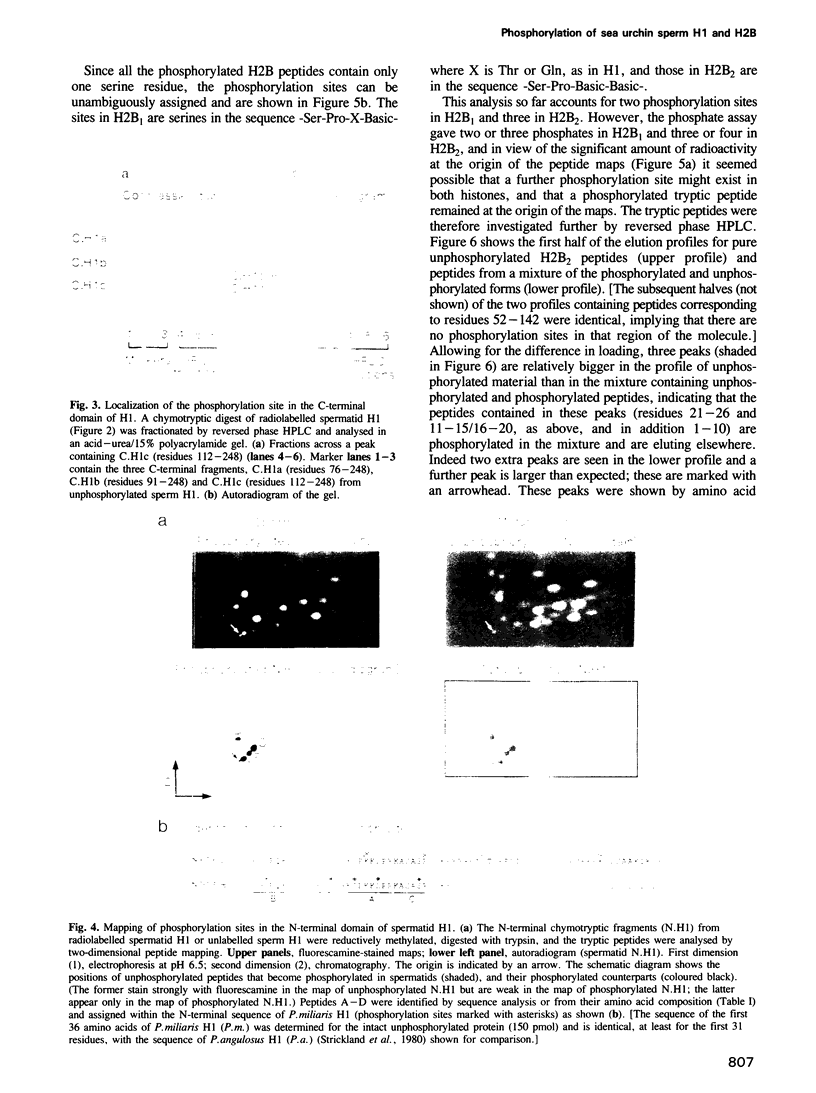
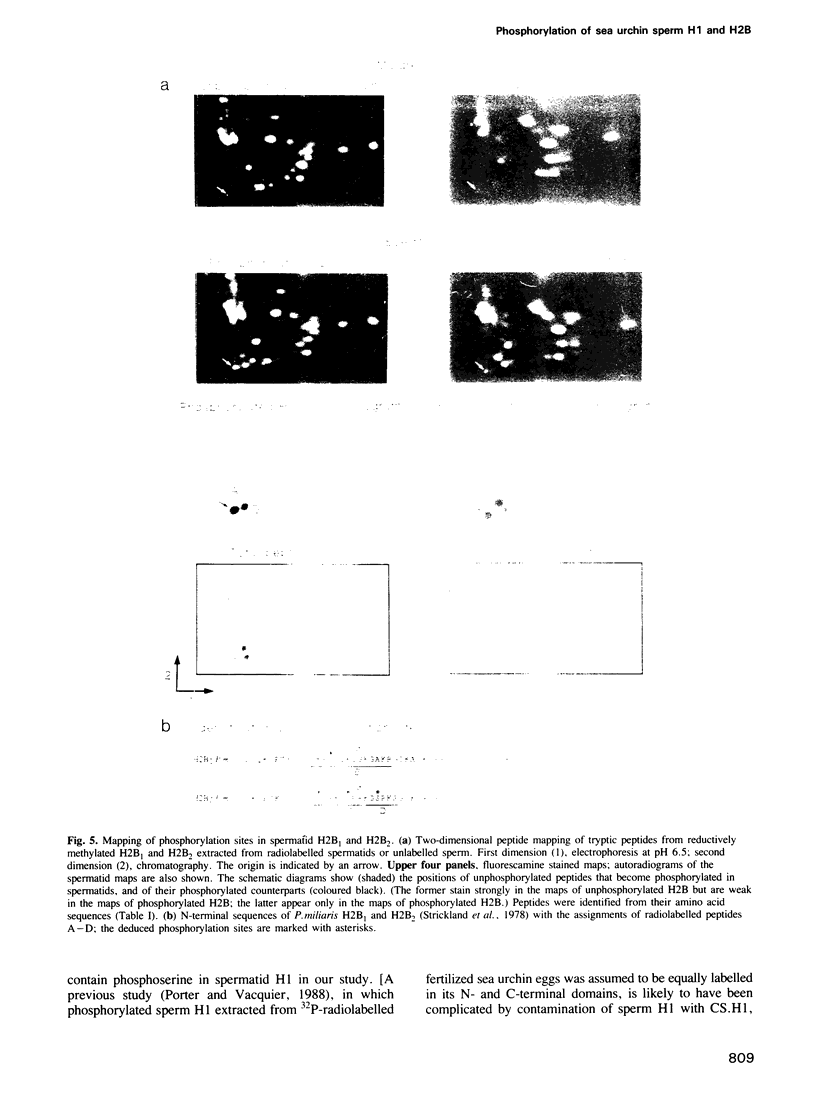
Abstract
Sea urchin sperm-specific histones H1 and H2B have distinctive N-terminal, and in the case of H1 also C-terminal, domains containing repeats of a basic motif (-Ser-Pro-Lys/Arg-Lys/Arg- or a closely related sequence). The histones in spermatids (the precursors of sperm) are phosphorylated, and the unphosphorylated histones of mature sperm are rephosphorylated upon fertilization. These changes correlate with finely tuned changes in chromatin packing in the nucleus, and the domains responsible are evidently the N-terminal domains. We show that in spermatids there are six tandemly repeated phosphorylation sites in the N-terminal domain of H1 (a typical cAMP dependent protein kinase site is not phosphorylated) and that H2B is phosphorylated in the N-terminal domain at two or three sites in the case of H2B1 and four sites in H2B2. The consensus sequence for phosphorylation is -Ser-Pro-X-Lys/Arg-, where X is Thr, Gln, Lys or Arg. There is an additional phosphorylated site in the C-terminal domain of H1 but most (or possibly all) copies of the consensus motif, which are here dispersed, are not phosphorylated. The negative charge introduced upon phosphorylation would be expected to weaken or abolish electrostatic interaction with DNA of this motif, which also occurs, and is phosphorylated, in somatic H1s.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradbury E. M., Inglis R. J., Matthews H. R. Control of cell division by very lysine rich histone (F1) phosphorylation. Nature. 1974 Feb 1;247(5439):257–261. doi: 10.1038/247257a0. [DOI] [PubMed] [Google Scholar]
- Briand G., Kmiecik D., Sautiere P., Wouters D., Borie-Loy O., Biserte G., Mazen A., Champagne M. Chicken erythrocyte histone H5. IV. Sequence of the carboxy-termined half of the molecule (96 residues) and complete sequence. FEBS Lett. 1980 Apr 7;112(2):147–151. doi: 10.1016/0014-5793(80)80167-0. [DOI] [PubMed] [Google Scholar]
- Buss J. E., Stull J. T. Measurement of chemical phosphate in proteins. Methods Enzymol. 1983;99:7–14. doi: 10.1016/0076-6879(83)99035-3. [DOI] [PubMed] [Google Scholar]
- Churchill M. E., Suzuki M. 'SPKK' motifs prefer to bind to DNA at A/T-rich sites. EMBO J. 1989 Dec 20;8(13):4189–4195. doi: 10.1002/j.1460-2075.1989.tb08604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisek L. J., Corden J. L. Phosphorylation of RNA polymerase by the murine homologue of the cell-cycle control protein cdc2. Nature. 1989 Jun 29;339(6227):679–684. doi: 10.1038/339679a0. [DOI] [PubMed] [Google Scholar]
