Abstract
The Bacillus subtilis FI DNA methyltransferase (M.BsuFI) modifies the outer cytosine of the DNA sequence CCGG, causing resistance against R.BsuFI and R.MspI restriction. The M.BsuFI gene was cloned and expressed in B.subtilis and Escherichia coli. As derived from the nucleotide sequence, the M.BsuFI protein has 409 amino acids, corresponding to a molecular mass of 46,918 daltons. Including these data we have compared the nucleotide and amino acid sequences of different CCGG recognizing enzymes. These analyses showed that M.BsuFI is highly related to two other CCGG specific methyltransferases, M.MspI and M.HpaII, which were isolated from Gram-negative bacteria. Between M.BsuFI and M.MspI the sequence similarity is particularly significant in a region, which has been postulated to contain the target recognition domains (TRDs) of cytosine-specific DNA methyltransferases. Apparently M.BsuFI and M.MspI, derived from phylogenetic distant organisms, use highly conserved structural elements for the recognition of the CCGG target sequence. In contrast the very same region of M.HpaII is quite different from those of M.BsuFI and M.MspI. We attribute this difference to the different targeting of methylation within the sequence CCGG, where M.HpaII methylates the inner, M.BsuFI/M.MspI the outer cytosine. Also the CCGG recognizing TRD of the multispecific B.subtilis phage SPR Mtase is distinct from that of the host enzyme, possibly indicating different requirements for TRDs operative in mono- and multispecific enzymes.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balganesh T. S., Reiners L., Lauster R., Noyer-Weidner M., Wilke K., Trautner T. A. Construction and use of chimeric SPR/phi 3T DNA methyltransferases in the definition of sequence recognizing enzyme regions. EMBO J. 1987 Nov;6(11):3543–3549. doi: 10.1002/j.1460-2075.1987.tb02681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens B., Noyer-Weidner M., Pawlek B., Lauster R., Balganesh T. S., Trautner T. A. Organization of multispecific DNA methyltransferases encoded by temperate Bacillus subtilis phages. EMBO J. 1987 Apr;6(4):1137–1142. doi: 10.1002/j.1460-2075.1987.tb04869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal N., Kleinschmidt A. K., Spatz H. C., Trautner T. A. Physical properties of the DNA of bacteriophage SP50. Mol Gen Genet. 1967;100(1):39–55. doi: 10.1007/BF00425774. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
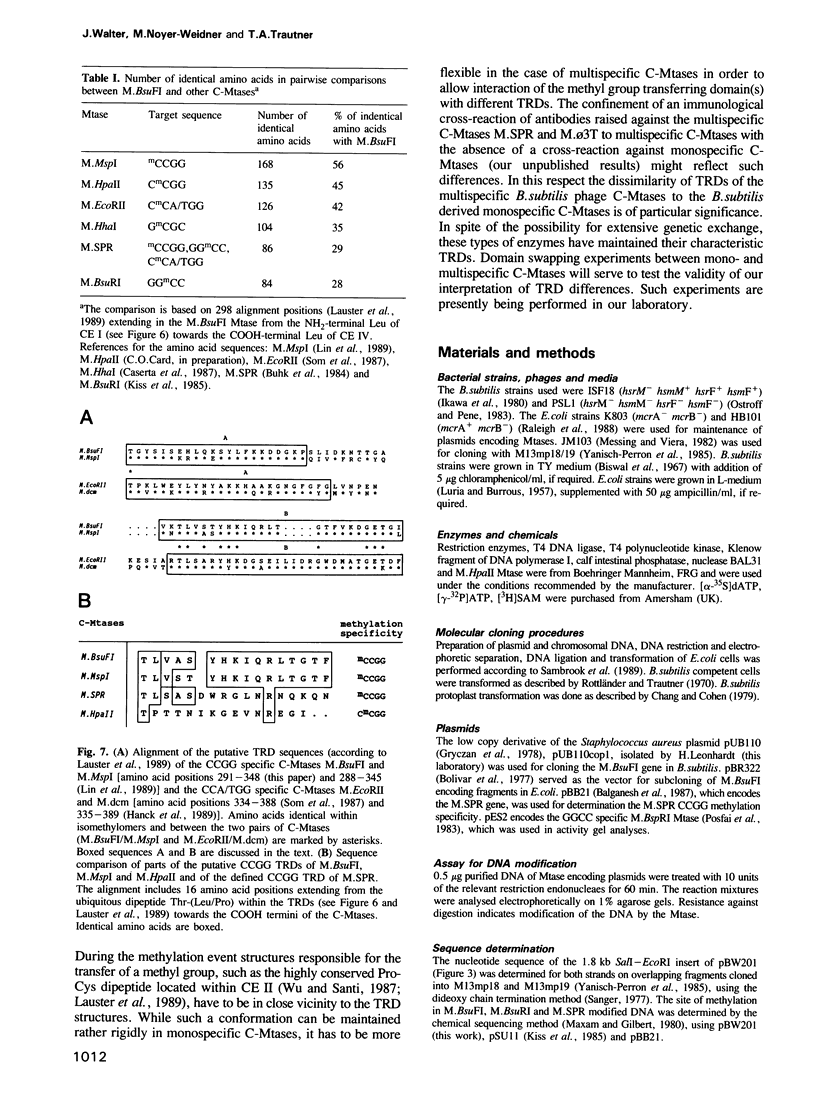
- Buhk H. J., Behrens B., Tailor R., Wilke K., Prada J. J., Günthert U., Noyer-Weidner M., Jentsch S., Trautner T. A. Restriction and modification in Bacillus subtilis: nucleotide sequence, functional organization and product of the DNA methyltransferase gene of bacteriophage SPR. Gene. 1984 Jul-Aug;29(1-2):51–61. doi: 10.1016/0378-1119(84)90165-3. [DOI] [PubMed] [Google Scholar]
- Caserta M., Zacharias W., Nwankwo D., Wilson G. G., Wells R. D. Cloning, sequencing, in vivo promoter mapping, and expression in Escherichia coli of the gene for the HhaI methyltransferase. J Biol Chem. 1987 Apr 5;262(10):4770–4777. [PubMed] [Google Scholar]
- Cedar H. DNA methylation and gene activity. Cell. 1988 Apr 8;53(1):3–4. doi: 10.1016/0092-8674(88)90479-5. [DOI] [PubMed] [Google Scholar]
