor alpha positive (ER+) breast cancers have a better prognosis than triple-negative breast cancers (TNBC), that by definition lack ER, progesterone receptor (PR), and amplification of human epidermal growth factor receptor 2 (HER2) [12–14]. Our group previously published that Dicer protein was higher in ER+ versus TNBC cell lines, and that increasing microRNA 200c (miR-200c) in TNBC cell lines to levels found in ER+ lines resulted in dramatically increased Dicer protein, as well as increased expression of the miRNAs characteristically higher in ER+ cell lines [15].
To validate our findings in clinical specimens of ER+ and TNBC, we evaluated the performance of Dicer antibodies for IHC on formalin-fixed paraffin-embedded (FFPE) specimens. Dicer monoclonal antibodies 13D6, 4A6 (made from the same immunogen as 13D6) (Abcam), and polyclonal antibodies ab5818 (Abcam) and HPA000694 (Sigma) were compared using western blot and IHC analyses. We also examined endogenous Dicer, exogenous Dicer from an expression vector, and manipulation of Dicer via restoration of miR-200c. Lastly, we tested ER+ and TNBC clinical specimens for Dicer protein by IHC. While the antibodies used performed similarly for western blot analyses, major differences were observed in cellular staining patterns of Dicer by IHC using the various antibodies, highlighting the necessity for rigorous antibody performance evaluation before drawing definitive conclusions regarding Dicer protein in breast or any other cancer, developmental state, or tissue. Based on results obtained with the highest performing Dicer antibody, we conclude that Dicer is significantly lower in TNBC than in ER+ breast cancer or adjacent non-involved breast epithelium.
2. Materials and Methods
2.1. Human Tissues
ER+ Breast Cancer: Postmenopausal women (n=25) with newly diagnosed ER+ breast cancer, grade 1–3, stage II/III were included in this study. The protocol (01-627) was approved by the Colorado Multiple Institutional Review Board (COMIRB) and informed consent was provided by all patients. Triple-Negative Breast Cancer: Patients (n=21) ranged in age from 19–72 years old with a mean age of 47.44 ± 12.03 years. All tumors were grade 3 and negative for ER, PR and HER2 (COMIRB protocol 04-0066).
2.2. Cell Culture
T47D breast cancer cells, which are ER+ and belong to the Luminal A subtype, were grown in Minimum Essential Medium (MEM), 5% fetal bovine serum (FBS), non-essential amino acids (NEAA), and insulin. The TNBC cell line BT549 was grown in Roswell Park Memorial Institute medium (RPMI), 10% FBS, and insulin. HEY ovarian cancer cells were grown in RPMI, 10% FBS, and L-glutamine. HEK293FT human embryonic kidney cells were grown in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% FBS. All cells were maintained at 37 degrees celsius (°C) and 5% carbon dioxide, and fingerprinted for authenticity using the Identifiler DNA profiling kit (ABI, Grand Island, NY, USA) at the University of Colorado Cancer Center Sequencing Core Facility.
2.3. miR-200c Inducible Cells
BT549 cells were transduced with a doxycycline (DOX) inducible lentiviral vector (pTRIPz) encoding the precursor sequence for miR-200c (pTRIPz-200c) and stably selected using puromycin. A clone of BT549-TripZ-200c, demonstrating robust expression of miR-200c upon induction with little background, was used in all subsequent experiments. BT549-TripZ-200c cells were plated at a density of 8 × 105 cells per 10cm dish, and miR-200c expression was induced with 1 μg/mL DOX for 48 hours.
2.4. miR-200c Inducible Xenograft Tumors
These methods are previously described [16].
2.5. Antibodies
Primary antibodies to Dicer were optimized and used at the following concentrations: mouse monoclonal 13D6 (1:25 for IHC) and 4A6 (made from the same immunogen as 13D6, and selected over 13D6) (1:25 for IHC, and 1:50 for western blot (WB)), rabbit polyclonal ab5818 (1:50 for IHC and WB) (Abcam, Cambridge, MA, USA) (note: ab5818 is currently sold as PA5-19437 ThermoFisher, Grand Island, NY, USA), and rabbit polyclonal HPA000694 (1:50 for IHC, 1:500 for WB) (Sigma-Aldrich, St. Louis, MO, USA). Additional antibodies used for western blot include Topoisomerase I (TOPOI) (C-21) (Santa Cruz Biotechnology Inc., Dallas, TX, USA), α-tubulin clone B-5-1-2, and Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) (Sigma).
2.6. Blocking Peptide
A blocking peptide was commercially available for antibody ab5818 (Abcam ab24556). Peptide at 10× the concentration of antibody was incubated with antibody ab5818 rotating overnight at 4°C prior to use on tissue or western blot.
2.7. Immunohistochemistry
Sections were cut at 4μm and heat immobilized for 1 hour at 60°C on charged glass slides. Sections were deparaffinized in three changes of xylene and a series of graded ethanols. Heat induced epitope retrieval was performed in a Decloaking Chamber (Biocare Medical, Concord, CA, USA) at 125°C for 5 min. in 10mM citrate buffer pH 6.0. Sections were blocked in 3% hydrogen peroxide followed by 2.5% normal horse serum before incubation for 1 hour with primary antibody. Tris buffered saline with Tween (0.05%) (TBST) was used for all washes, and the Vectastain Universal Elite ABC horseradish peroxidase (HRP) kit (Vector Laboratories, Burlingame, CA, USA) was used for antibody detection, followed by 3,3′-diaminobenzidine (Dako, Carpinteria, CA, USA) for chromagen visualization.
2.8. IHC Score
All clinical breast cancer slides were analyzed by a pathologist (PJ) for intensity (0–4 scale) and % positive cells stained which were multiplied to obtain a score.
2.9. Dicer Overexpression
HEK293FT cells were plated at 7.5 × 105 cells per 10cm dish. The following day, cells were transfected with media plus transfection reagent without DNA (mock) or 2μg of pCMV-Entry plasmid containing the 5769 base pair open reading frame of Human Dicer1, ribonuclease type III, transcript variant 2 (TruORF® OriGene Technologies, Rockville, MD, USA) using Lipofectamine 2000 (Invitrogen, Grand Island, NY, USA). After 48h, cells were harvested for protein analysis.