- Clark D. J., Hill C. S., Martin S. R., Thomas J. O. Alpha-helix in the carboxy-terminal domains of histones H1 and H5. EMBO J. 1988 Jan;7(1):69–75. doi: 10.1002/j.1460-2075.1988.tb02784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green G. R., Poccia D. L. Interaction of sperm histone variants and linker DNA during spermiogenesis in the sea urchin. Biochemistry. 1988 Jan 26;27(2):619–625. doi: 10.1021/bi00402a019. [DOI] [PubMed] [Google Scholar]
- Green G. R., Poccia D. L. Phosphorylation of sea urchin sperm H1 and H2B histones precedes chromatin decondensation and H1 exchange during pronuclear formation. Dev Biol. 1985 Mar;108(1):235–245. doi: 10.1016/0012-1606(85)90026-0. [DOI] [PubMed] [Google Scholar]
- Gurley L. R., D'Anna J. A., Barham S. S., Deaven L. L., Tobey R. A. Histone phosphorylation and chromatin structure during mitosis in Chinese hamster cells. Eur J Biochem. 1978 Mar;84(1):1–15. doi: 10.1111/j.1432-1033.1978.tb12135.x. [DOI] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Hill C. S., Martin S. R., Thomas J. O. A stable alpha-helical element in the carboxy-terminal domain of free and chromatin-bound histone H1 from sea urchin sperm. EMBO J. 1989 Sep;8(9):2591–2599. doi: 10.1002/j.1460-2075.1989.tb08398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake R. S., Salzman N. P. Occurrence and properties of a chromatin-associated F1-histone phosphokinase in mitotic Chinese hamster cells. Biochemistry. 1972 Dec 5;11(25):4817–4826. doi: 10.1021/bi00775a027. [DOI] [PubMed] [Google Scholar]
- Lambert S. F., Thomas J. O. Lysine-containing DNA-binding regions on the surface of the histone octamer in the nucleosome core particle. Eur J Biochem. 1986 Oct 1;160(1):191–201. doi: 10.1111/j.1432-1033.1986.tb09957.x. [DOI] [PubMed] [Google Scholar]
- Langan T. A., Gautier J., Lohka M., Hollingsworth R., Moreno S., Nurse P., Maller J., Sclafani R. A. Mammalian growth-associated H1 histone kinase: a homolog of cdc2+/CDC28 protein kinases controlling mitotic entry in yeast and frog cells. Mol Cell Biol. 1989 Sep;9(9):3860–3868. doi: 10.1128/mcb.9.9.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmore J. P., Paulson J. R. Low angle x-ray diffraction studies of chromatin structure in vivo and in isolated nuclei and metaphase chromosomes. J Cell Biol. 1983 Apr;96(4):1120–1131. doi: 10.1083/jcb.96.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macleod A. R., Wong N. C., Dixon G. H. The amino-acid sequence of trout-testis histone H1. Eur J Biochem. 1977 Aug 15;78(1):281–291. doi: 10.1111/j.1432-1033.1977.tb11739.x. [DOI] [PubMed] [Google Scholar]
