Abstract
The introduction of activated c-myc and v-myc genes into a variety of non-established and established cells results in the suppression of endogenous c-myc expression. As measured in Rat-1 fibroblasts, the suppression occurs at the level of transcriptional initiation. Moreover, the extent of the down-regulation is proportional to the cellular concentration of c-myc protein, and the critical concentration range in which the endogenous c-myc RNA is effectively suppressed corresponds to that found in non-transformed cells. In addition, the autoregulatory mechanism is not only dependent on c-myc protein, but also requires additional trans-acting factors. These results support a role for c-myc in the regulation of cellular gene transcription and suggest that a negative feedback mechanism can act as a homeostatic regulator of c-myc expression in vivo.
Full text
PDF



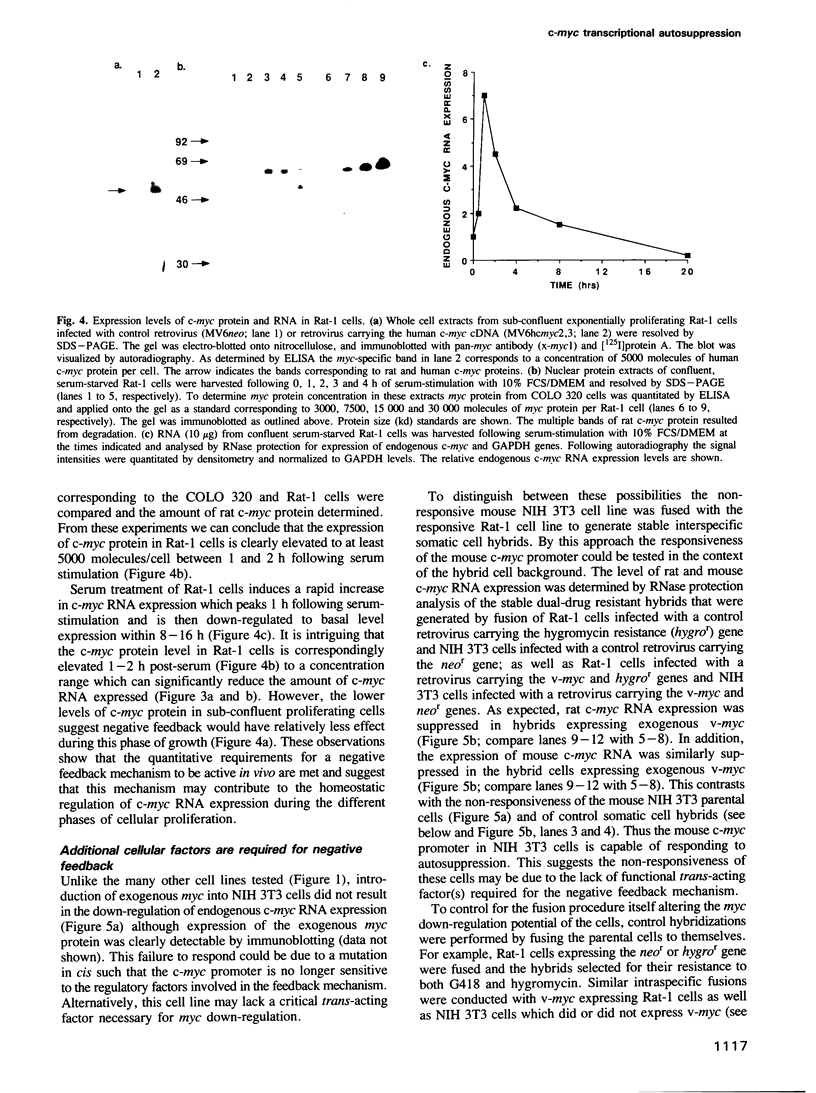
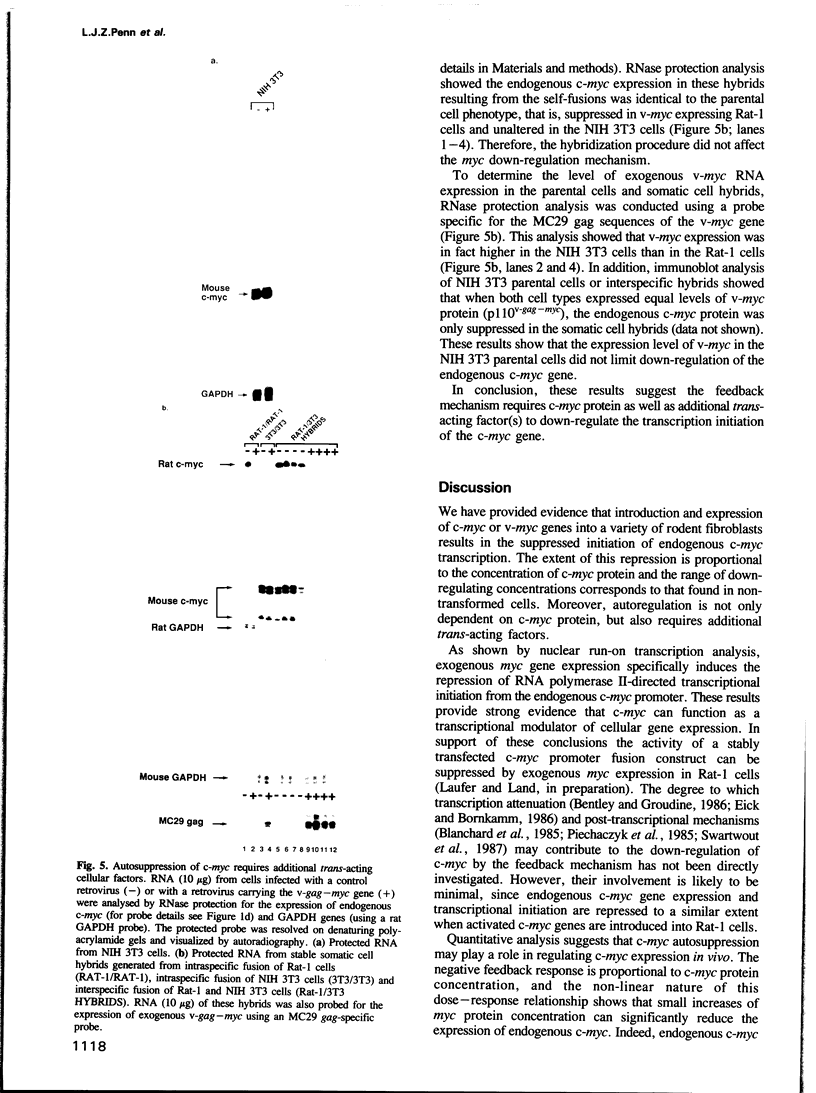



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aaronson S. A., Todaro G. J. Development of 3T3-like lines from Balb-c mouse embryo cultures: transformation susceptibility to SV40. J Cell Physiol. 1968 Oct;72(2):141–148. doi: 10.1002/jcp.1040720208. [DOI] [PubMed] [Google Scholar]
- Adams J. M., Gerondakis S., Webb E., Corcoran L. M., Cory S. Cellular myc oncogene is altered by chromosome translocation to an immunoglobulin locus in murine plasmacytomas and is rearranged similarly in human Burkitt lymphomas. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1982–1986. doi: 10.1073/pnas.80.7.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams J. M., Harris A. W., Pinkert C. A., Corcoran L. M., Alexander W. S., Cory S., Palmiter R. D., Brinster R. L. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985 Dec 12;318(6046):533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986 Jun 12;321(6071):702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- Bernard O., Cory S., Gerondakis S., Webb E., Adams J. M. Sequence of the murine and human cellular myc oncogenes and two modes of myc transcription resulting from chromosome translocation in B lymphoid tumours. EMBO J. 1983;2(12):2375–2383. doi: 10.1002/j.1460-2075.1983.tb01749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard J. M., Piechaczyk M., Dani C., Chambard J. C., Franchi A., Pouyssegur J., Jeanteur P. c-myc gene is transcribed at high rate in G0-arrested fibroblasts and is post-transcriptionally regulated in response to growth factors. Nature. 1985 Oct 3;317(6036):443–445. doi: 10.1038/317443a0. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Cleveland J. L., Huleihel M., Bressler P., Siebenlist U., Akiyama L., Eisenman R. N., Rapp U. R. Negative regulation of c-myc transcription involves myc family proteins. Oncogene Res. 1988;3(4):357–375. [PubMed] [Google Scholar]