2.10. Western Blot
The NE-PER Nuclear and Cytoplasmic Extraction kit (ThermoFisher) was used for nuclear/cytosolic protein extraction. Whole cell protein extracts were prepared using Radioimmunoprecipitation assay (RIPA) lysis buffer, and 50 or 90μg of protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene fluoride (PVDF) membrane. 3% bovine serum albumin in TBST (0.1% Tween) was used for all blocking and antibody dilution. Secondary antibodies used were goat anti-mouse or rabbit Alexa Fluor® 680 (Molecular Probes, Grand Island, NY, USA). All blots were scanned using an Odyssey infrared imager (LI-COR Biosciences, Lincoln, NE, USA).
2.11. qRT-PCR
Total RNA was collected using the RNeasy Plus kit (Qiagen, Valencia, CA, USA). Mature miR-200c (#002300) and U6 small nuclear RNA (#001973, used as a house-keeping gene for normalization) were reverse transcribed using the TaqMan microRNA Reverse Transcription Kit and expression levels were analyzed using TaqMan Universal PCR Master Mix from Life Sciences and an Applied Biosystems 7900HT qRT-PCR machine (ThermoFisher).
2.12. Statistics
The Mann-Whitney non-parametric t-test was used to compare unpaired groups of data, and a paired t-test was used in cases where data were paired. Both analyses were two-tailed.
2.13. Bioinformatics
Dicer gene expression data was obtained from www.oncomine.org, using the METABRIC dataset [17]. A total of 1950 patients were included in this analysis, with 440 patients having ER− tumors and 1510 having ER+ tumors.
3. Results
3.1. Dicer Antibody Characteristics
We initially tested 4 different antibodies to human Dicer (Fig. 1). Mouse monoclonal antibody 13D6 (Abcam) [8, 9, 11, 18, 19] and 4A6 (Abcam), are made from the same immunogen, which is proprietary but contains the central region of Dicer (Clonegene, Hartford, CT, USA) (Fig. 1). Since antibody 4A6 had stronger IHC staining compared to 13D6 (not shown) it was therefore selected for further analysis. Rabbit polyclonal antibody HPA000694 (Sigma) was selected due to its extensive use on various tissues by the Human Protein Atlas project (www.proteinatlas.org). HPA000694 targets the Piwi Argonaut Zwille (PAZ) domain (Fig. 1). Rabbit polyclonal antibody ab5818 (Abcam) was selected because it was one of the few Dicer antibodies with a commercially available blocking peptide when this project was initiated. Ab5818 targets a sequence that is 3′ to the PAZ domain and contains the first 5 residues of the Ribonuclease IIIa domain (Fig. 1).
Figure 1. Dicer antibody overview.
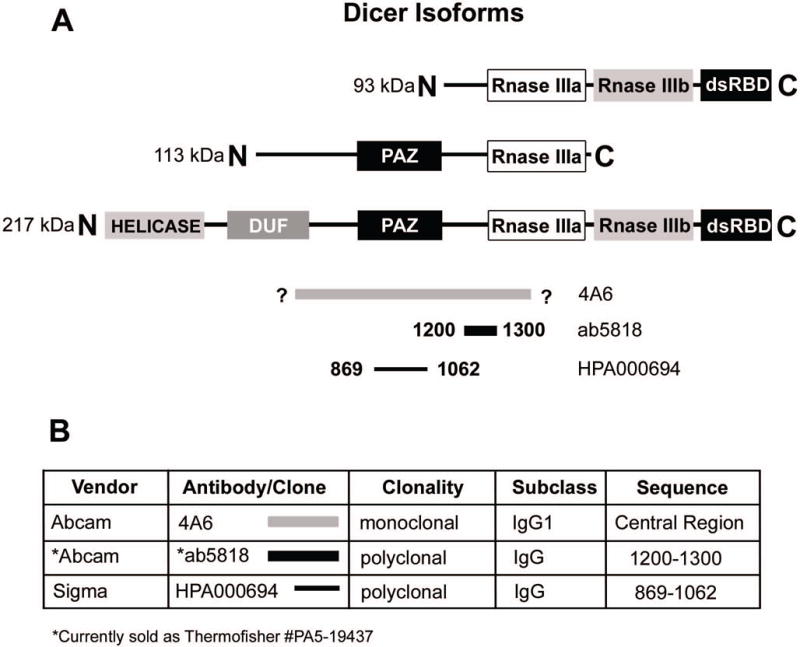
A) Schematic of Dicer protein, its isoforms, and antibody immunogen sequence homology. HELICASE = Helicase domain, DUF = Domain of unknown function, PAZ = PAZ domain, Rnase III (a and b) = ribonuclease III c terminal domain(s), dsRBD = double stranded RNA binding domain. 93kD and 113kD = isoforms according to Grelier et al. 217kD = full length Dicer protein. Gray line: Clone 4A6, sequence proprietary/unknown corresponding to the central region of Dicer, bold black line: ab5818, residues 1200–1300 including the Rnase IIIa domain, thin black line: HPA000694, residues 869–1062, including the PAZ domain. B) Summary of antibody information.
3.2. Antibody Comparison of Endogenous Dicer Protein from Breast Cancer Cell Lysates
To examine specificity of the above antibodies, we performed a side-by-side comparison by western blot analysis. T47D and BT549 whole cell lysates were loaded in triplicate on an SDS PAGE gel, transferred to PVDF, and probed separately with ab5818, 4A6, and HPA000694. All three antibodies showed higher expression of Dicer in T47D (an ER+ breast cancer cell line) versus BT549 (a basal-like TNBC cell line), as previously reported [15]. 4A6 had a strong signal for Dicer with minimal cross-reactivity (fewer lower molecular weight bands) compared to the other antibodies. Both antibodies ab5818 and HPA000694 had more non-specific bands than 4A6, but they also showed clear Dicer signal, with HPA000694 demonstrating the strongest Dicer signal of all three antibodies at the same exposure (Fig. 2).
Figure 2. Antibody comparison of Dicer expression in ER+ versus triple-negative breast cancer cell lines.
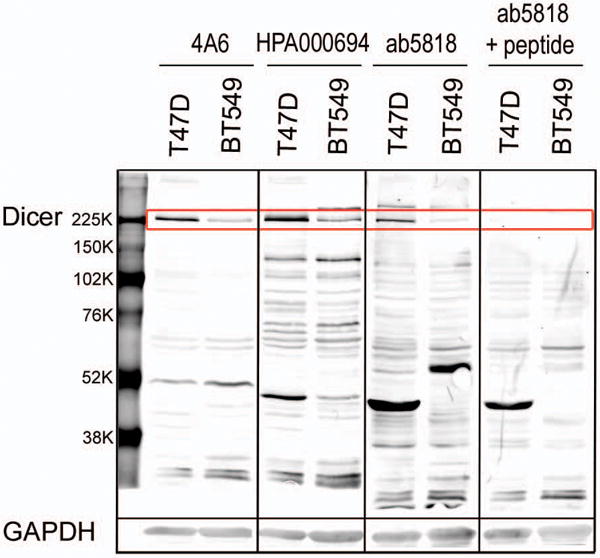
All blots were scanned at similar intensity setting. Whole cell lysate of T47D (ER+) and BT549 (triple-negative) breast cancer cell lines probed for Dicer with 4A6, HPA000694, ab5818, and ab5818+10× blocking peptide (Dicer 217kD band highlighted in red box). GAPDH loading control corresponding to the above membrane 35 kD.