- Marion C., Martinage A., Tirard A., Roux B., Daune M., Mazen A. Histone phosphorylation in native chromatin induces local structural changes as probed by electric birefringence. J Mol Biol. 1985 Nov 20;186(2):367–379. doi: 10.1016/0022-2836(85)90111-1. [DOI] [PubMed] [Google Scholar]
- Meyer H. E., Hoffmann-Posorske E., Korte H., Heilmeyer L. M., Jr Sequence analysis of phosphoserine-containing peptides. Modification for picomolar sensitivity. FEBS Lett. 1986 Aug 11;204(1):61–66. doi: 10.1016/0014-5793(86)81388-6. [DOI] [PubMed] [Google Scholar]
- Nurse P., Bissett Y. Gene required in G1 for commitment to cell cycle and in G2 for control of mitosis in fission yeast. Nature. 1981 Aug 6;292(5823):558–560. doi: 10.1038/292558a0. [DOI] [PubMed] [Google Scholar]
- Panyim S., Chalkley R. High resolution acrylamide gel electrophoresis of histones. Arch Biochem Biophys. 1969 Mar;130(1):337–346. doi: 10.1016/0003-9861(69)90042-3. [DOI] [PubMed] [Google Scholar]
- Poccia D. L., Green G. R. Nuclei and chromosomal proteins. Methods Cell Biol. 1986;27:153–174. doi: 10.1016/s0091-679x(08)60347-7. [DOI] [PubMed] [Google Scholar]
- Poccia D. L., Simpson M. V., Green G. R. Transitions in histone variants during sea urchin spermatogenesis. Dev Biol. 1987 Jun;121(2):445–453. doi: 10.1016/0012-1606(87)90181-3. [DOI] [PubMed] [Google Scholar]
- Poccia D., Salik J., Krystal G. Transitions in histone variants of the male pronucleus following fertilization and evidence for a maternal store of cleavage-stage histones in the sera urchin egg. Dev Biol. 1981 Mar;82(2):287–296. doi: 10.1016/0012-1606(81)90452-8. [DOI] [PubMed] [Google Scholar]
- Porter D. C., Moy G. W., Vacquier V. D. The amino terminal sequence of sea urchin sperm histone H1 and its phosphorylation by egg cytosol. Comp Biochem Physiol B. 1989;92(2):381–384. doi: 10.1016/0305-0491(89)90296-4. [DOI] [PubMed] [Google Scholar]
- Porter D. C., Vacquier V. D. Extraction of phosphorylated sperm specific histone H1 from sea urchin eggs: analysis of phosphopeptide maps. Biochem Biophys Res Commun. 1988 Mar 30;151(3):1200–1204. doi: 10.1016/s0006-291x(88)80493-5. [DOI] [PubMed] [Google Scholar]
- Shenoy S., Choi J. K., Bagrodia S., Copeland T. D., Maller J. L., Shalloway D. Purified maturation promoting factor phosphorylates pp60c-src at the sites phosphorylated during fibroblast mitosis. Cell. 1989 Jun 2;57(5):763–774. doi: 10.1016/0092-8674(89)90791-5. [DOI] [PubMed] [Google Scholar]
- Simpson M. V., Poccia D. Sea urchin testicular cells evaluated by fluorescence microscopy of unfixed tissue. Gamete Res. 1987 Jun;17(2):131–144. doi: 10.1002/mrd.1120170205. [DOI] [PubMed] [Google Scholar]