- Clynes R., Wax J., Stanton L. W., Smith-Gill S., Potter M., Marcu K. B. Rapid induction of IgM-secreting murine plasmacytomas by pristane and an immunoglobulin heavy-chain promoter/enhancer-driven c-myc/v-Ha-ras retrovirus. Proc Natl Acad Sci U S A. 1988 Aug;85(16):6067–6071. doi: 10.1073/pnas.85.16.6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole M. D. The myc oncogene: its role in transformation and differentiation. Annu Rev Genet. 1986;20:361–384. doi: 10.1146/annurev.ge.20.120186.002045. [DOI] [PubMed] [Google Scholar]
- Coppola J. A., Cole M. D. Constitutive c-myc oncogene expression blocks mouse erythroleukaemia cell differentiation but not commitment. Nature. 1986 Apr 24;320(6064):760–763. doi: 10.1038/320760a0. [DOI] [PubMed] [Google Scholar]
- Dang C. V., McGuire M., Buckmire M., Lee W. M. Involvement of the 'leucine zipper' region in the oligomerization and transforming activity of human c-myc protein. Nature. 1989 Feb 16;337(6208):664–666. doi: 10.1038/337664a0. [DOI] [PubMed] [Google Scholar]
- Davis R. L., Weintraub H., Lassar A. B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987 Dec 24;51(6):987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Dean M., Cleveland J. L., Rapp U. R., Ihle J. N. Role of myc in the abrogation of IL3 dependence of myeloid FDC-P1 cells. Oncogene Res. 1987 Aug;1(3):279–296. [PubMed] [Google Scholar]
- Dildrop R., Ma A., Zimmerman K., Hsu E., Tesfaye A., DePinho R., Alt F. W. IgH enhancer-mediated deregulation of N-myc gene expression in transgenic mice: generation of lymphoid neoplasias that lack c-myc expression. EMBO J. 1989 Apr;8(4):1121–1128. doi: 10.1002/j.1460-2075.1989.tb03482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitrovsky E., Kuehl W. M., Hollis G. F., Kirsch I. R., Bender T. P., Segal S. Expression of a transfected human c-myc oncogene inhibits differentiation of a mouse erythroleukaemia cell line. Nature. 1986 Aug 21;322(6081):748–750. doi: 10.1038/322748a0. [DOI] [PubMed] [Google Scholar]
- Dugaiczyk A., Haron J. A., Stone E. M., Dennison O. E., Rothblum K. N., Schwartz R. J. Cloning and sequencing of a deoxyribonucleic acid copy of glyceraldehyde-3-phosphate dehydrogenase messenger ribonucleic acid isolated from chicken muscle. Biochemistry. 1983 Mar 29;22(7):1605–1613. doi: 10.1021/bi00276a013. [DOI] [PubMed] [Google Scholar]
- Dunnick W., Baumgartner J., Fradkin L., Schultz C., Szurek P. Methylation of plasmacytoma c-myc genes. Gene. 1985;39(2-3):287–292. doi: 10.1016/0378-1119(85)90325-7. [DOI] [PubMed] [Google Scholar]
- Dunnick W., Shell B. E., Dery C. DNA sequences near the site of reciprocal recombination between a c-myc oncogene and an immunoglobulin switch region. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7269–7273. doi: 10.1073/pnas.80.23.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eick D., Bornkamm G. W. Transcriptional arrest within the first exon is a fast control mechanism in c-myc gene expression. Nucleic Acids Res. 1986 Nov 11;14(21):8331–8346. doi: 10.1093/nar/14.21.8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan G. I., Lewis G. K., Ramsay G., Bishop J. M. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985 Dec;5(12):3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan G. I., Moore J. P., Ibson J. M., Waters C. M., Hancock D. C., Littlewood T. D. Immunological probes in the analysis of myc protein expression. Curr Top Microbiol Immunol. 1988;141:189–201. doi: 10.1007/978-3-642-74006-0_26. [DOI] [PubMed] [Google Scholar]