The synthetic peptide specific for ab5818, when incubated at a concentration of 10 times the primary antibody, effectively blocked Dicer signal of both ER+ (T47D) and TNBC (BT549) lysates Fig. 2).
3.3. Antibody Comparison of Overexpressed Dicer
To further test specificity of the antibodies for Dicer we introduced a Dicer cDNA expression vector to cells. HEK293FT cells were transiently transfected for 48h with either media plus transfection reagent (mock) or the TruORF® pCMV-Entry plasmid containing Human Dicer1 (ribonuclease type III, transcript variant 2). Whole cell extracts were loaded in triplicate and western blot analyses with the three antibodies were performed. All antibodies detected higher amounts of Dicer in extract from Dicer overexpressing cells compared to mock transfected cells (Fig. 3), although each antibody demonstrated unique, fairly strong recognition of various non-specific bands. 4A6 had the least amount of non-specific binding, although there was a strong, likely non-specific signal at 50kD. HPA000694 and ab5818 had many non-specific bands, although they both had only one relatively intense non-specific signal at ~145 kD and ~60 kD respectively (Fig. 3.). It should be noted that these bands are assumed to be non-specific because they appear at equal levels in both the mock and Dicer transfected cells and do not correspond with reported molecular weights of documented Dicer isoforms [18].
Figure 3. Dicer overexpression is well detected by all three antibodies.
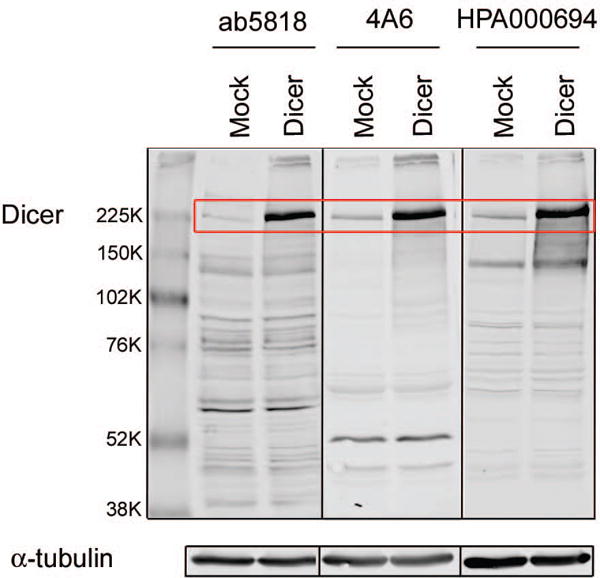
Whole cell extracts of HEK293FT cells mock transfected or transfected with a Dicer cDNA expression plasmid. Dicer 217kD band highlighted in red box. α-tubulin (50kD) was used as a loading control. All blots were scanned at similar intensity.
3.4. Antibody Comparison of Endogenous Dicer in Nuclear versus Cytosolic Cellular Fractions
In the literature, the predominant location and activity of Dicer is in the cytoplasm [20, 21]; however, nuclear Dicer has been reported [19, 22]. Consequently we sought to determine if the antibodies would detect Dicer predominately in the cytoplasmic or nuclear fraction of breast cancer cells. Nuclear and cytosolic extracts were prepared from T47D cells (n=2 biological replicates for each), loaded in triplicate and membranes were probed for Dicer using ab5818, 4A6, or HPA000694 (Fig. 4a). Antibodies recognizing human Topoisomerase I (TOPOI), and α-Tubulin, were used to assess the purity of nuclear and cytosolic fractions respectively. All three antibodies showed higher Dicer expression in the cytosolic fractions compared to the nuclear fractions (Fig. 4a).
Figure 4. Antibody detection of Dicer in nuclear and cytoplasmic fractions and by IHC.
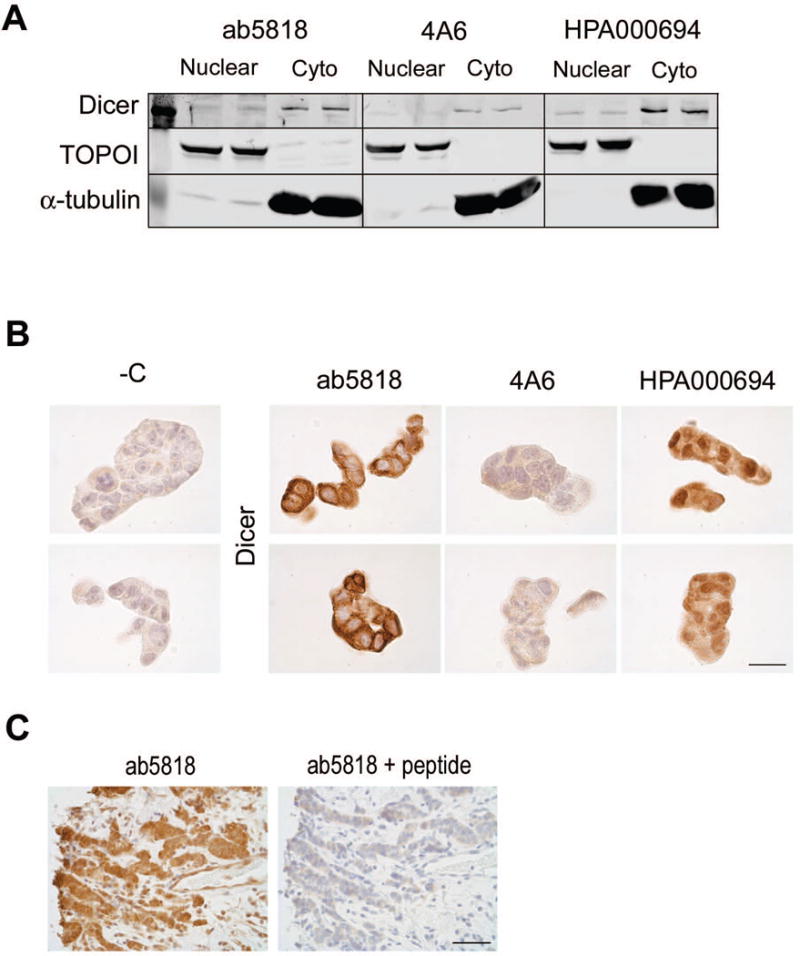
A) T47D cell extracts showing Dicer expression in the nuclear versus cytoplasmic fraction with all three antibodies. TOPOI used as a control for nuclear protein. α-tubulin used as a control for cytoplasmic protein. B) T47D cells, FFPE and stained for Dicer with the three antibodies. Bar = 25μm. C) ER+ Breast cancer stained for Dicer with ab5818 alone (left) and with ab5818+10× blocking peptide (right). Bar = 50μm.