- Chang S., Cohen S. N. High frequency transformation of Bacillus subtilis protoplasts by plasmid DNA. Mol Gen Genet. 1979 Jan 5;168(1):111–115. doi: 10.1007/BF00267940. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Contente S., Dubnau D. Characterization of Staphylococcus aureus plasmids introduced by transformation into Bacillus subtilis. J Bacteriol. 1978 Apr;134(1):318–329. doi: 10.1128/jb.134.1.318-329.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günthert U., Freund M., Trautner T. A. Restriction and modification in Bacillus subtilis: two DNA methyltransferases with BsuRI specificity. I. Purification and physical properties. J Biol Chem. 1981 Sep 10;256(17):9340–9345. [PubMed] [Google Scholar]
- Günthert U., Reiners L. Bacillus subtilis phage SPR codes for a DNA methyltransferase with triple sequence specificity. Nucleic Acids Res. 1987 May 11;15(9):3689–3702. doi: 10.1093/nar/15.9.3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanck T., Gerwin N., Fritz H. J. Nucleotide sequence of the dcm locus of Escherichia coli K12. Nucleic Acids Res. 1989 Jul 25;17(14):5844–5844. doi: 10.1093/nar/17.14.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982 May;150(2):815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübscher U., Pedrali-Noy G., Knust-Kron B., Doerfler W., Spadari S. DNA methyltransferases: activity minigel analysis and determination with DNA covalently bound to a solid matrix. Anal Biochem. 1985 Nov 1;150(2):442–448. doi: 10.1016/0003-2697(85)90533-0. [DOI] [PubMed] [Google Scholar]
- Ikawa S., Shibata T., Ando T., Saito H. Genetic studies on site-specific endodeoxyribonucleases in Bacillus subtilis: multiple modification and restriction systems in transformants of Bacillus subtilis 168. Mol Gen Genet. 1980 Feb;177(3):359–368. doi: 10.1007/BF00271474. [DOI] [PubMed] [Google Scholar]
- Ikawa S., Shibata T., Matsumoto K., Iijima T., Saito H., Ando T. Chromosomal loci of genes controlling site-specific restriction endonucleases of Bacillus subtilis. Mol Gen Genet. 1981;183(1):1–6. doi: 10.1007/BF00270129. [DOI] [PubMed] [Google Scholar]
- Jentsch S., Günthert U., Trautner T. A. DNA methyltransferases affecting the sequence 5'CCGG. Nucleic Acids Res. 1981 Jun 25;9(12):2753–2759. doi: 10.1093/nar/9.12.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S. Restriction and modification in Bacillus subtilis: sequence specificities of restriction/modification systems BsuM, BsuE, and BsuF. J Bacteriol. 1983 Nov;156(2):800–808. doi: 10.1128/jb.156.2.800-808.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss A., Posfai G., Keller C. C., Venetianer P., Roberts R. J. Nucleotide sequence of the BsuRI restriction-modification system. Nucleic Acids Res. 1985 Sep 25;13(18):6403–6421. doi: 10.1093/nar/13.18.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LURIA S. E., BURROUS J. W. Hybridization between Escherichia coli and Shigella. J Bacteriol. 1957 Oct;74(4):461–476. doi: 10.1128/jb.74.4.461-476.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauster R., Trautner T. A., Noyer-Weidner M. Cytosine-specific type II DNA methyltransferases. A conserved enzyme core with variable target-recognizing domains. J Mol Biol. 1989 Mar 20;206(2):305–312. doi: 10.1016/0022-2836(89)90480-4. [DOI] [PubMed] [Google Scholar]