- Forrest D., Onions D., Lees G., Neil J. C. Altered structure and expression of c-myc in feline T-cell tumours. Virology. 1987 May;158(1):194–205. doi: 10.1016/0042-6822(87)90253-4. [DOI] [PubMed] [Google Scholar]
- Groudine M., Peretz M., Weintraub H. Transcriptional regulation of hemoglobin switching in chicken embryos. Mol Cell Biol. 1981 Mar;1(3):281–288. doi: 10.1128/mcb.1.3.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K., Makino R., Kawamura H., Arisawa A., Yoneda K. Characterization of rat c-myc and adjacent regions. Nucleic Acids Res. 1987 Aug 25;15(16):6419–6436. doi: 10.1093/nar/15.16.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keath E. J., Caimi P. G., Cole M. D. Fibroblast lines expressing activated c-myc oncogenes are tumorigenic in nude mice and syngeneic animals. Cell. 1984 Dec;39(2 Pt 1):339–348. doi: 10.1016/0092-8674(84)90012-6. [DOI] [PubMed] [Google Scholar]
- Kelekar A., Cole M. D. Immortalization by c-myc, H-ras, and Ela oncogenes induces differential cellular gene expression and growth factor responses. Mol Cell Biol. 1987 Nov;7(11):3899–3907. doi: 10.1128/mcb.7.11.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly K., Siebenlist U. The regulation and expression of c-myc in normal and malignant cells. Annu Rev Immunol. 1986;4:317–338. doi: 10.1146/annurev.iy.04.040186.001533. [DOI] [PubMed] [Google Scholar]
- Kirschmeier P. T., Housey G. M., Johnson M. D., Perkins A. S., Weinstein I. B. Construction and characterization of a retroviral vector demonstrating efficient expression of cloned cDNA sequences. DNA. 1988 Apr;7(3):219–225. doi: 10.1089/dna.1988.7.219. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Land H., Chen A. C., Morgenstern J. P., Parada L. F., Weinberg R. A. Behavior of myc and ras oncogenes in transformation of rat embryo fibroblasts. Mol Cell Biol. 1986 Jun;6(6):1917–1925. doi: 10.1128/mcb.6.6.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land H., Parada L. F., Weinberg R. A. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983 Aug 18;304(5927):596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988 Jun 24;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Lania L., Gandini-Attardi D., Griffiths M., Cooke B., De Cicco D., Fried M. The polyoma virus 100K large T-antigen is not required for the maintenance of transformation. Virology. 1980 Feb;101(1):217–232. doi: 10.1016/0042-6822(80)90497-3. [DOI] [PubMed] [Google Scholar]
- Leder P., Battey J., Lenoir G., Moulding C., Murphy W., Potter H., Stewart T., Taub R. Translocations among antibody genes in human cancer. Science. 1983 Nov 18;222(4625):765–771. doi: 10.1126/science.6356357. [DOI] [PubMed] [Google Scholar]