3.5. Antibody Comparison by IHC on FFPE Cells
To evaluate the performance of these antibodies for IHC, T47D cells were FFPE and stained for Dicer (Fig. 4b). All antibodies were optimized using a standardized HRP IHC protocol (5 min at 125°C heat-induced epitope retrieval in 10mM citrate buffer at pH 6.0). We chose the Vectastain Universal Elite ABC kit for IHC optimization because it detects all immunoglobulin G (IgG) classes of primary mouse or rabbit antibodies and has minimal background when used with human tissue (data not shown). Compared to the negative control (-C; no primary antibody, Fig. 4b), antibody ab5818 had strong cytoplasmic staining and 4A6 had weaker cytoplasmic staining, each with minimal staining in the nuclei (Fig. 4b). In contrast, HPA000694 demonstrated strong nuclear staining by IHC. However, when this antibody was used for detection of Dicer in nuclear versus cytosolic lysates it predominately detected Dicer in the cytoplasmic fraction (Fig 4a, b). One possible explanation for this discrepancy is that HPA000694 is recognizing a conformational epitope in the native conditions of the FFPE that is not present under the denaturing and reducing conditions used for the SDS-PAGE gel. To further test the specificity of ab5818 we used the blocking peptide specific to ab5818 which effectively blocked Dicer staining in an ER+ clinical sample (Fig. 4c).
3.6. Antibody Comparison with miR-200c Manipulation
We and others have previously reported that reduced Dicer expression is connected with a global down-regulation of the miRNAome in TNBC and ovarian cancer [15, 23], and that Dicer and the more epithelial mRNAome are increased by restoring miR-200c to poorly differentiated TNBC [15]. Here we use a doxycyline (DOX)-inducible expression vector for miR-200c in both BT549 TNBC cells and HEY ovarian cancer cells to compare the ability of ab5818 and HPA000694 to detect increased endogenous Dicer. BT549 cells stably containing the DOX-inducible miR-200c expression vector were treated with or without DOX for 48h, and cells were harvested for RNA/qRT-PCR and FFPE. DOX-induced cells had a 350-fold increase in miR-200c relative to un-induced cells (p<0.0001 unpaired t-test) (Fig. 5a left). BT549 −/+ DOX FFPE cell pellets were stained for Dicer using ab5818 and HPA000694. Ab5818 was able to detect an increase in cytoplasmic Dicer in the DOX-induced cells compared to un-induced cells (Fig. 5a right, top panel). HPA000694 on the other hand, detected minimal difference between BT549 cells treated with or without DOX, and demonstrated both nuclear and cytoplasmic staining (Fig. 5a right, bottom panel). Likewise, in HEY ovarian cancer cells stably containing the DOX-inducible expression vector for miR-200c grown as xenografts in mice treated with or without DOX for 21 days, the detection of Dicer protein differed when using ab5818 versus HPA000694 (Fig. 5b). Specifically, antibody ab5818 had clean moderate cytoplasmic staining, with higher intensity staining in the miR-200c expressing tumors compared to the non-miR-200c expressing controls (Fig. 5b, left). However, again, as seen in the breast cancer cells, HPA000694 had high nuclear and cytoplasmic signal as well as high background in all tumors regardless of miR-200c expression (Fig. 5b right), further confirming the lack of specificity of this antibody by IHC. HPA000694 does show an increase in Dicer with induction of miR-200c by immunoblot analysis [15].
Figure 5. Antibody IHC comparison with miR-200c manipulation in breast cancer cells and ovarian cancer xenografts.
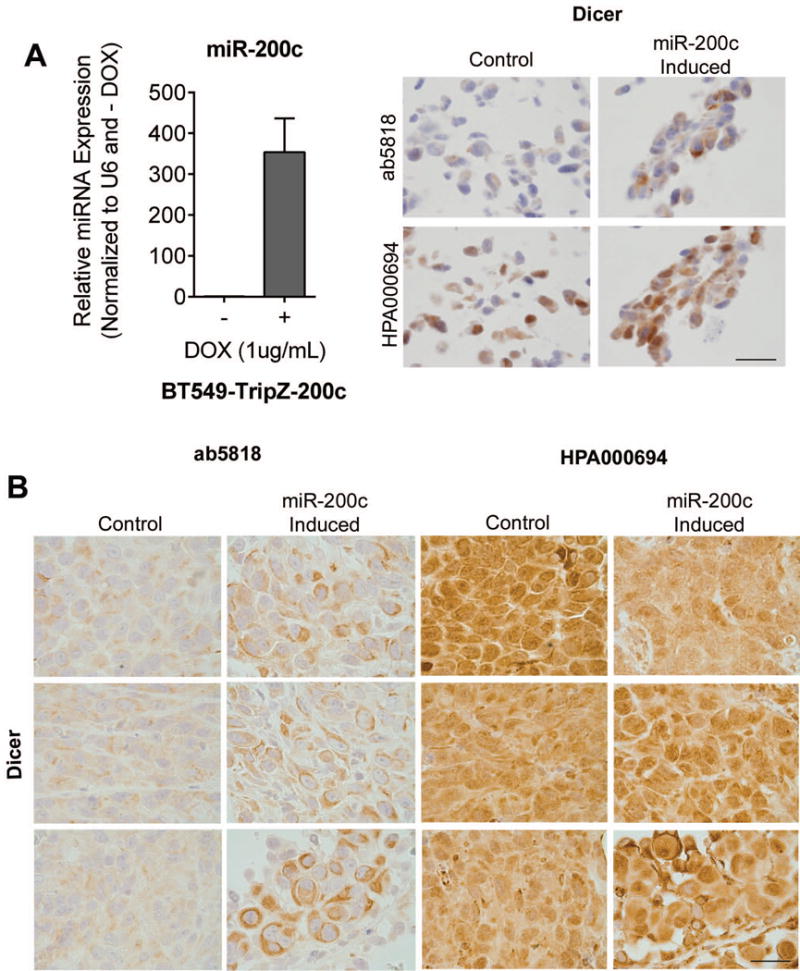
BT549-TRIPz-200c cells were untreated (Control) or DOX treated (miR-200c induced) for 48h. A left) relative qRT-PCR for miR-200c. A right) IHC for Dicer using ab5818 and HPA000694, bar = 25μm. B) HEYTGL xenograft tumors with DOX-inducible miR200c. Untreated (Control) or DOX treated (miR-200c Induced) with 2g/L DOX for 21 days, stained for Dicer using ab5818 and HPA000694. Bar = 25μm.