- Lin P. M., Lee C. H., Roberts R. J. Cloning and characterization of the genes encoding the MspI restriction modification system. Nucleic Acids Res. 1989 Apr 25;17(8):3001–3011. doi: 10.1093/nar/17.8.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M. B., Smith H. O. Specificity of Hpa II and Hae III DNA methylases. Nucleic Acids Res. 1977 Dec;4(12):4211–4221. doi: 10.1093/nar/4.12.4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Moran C. P., Jr, Lang N., LeGrice S. F., Lee G., Stephens M., Sonenshein A. L., Pero J., Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186(3):339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- Nelson M., McClelland M. The effect of site-specific methylation on restriction-modification enzymes. Nucleic Acids Res. 1987;15 (Suppl):r219–r230. doi: 10.1093/nar/15.suppl.r219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori H., Tomizawa J. I., Maxam A. M. Detection of 5-methylcytosine in DNA sequences. Nucleic Acids Res. 1978 May;5(5):1479–1485. doi: 10.1093/nar/5.5.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pósfai G., Kiss A., Erdei S., Pósfai J., Venetianer P. Structure of the Bacillus sphaericus R modification methylase gene. J Mol Biol. 1983 Nov 5;170(3):597–610. doi: 10.1016/s0022-2836(83)80123-5. [DOI] [PubMed] [Google Scholar]
- Raleigh E. A., Murray N. E., Revel H., Blumenthal R. M., Westaway D., Reith A. D., Rigby P. W., Elhai J., Hanahan D. McrA and McrB restriction phenotypes of some E. coli strains and implications for gene cloning. Nucleic Acids Res. 1988 Feb 25;16(4):1563–1575. doi: 10.1093/nar/16.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff S. W., Luh J., Ganesan A. T., Behrens B., Thompson R., Montenegro M. A., Morelli G., Trautner T. A. The genome of Bacillus subtilis phage SPP1: the arrangement of restriction endonuclease generated fragments. Mol Gen Genet. 1979 Jan 10;168(2):165–172. doi: 10.1007/BF00431442. [DOI] [PubMed] [Google Scholar]
- Roberts R. J. Restriction enzymes and their isoschizomers. Nucleic Acids Res. 1988;16 (Suppl):r271–r313. doi: 10.1093/nar/16.suppl.r271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Rottländer E., Trautner T. A. Genetic and transfection studies with B, subtilis phage SP 50. I. Phage mutants with restricted growth on B. subtilis strain 168. Mol Gen Genet. 1970;108(1):47–60. doi: 10.1007/BF00343184. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatko B. E., Croft R., Moran L. S., Wilson G. G. Cloning and analysis of the HaeIII and HaeII methyltransferase genes. Gene. 1988 Dec 25;74(1):45–50. doi: 10.1016/0378-1119(88)90248-x. [DOI] [PubMed] [Google Scholar]
- Som S., Bhagwat A. S., Friedman S. Nucleotide sequence and expression of the gene encoding the EcoRII modification enzyme. Nucleic Acids Res. 1987 Jan 12;15(1):313–332. doi: 10.1093/nar/15.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Trautner T. A., Balganesh T. S., Pawlek B. Chimeric multispecific DNA methyltransferases with novel combinations of target recognition. Nucleic Acids Res. 1988 Jul 25;16(14A):6649–6658. doi: 10.1093/nar/16.14.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walder R. Y., Langtimm C. J., Chatterjee R., Walder J. A. Cloning of the MspI modification enzyme. The site of modification and its effects on cleavage by MspI and HpaII. J Biol Chem. 1983 Jan 25;258(2):1235–1241. [PubMed] [Google Scholar]
- Wilke K., Rauhut E., Noyer-Weidner M., Lauster R., Pawlek B., Behrens B., Trautner T. A. Sequential order of target-recognizing domains in multispecific DNA-methyltransferases. EMBO J. 1988 Aug;7(8):2601–2609. doi: 10.1002/j.1460-2075.1988.tb03110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. C., Santi D. V. Kinetic and catalytic mechanism of HhaI methyltransferase. J Biol Chem. 1987 Apr 5;262(10):4778–4786. [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]