- Lee W. M., Schwab M., Westaway D., Varmus H. E. Augmented expression of normal c-myc is sufficient for cotransformation of rat embryo cells with a mutant ras gene. Mol Cell Biol. 1985 Dec;5(12):3345–3356. doi: 10.1128/mcb.5.12.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linial M., Gunderson N., Groudine M. Enhanced transcription of c-myc in bursal lymphoma cells requires continuous protein synthesis. Science. 1985 Dec 6;230(4730):1126–1132. doi: 10.1126/science.2999973. [DOI] [PubMed] [Google Scholar]
- Littlewood T. D., Hancock D. C., Evan G. I. Characterization of a heat shock-induced insoluble complex in the nuclei of cells. J Cell Sci. 1987 Aug;88(Pt 1):65–72. doi: 10.1242/jcs.88.1.65. [DOI] [PubMed] [Google Scholar]
- Lombardi L., Newcomb E. W., Dalla-Favera R. Pathogenesis of Burkitt lymphoma: expression of an activated c-myc oncogene causes the tumorigenic conversion of EBV-infected human B lymphoblasts. Cell. 1987 Apr 24;49(2):161–170. doi: 10.1016/0092-8674(87)90556-3. [DOI] [PubMed] [Google Scholar]
- Mango S. E., Schuler G. D., Steele M. E., Cole M. D. Germ line c-myc is not down-regulated by loss or exclusion of activating factors in myc-induced macrophage tumors. Mol Cell Biol. 1989 Aug;9(8):3482–3490. doi: 10.1128/mcb.9.8.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann R., Mulligan R. C., Baltimore D. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell. 1983 May;33(1):153–159. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- Marcu K. B. Regulation of expression of the c-myc proto-oncogene. Bioessays. 1987 Jan;6(1):28–32. doi: 10.1002/bies.950060108. [DOI] [PubMed] [Google Scholar]
- Moore J. P., Hancock D. C., Littlewood T. D., Evan G. I. A sensitive and quantitative enzyme-linked immunosorbence assay for the c-myc and N-myc oncoproteins. Oncogene Res. 1987;2(1):65–80. [PubMed] [Google Scholar]
- Morse H. C., 3rd, Hartley J. W., Fredrickson T. N., Yetter R. A., Majumdar C., Cleveland J. L., Rapp U. R. Recombinant murine retroviruses containing avian v-myc induce a wide spectrum of neoplasms in newborn mice. Proc Natl Acad Sci U S A. 1986 Sep;83(18):6868–6872. doi: 10.1073/pnas.83.18.6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989 Mar 10;56(5):777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Vaessin H., Caudy M., Jan L. Y., Jan Y. N., Cabrera C. V., Buskin J. N., Hauschka S. D., Lassar A. B. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989 Aug 11;58(3):537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- Nepveu A., Marcu K. B. Intragenic pausing and anti-sense transcription within the murine c-myc locus. EMBO J. 1986 Nov;5(11):2859–2865. doi: 10.1002/j.1460-2075.1986.tb04580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K., Murray J. M. The mechanism of inactivation of the normal c-myc gene locus in human Burkitt lymphoma cells. Oncogene. 1988 May;2(5):493–498. [PubMed] [Google Scholar]