3.7. Antibody Comparison in Clinical Breast Specimens
We next tested all three antibodies in primary human breast cancer specimens (n=5 ER+ and n=5 TNBC). Although anti-Dicer clone 4A6 was sensitive and specific on immunoblots (Fig. 2–4) it showed weak cytoplasmic staining by IHC at concentrations as high as 1:25, for 1 out of 5 ER+ breast cancers tested, and no staining in TNBCs (Fig 6a, b). Ab5818 showed moderate to strong cytoplasmic staining in all ER+ breast cancers tested, with weaker cytoplasmic staining in TNBC cases (Fig. 6a, b). HPA000694 showed moderate to strong nuclear and cytoplasmic staining in all ER+ breast cancers, and moderate to weak nuclear and cytoplasmic staining in TNBC (Fig. 6a, b). In summary, ab5818 showed the greatest difference between ER+ and ER− (TNBC) and consistently stained the cytoplasm, while HPA000694 showed a less dramatic difference in the ER+ as compared to TNBC cases, and again appeared to stain both the nuclear and cytoplasmic compartments.
Figure 6. Dicer expression in ER+ versus TNBC clinical specimens as detected by IHC with the three different antibodies.
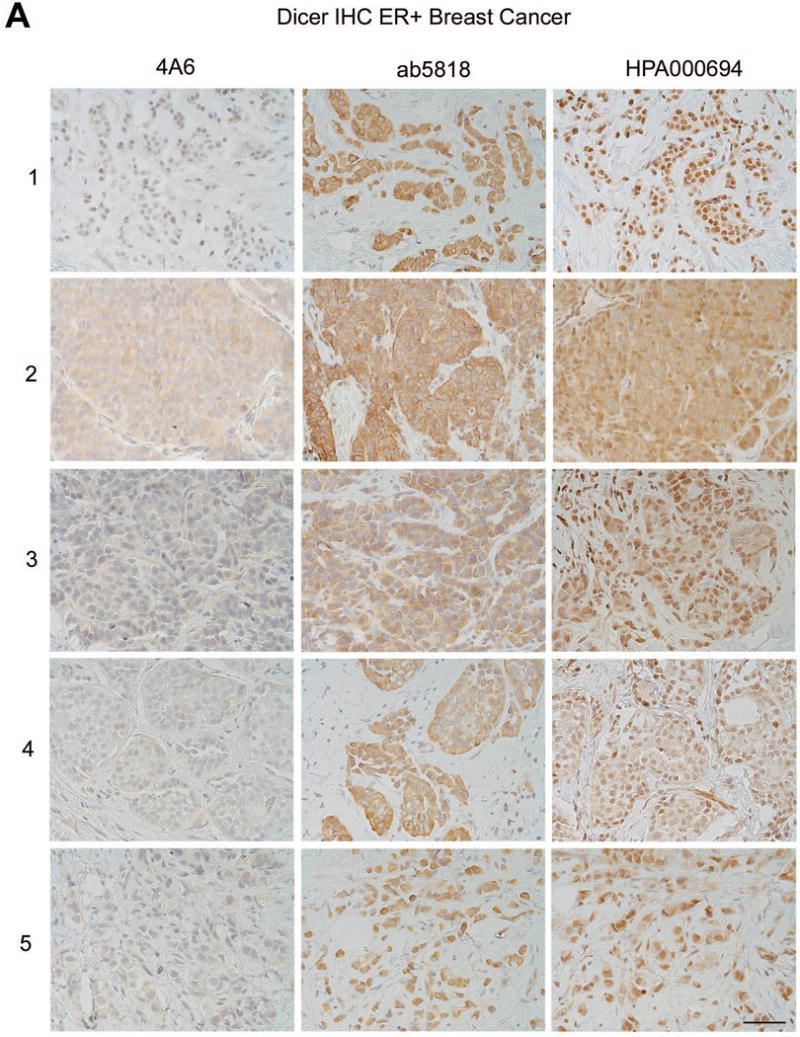
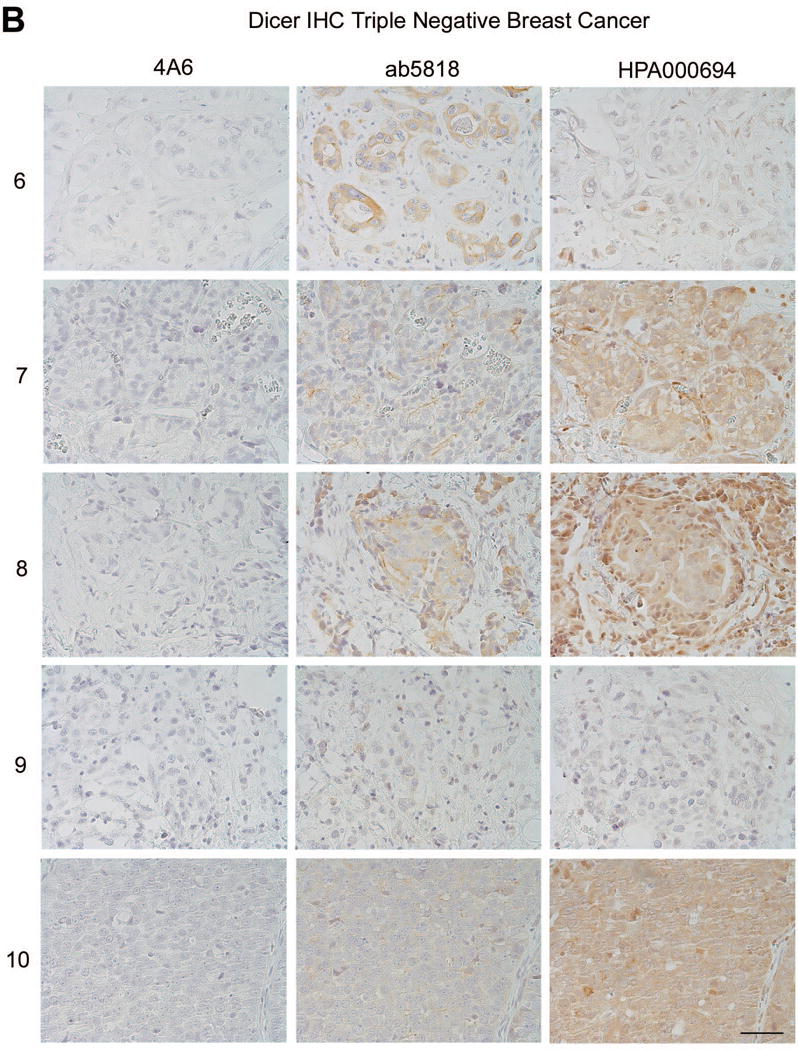
A) ER+ breast cancers each stained with Dicer antibodies 4A6, ab5818, and HPA000694, n=5 bar = 50μm. B) Triple-negative breast cancers each stained with Dicer antibodies 4A6, ab5818, and HPA000694, n=5, bar = 50μm. Numbers = de-identified patient cases, each used in triplicate for comparison between the three antibodies.