- Strickland M., Strickland W. N., Brandt W. F., von Holt C. The partial amino acid sequences of the two H2B histones from sperm of the sea urchin Psammechinus miliaris. Biochim Biophys Acta. 1978 Sep 26;536(1):289–297. doi: 10.1016/0005-2795(78)90076-4. [DOI] [PubMed] [Google Scholar]
- Strickland W. N., Strickland M., Brandt W. F., Von Holt C., Lehmann A., Wittmann-Liebold B. The primary structure of histone H1 from sperm of the sea urchin Parechinus angulosus. 2. Sequence of the C-terminal CNBr peptide and the entire primary structure. Eur J Biochem. 1980 Mar;104(2):567–578. doi: 10.1111/j.1432-1033.1980.tb04460.x. [DOI] [PubMed] [Google Scholar]
- Sung M. T., Freedlender E. F. Sites of in vivo phosphorylation of histone H5. Biochemistry. 1978 May 16;17(10):1884–1890. doi: 10.1021/bi00603a013. [DOI] [PubMed] [Google Scholar]
- Suzuki M. SPKK, a new nucleic acid-binding unit of protein found in histone. EMBO J. 1989 Mar;8(3):797–804. doi: 10.1002/j.1460-2075.1989.tb03440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M. SPXX, a frequent sequence motif in gene regulatory proteins. J Mol Biol. 1989 May 5;207(1):61–84. doi: 10.1016/0022-2836(89)90441-5. [DOI] [PubMed] [Google Scholar]
- Taylor S. S. The in vitro phosphorylation of chromatin by the catalytic subunit of cAMP-dependent protein kinase. J Biol Chem. 1982 Jun 10;257(11):6056–6063. [PubMed] [Google Scholar]
- Thomas J. O. Chemical radiolabeling of lysines that interact strongly with DNA in chromatin. Methods Enzymol. 1989;170:369–385. doi: 10.1016/0076-6879(89)70057-4. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Kornberg R. D. The study of histone--histone associations by chemical cross-linking. Methods Cell Biol. 1978;18:429–440. [PubMed] [Google Scholar]
- Thomas J. O., Rees C., Butler P. J. Salt-induced folding of sea urchin sperm chromatin. Eur J Biochem. 1986 Jan 15;154(2):343–348. doi: 10.1111/j.1432-1033.1986.tb09403.x. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Wilson C. M. Selective radiolabelling and identification of a strong nucleosome binding site on the globular domain of histone H5. EMBO J. 1986 Dec 20;5(13):3531–3537. doi: 10.1002/j.1460-2075.1986.tb04679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner T. E., Hartford J. B., Serra M., Vandegrift V., Sung M. T. Phosphorylation and dephosphorylation of histone (V (H5): controlled condensation of avian erythrocyte chromatin. Appendix: Phosphorylation and dephosphorylation of histone H5. II. Circular dichroic studies. Biochemistry. 1977 Jan 25;16(2):286–290. doi: 10.1021/bi00621a020. [DOI] [PubMed] [Google Scholar]