- Nishikura K., ar-Rushdi A., Erikson J., Watt R., Rovera G., Croce C. M. Differential expression of the normal and of the translocated human c-myc oncogenes in B cells. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4822–4826. doi: 10.1073/pnas.80.15.4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn L. J., Laufer E. M., Land H. C-MYC: evidence for multiple regulatory functions. Semin Cancer Biol. 1990 Feb;1(1):69–80. [PubMed] [Google Scholar]
- Piechaczyk M., Yang J. Q., Blanchard J. M., Jeanteur P., Marcu K. B. Posttranscriptional mechanisms are responsible for accumulation of truncated c-myc RNAs in murine plasma cell tumors. Cell. 1985 Sep;42(2):589–597. doi: 10.1016/0092-8674(85)90116-3. [DOI] [PubMed] [Google Scholar]
- Potter M., Mushinski J. F., Mushinski E. B., Brust S., Wax J. S., Wiener F., Babonits M., Rapp U. R., Morse H. C., 3rd Avian v-myc replaces chromosomal translocation in murine plasmacytomagenesis. Science. 1987 Feb 13;235(4790):787–789. doi: 10.1126/science.3810165. [DOI] [PubMed] [Google Scholar]
- Prendergast G. C., Ziff E. B. DNA-binding motif. Nature. 1989 Oct 5;341(6241):392–392. doi: 10.1038/341392a0. [DOI] [PubMed] [Google Scholar]
- Quinn L. A., Moore G. E., Morgan R. T., Woods L. K. Cell lines from human colon carcinoma with unusual cell products, double minutes, and homogeneously staining regions. Cancer Res. 1979 Dec;39(12):4914–4924. [PubMed] [Google Scholar]
- Rapp U. R., Cleveland J. L., Brightman K., Scott A., Ihle J. N. Abrogation of IL-3 and IL-2 dependence by recombinant murine retroviruses expressing v-myc oncogenes. Nature. 1985 Oct 3;317(6036):434–438. doi: 10.1038/317434a0. [DOI] [PubMed] [Google Scholar]
- Reddy E. P., Reynolds R. K., Watson D. K., Schultz R. A., Lautenberger J., Papas T. S. Nucleotide sequence analysis of the proviral genome of avian myelocytomatosis virus (MC29). Proc Natl Acad Sci U S A. 1983 May;80(9):2500–2504. doi: 10.1073/pnas.80.9.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznikoff C. A., Brankow D. W., Heidelberger C. Establishment and characterization of a cloned line of C3H mouse embryo cells sensitive to postconfluence inhibition of division. Cancer Res. 1973 Dec;33(12):3231–3238. [PubMed] [Google Scholar]
- Rosenbaum H., Webb E., Adams J. M., Cory S., Harris A. W. N-myc transgene promotes B lymphoid proliferation, elicits lymphomas and reveals cross-regulation with c-myc. EMBO J. 1989 Mar;8(3):749–755. doi: 10.1002/j.1460-2075.1989.tb03435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz R. C., Stanton L. W., Riley S. C., Marcu K. B., Witte O. N. Synergism of v-myc and v-Ha-ras in the in vitro neoplastic progression of murine lymphoid cells. Mol Cell Biol. 1986 Sep;6(9):3221–3231. doi: 10.1128/mcb.6.9.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seif R., Cuzin F. Temperature-sensitive growth regulation in one type of transformed rat cells induced by the tsa mutant of polyoma virus. J Virol. 1977 Dec;24(3):721–728. doi: 10.1128/jvi.24.3.721-728.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih C. K., Linial M., Goodenow M. M., Hayward W. S. Nucleotide sequence 5' of the chicken c-myc coding region: localization of a noncoding exon that is absent from myc transcripts in most avian leukosis virus-induced lymphomas. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4697–4701. doi: 10.1073/pnas.81.15.4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U., Hennighausen L., Battey J., Leder P. Chromatin structure and protein binding in the putative regulatory region of the c-myc gene in Burkitt lymphoma. Cell. 1984 Jun;37(2):381–391. doi: 10.1016/0092-8674(84)90368-4. [DOI] [PubMed] [Google Scholar]