3.8. Dicer in Breast Cancer and Normal Breast Epithelium
Antibody ab5818 demonstrated both sensitivity and specificity for Dicer by IHC and WB in human breast cancer specimens, cells, and xenografts. Therefore we chose antibody ab5818 to assess Dicer protein by IHC in a larger cohort of ER+ (n=25) and TNBC (n=21) samples. A pathologist (PJ) scored these tissues for Dicer expression in both tumor and adjacent normal epithelium. All ER+ tumors showed moderate to strong cytoplasmic staining for Dicer, while all triple-negative tumors showed much weaker cytoplasmic staining compared to the ER+ cohort (Fig. 7a, b). ER+ tumors had an average score of 266.2 for Dicer expression while TNBC had an average Dicer score of 88.9 (Fig. 8a, p<0.0001 unpaired t-test). These data were also significant (p<0.0001 unpaired t-test) when plotted as either intensity or % positive cells alone (data not shown).
Figure 7. Differential expression of Dicer in ER+ versus TNBC as detected by ab5818 IHC.
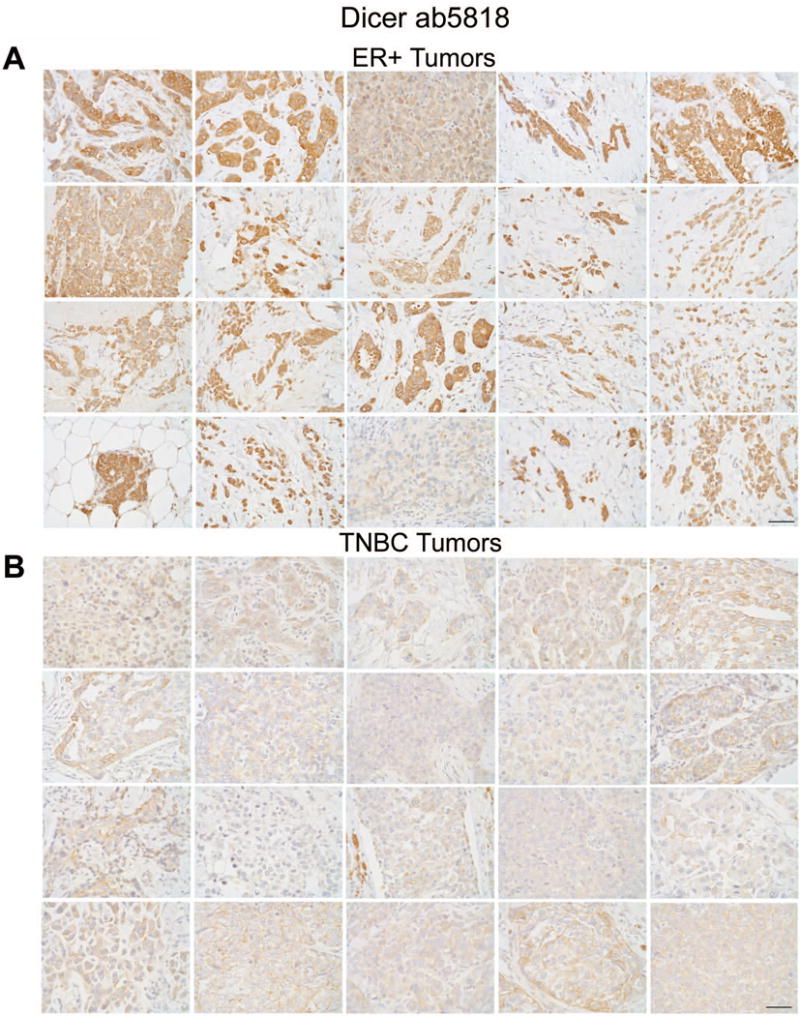
20 representative images of A) ER+ and B) TNBC specimens stained for Dicer using ab5818, bar = 50μm.
Figure 8. Quantification of Dicer expression in breast cancer and in adjacent normal breast.
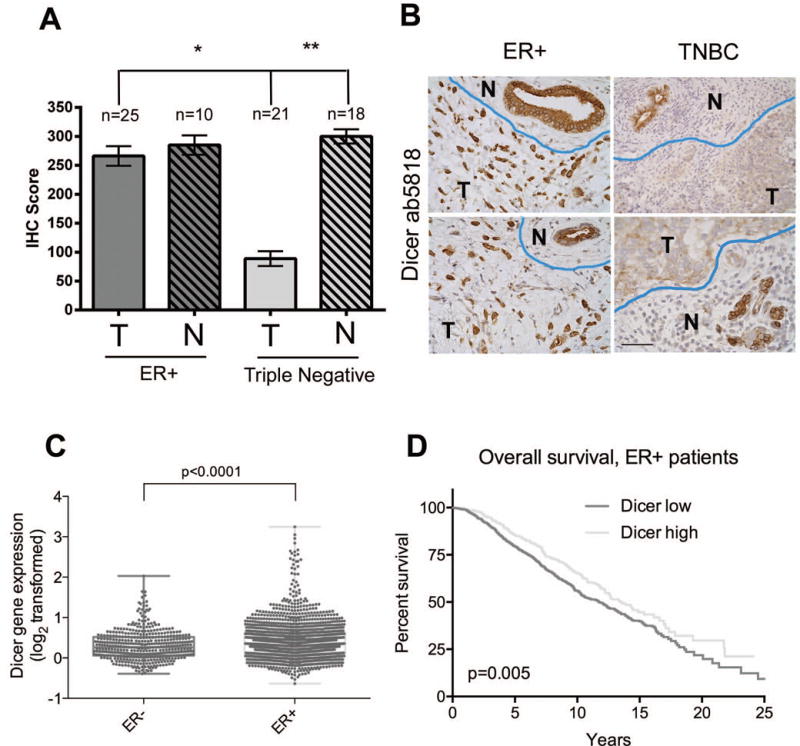
A) ER+ and triple-negative breast cancers and adjacent normal (if present in the tissue section) were stained for Dicer using ab5818, and were scored for Dicer expression by a pathologist (IHC score = intensity rating on a scale of 0–4 multiplied by the percent positive). *p<0.0001, unpaired t-test. **p<0.0001, paired t-test, n=18 pairs. B) Representative images of tumor (T) and adjacent normal ducts (N) defined by blue line in ER+ and TNBC stained for Dicer using ab5818, bar = 50μm. C) Dicer mRNA expression from the METABRIC breast cancer data set [17] in ER− versus ER+ breast cancers, p<0.0001, unpaired t-test. (D) Kaplan-Meier analysis of overall survival (n=1508) of patients with ER+ breast cancer in the METABRIC dataset, based on Dicer1 relative mRNA expression levels. Cutoff for high versus low Dicer1 expression was top 25% versus bottom 75%.
Interestingly, non-involved ‘normal’ epithelium adjacent to either ER+ or TNBC showed consistently high expression for Dicer (an average score of 285 for ER+ adjacent normal, and 300 for triple-negative adjacent normal). Dicer expression was not significantly different for ER+ tumor versus adjacent normal (p = 0.6283), however, in a paired t-test (n=18 pairs) between triple-negative tumor and its adjacent normal epithelium, Dicer was significantly lower in tumor versus normal p<0.0001 (Fig. 8a, b).
4. Discussion
Reports of Dicer expression in breast cancer have been variable. By qRT-PCR, Dicer1 expression was lower in TNBC versus normal breast [10, 24]. However, a third group did not conclude a difference in Dicer1 mRNA between TNBC and normal breast [25]. Other studies indicate that decreased Dicer1 was associated with breast tumor progression and metastasis [18], but there is also contrasting evidence of elevated Dicer1 in tamoxifen-resistant metastatic breast tumors [26]. These inconsistencies even occur within studies where groups have compared Dicer mRNA and protein. For example, Grelier et al report that lower Dicer1 mRNA expression was associated with lower metastasis-free survival, but that protein levels (using Dicer antibody 13D6) were not predictive of survival, and Martello et al report that breast tumors with less intense Dicer staining (using an unspecified Dicer antibody) were associated with metastatic disease, but they did not detect this at the mRNA level [18, 27]. It should be noted that in both cases these studies used different cohorts to compare Dicer mRNA and protein. It would be ideal to compare Dicer mRNA and protein within the same cohort, and two studies performed qRT-PCR on FFPE tissue, but they claim that Dicer evaluation by IHC was problematic or that the Dicer antibodies were suboptimal, and therefore did not show their IHC results [10, 24].
To help clarify this issue, we tested three Dicer antibodies: Abcam ab5818, Abcam 4A6, and Sigma HPA000694 by western blot and IHC comparing ER+ and TNBC, where we have already reported distinct differences in Dicer protein expression between cell lines [15]. All three antibodies detected higher Dicer expression in ER+ breast cancer versus TNBC by western blot, but had very different staining patterns by IHC (intensity levels, percent positivity, and nuclear/cytoplasmic localization) when the exact same tumor samples were compared. Importantly, unlike the other two antibodies, antibody HPA000694 detected high levels of Dicer in the nucleus of T47D cells (Fig. 4b), BT549 cells (with or without miR-200c induction) (Fig. 5a), and in breast cancer (Fig. 6) by IHC.
Small punctate amounts of Dicer have been reported in the nucleus of HeLa [22] and HEK293 [19] cells using confocal microscopy with either a non-commercially available antibody (12B5/4C6), or antibody 13D6, respectively. In the case of the former, digitonin treatment was needed to wash out the cytoplasm in order to visualize Dicer nuclear staining. Most of the literature using bright field microscopy report cytoplasmic Dicer expression in breast cancer by IHC with the exception of one group that exhibited strong nuclear Dicer staining in TNBC by IHC, but the anti-Dicer antibody used for this staining was not completely described [25].
Dicer lacks a canonical nuclear localization signal (NLS) but has been shown to associate with a component of the nuclear pore complex [22]. Other groups have shown that Dicer can shuttle to the nucleus and that the double-stranded RNA binding domain (dsRBD) of Dicer mediates nuclear localization [28, 29]. There are three reported isoforms of Dicer [18]. The full-length isoform is 1922aa (217kD) and additional isoforms include one lacking the helicase and dsRBD (997aa, 113kD), and one that lacks the helicase and PAZ domain (820aa, 93kD) (Fig. 1). Antibody 13D6 recognizes all three isoforms of Dicer [18]. 4A6 by virtue of being made against the same immunogen as 13D6 should recognize all three isoforms as well. However, since 4A6 is monoclonal it is possible that it might not detect the smallest isoform. Ab5818 should recognize all three isoforms considering its target epitope, and HPA000694 should only recognize the two larger isoforms that contain the PAZ domain, one of which is missing the dsRBD. Therefore, the fact that HPA000694 shows nuclear staining cannot be explained by its recognition of a unique isoform that cannot be detected by the other antibodies.
In addition to consistent staining patterns between western blot and IHC, ab5818 had stronger intensity by IHC than that of 4A6, which was weak in our tissue samples at concentrations as high as 1:25 (Fig 6). ab5818 showed clean, distinctive cytoplasmic staining for Dicer that increased with the induction of miR-200c (Fig. 5) which was consistent with our previous findings [15]. Therefore, we decided to use ab5818 for further Dicer IHC analysis of breast cancer specimens. We observed significant differences in Dicer expression between ER+ and TNBC with Dicer being significantly higher in the more well-differentiated ER+ disease (p<0.0001), as well as significantly lower Dicer in TNBC compared to adjacent non-involved epithelium (Fig. 8a, b), consistent with previous reports [9, 10]. These results are also consistent with results reported at the mRNA level in the METABRIC dataset [17] where Dicer1 mRNA was significantly higher in ER+ versus ER− breast cancer tumors (Fig. 8c). Interestingly, ER+ breast cancer patients in the METABRIC dataset [17] whose tumors had higher Dicer1 mRNA expression had significantly longer overall survival (Fig. 8d), and disease-free survival (data not shown) compared to those whose tumors had lower Dicer1.