- Stewart T. A., Pattengale P. K., Leder P. Spontaneous mammary adenocarcinomas in transgenic mice that carry and express MTV/myc fusion genes. Cell. 1984 Oct;38(3):627–637. doi: 10.1016/0092-8674(84)90257-5. [DOI] [PubMed] [Google Scholar]
- Stone J., de Lange T., Ramsay G., Jakobovits E., Bishop J. M., Varmus H., Lee W. Definition of regions in human c-myc that are involved in transformation and nuclear localization. Mol Cell Biol. 1987 May;7(5):1697–1709. doi: 10.1128/mcb.7.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartwout S. G., Preisler H., Guan W. D., Kinniburgh A. J. Relatively stable population of c-myc RNA that lacks long poly(A). Mol Cell Biol. 1987 Jun;7(6):2052–2058. doi: 10.1128/mcb.7.6.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TODARO G. J., GREEN H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol. 1963 May;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapscott S. J., Davis R. L., Thayer M. J., Cheng P. F., Weintraub H., Lassar A. B. MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science. 1988 Oct 21;242(4877):405–411. doi: 10.1126/science.3175662. [DOI] [PubMed] [Google Scholar]
- Thompson T. C., Southgate J., Kitchener G., Land H. Multistage carcinogenesis induced by ras and myc oncogenes in a reconstituted organ. Cell. 1989 Mar 24;56(6):917–930. doi: 10.1016/0092-8674(89)90625-9. [DOI] [PubMed] [Google Scholar]
- Tso J. Y., Sun X. H., Kao T. H., Reece K. S., Wu R. Isolation and characterization of rat and human glyceraldehyde-3-phosphate dehydrogenase cDNAs: genomic complexity and molecular evolution of the gene. Nucleic Acids Res. 1985 Apr 11;13(7):2485–2502. doi: 10.1093/nar/13.7.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varley J. M., Wainwright A. M., Brammar W. J. An unusual alteration in c-myc in tissue from a primary breast carcinoma. Oncogene. 1987;1(4):431–438. [PubMed] [Google Scholar]
- Villares R., Cabrera C. V. The achaete-scute gene complex of D. melanogaster: conserved domains in a subset of genes required for neurogenesis and their homology to myc. Cell. 1987 Jul 31;50(3):415–424. doi: 10.1016/0092-8674(87)90495-8. [DOI] [PubMed] [Google Scholar]
- Zerlin M., Julius M. A., Cerni C., Marcu K. B. Elevated expression of an exogenous c-myc gene is insufficient for transformation and tumorigenic conversion of established fibroblasts. Oncogene. 1987 Mar;1(1):19–27. [PubMed] [Google Scholar]
- Zinn K., DiMaio D., Maniatis T. Identification of two distinct regulatory regions adjacent to the human beta-interferon gene. Cell. 1983 Oct;34(3):865–879. doi: 10.1016/0092-8674(83)90544-5. [DOI] [PubMed] [Google Scholar]