Dicer is currently being examined as a prognostic indicator, however, the strikingly different results detected with different Dicer antibodies by IHC highlights the need for further optimization and standardization in order to implement use in a clinical setting. For ongoing research as well, the issue of antibody specificity, and why some antibodies detect nuclear Dicer, while others do not, will need to be further examined to insure proper interpretation of results regarding the complex actions of this important enzyme.
Acknowledgments
This work was supported by Department of Pathology funds and DOD Idea Expansion Grant BC122994 W81XWH-13-1-0222 to JKR. We acknowledge the use of the University of Colorado Cancer Center Shared Resources for DNA Sequencing and Analysis, and Tissue Procurement and Pathology, supported by the NIH/National Cancer Institute Cancer Core Support Grant P30 CA046934.
Footnotes
Disclosure/Conflict of Interest: The authors declare they have no conflict of interest. The authors have no financial disclosures to report.
References
- 1.Emde A, Hornstein E. miRNAs at the interface of cellular stress and disease. EMBO J. 2014;33:1428–37. doi: 10.15252/embj.201488142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–7. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 3.Kumar MS, Pester RE, Chen CY, et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23:2700–4. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lambertz I, Nittner D, Mestdagh P, et al. Monoallelic but not biallelic loss of Dicer1 promotes tumorigenesis in vivo. Cell Death Differ. 2010;17:633–41. doi: 10.1038/cdd.2009.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karube Y, Tanaka H, Osada H, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–5. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin MG, Payton JE, Link DC. Dicer and outcomes in patients with acute myeloid leukemia (AML) Leuk Res. 2009;33:e127. doi: 10.1016/j.leukres.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Merritt WM, Lin YG, Han LY, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641–50. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valencak J, Schmid K, Trautinger F, et al. High expression of Dicer reveals a negative prognostic influence in certain subtypes of primary cutaneous T cell lymphomas. J Dermatol Sci. 2011;64:185–90. doi: 10.1016/j.jdermsci.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 9.Khoshnaw SM, Rakha EA, Abdel-Fatah TM, et al. Loss of Dicer expression is associated with breast cancer progression and recurrence. Breast Cancer Res Treat. 2012;135:403–13. doi: 10.1007/s10549-012-2169-3. [DOI] [PubMed] [Google Scholar]
- 10.Avery-Kiejda KA, Braye SG, Forbes JF, Scott RJ. The expression of Dicer and Drosha in matched normal tissues, tumours and lymph node metastases in triple negative breast cancer. BMC Cancer. 2014;14:253. doi: 10.1186/1471-2407-14-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caffrey E, Ingoldsby H, Wall D, et al. Prognostic significance of deregulated dicer expression in breast cancer. PLoS One. 2013;8:e83724. doi: 10.1371/journal.pone.0083724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu Z, Fan C, Oh DS, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pazaiti A, Fentiman IS. Basal phenotype breast cancer: implications for treatment and prognosis. Womens Health (Lond Engl) 2011;7:181–202. doi: 10.2217/whe.11.5. [DOI] [PubMed] [Google Scholar]
- 14.Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Mol Oncol. 2011;5:5–23. doi: 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cochrane DR, Cittelly DM, Howe EN, et al. MicroRNAs link estrogen receptor alpha status and Dicer levels in breast cancer. Horm Cancer. 2010;1:306–19. doi: 10.1007/s12672-010-0043-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cittelly DM, Dimitrova I, Howe EN, et al. Restoration of miR-200c to ovarian cancer reduces tumor burden and increases sensitivity to paclitaxel. Mol Cancer Ther. 2012;11:2556–65. doi: 10.1158/1535-7163.MCT-12-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Curtis C, Shah SP, Chin SF, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–52. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grelier G, Voirin N, Ay AS, et al. Prognostic value of Dicer expression in human breast cancers and association with the mesenchymal phenotype. Br J Cancer. 2009;101:673–83. doi: 10.1038/sj.bjc.6605193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White E, Schlackow M, Kamieniarz-Gdula K, Proudfoot NJ, Gullerova M. Human nuclear Dicer restricts the deleterious accumulation of endogenous double-stranded RNA. Nat Struct Mol Biol. 2014;21:552–9. doi: 10.1038/nsmb.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–24. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 21.Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annu Rev Biophys. 2013;42:217–39. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ando Y, Tomaru Y, Morinaga A, et al. Nuclear pore complex protein mediated nuclear localization of dicer protein in human cells. PLoS One. 2011;6:e23385. doi: 10.1371/journal.pone.0023385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faggad A, Budczies J, Tchernitsa O, et al. Prognostic significance of Dicer expression in ovarian cancer-link to global microRNA changes and oestrogen receptor expression. J Pathol. 2010;220:382–91. doi: 10.1002/path.2658. [DOI] [PubMed] [Google Scholar]
- 24.Dedes KJ, Natrajan R, Lambros MB, et al. Down-regulation of the miRNA master regulators Drosha and Dicer is associated with specific subgroups of breast cancer. Eur J Cancer. 2011;47:138–50. doi: 10.1016/j.ejca.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 25.Passon N, Gerometta A, Puppin C, et al. Expression of Dicer and Drosha in triple-negative breast cancer. J Clin Pathol. 2012;65:320–6. doi: 10.1136/jclinpath-2011-200496. [DOI] [PubMed] [Google Scholar]
- 26.Selever J, Gu G, Lewis MT, et al. Dicer-mediated upregulation of BCRP confers tamoxifen resistance in human breast cancer cells. Clin Cancer Res. 2011;17:6510–21. doi: 10.1158/1078-0432.CCR-11-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martello G, Rosato A, Ferrari F, et al. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141:1195–207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 28.Doyle M, Badertscher L, Jaskiewicz L, et al. The double-stranded RNA binding domain of human Dicer functions as a nuclear localization signal. RNA. 2013;19:1238–52. doi: 10.1261/rna.039255.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emmerth S, Schober H, Gaidatzis D, Roloff T, Jacobeit K, Buhler M. Nuclear retention of fission yeast dicer is a prerequisite for RNAi-mediated heterochromatin assembly. Dev Cell. 2010;18:102–13. doi: 10.1016/j.devcel.2009.11.011. [DOI] [PubMed] [Google Scholar]


