Abstract
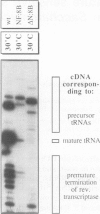
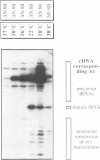
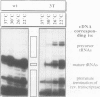
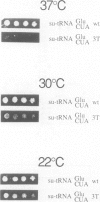
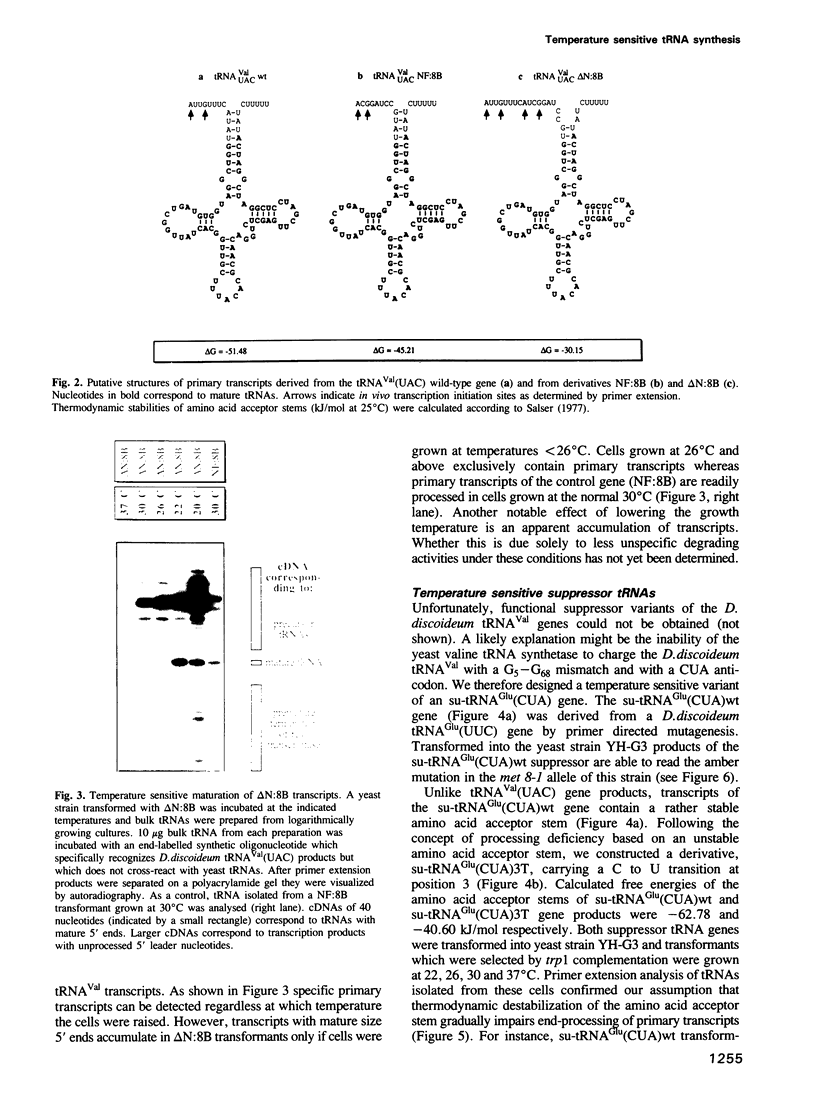
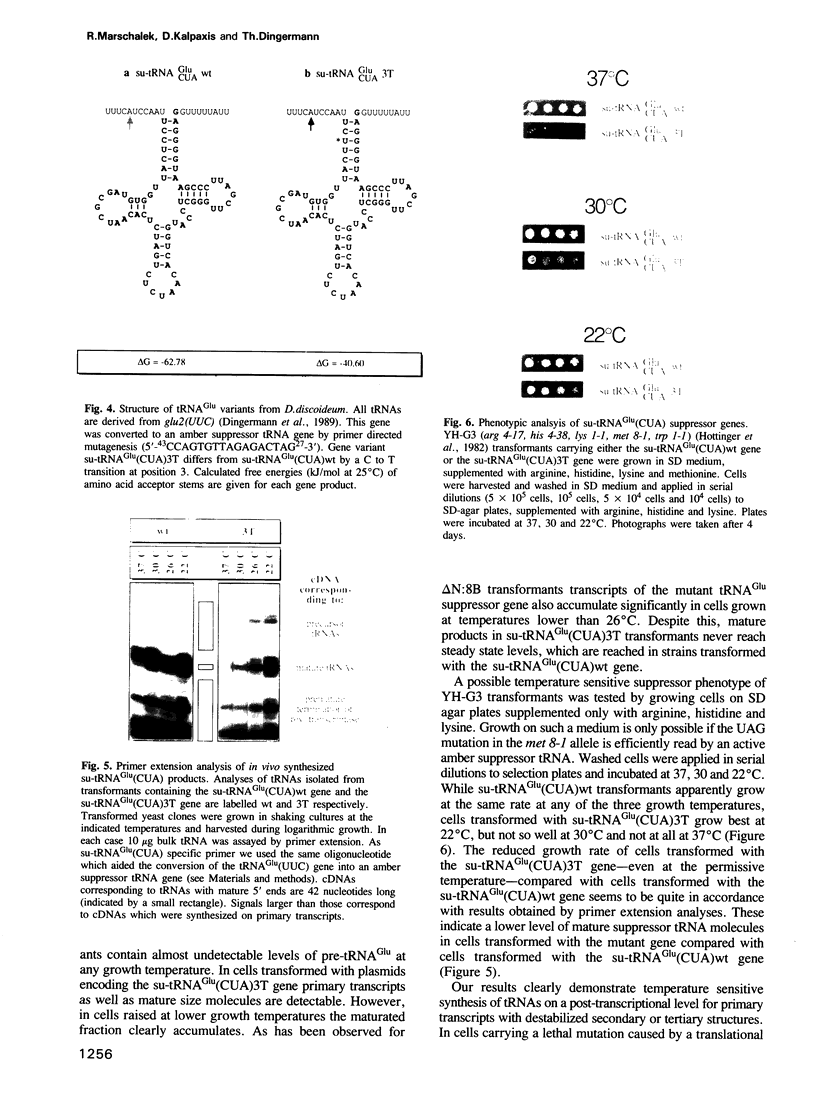
Dictyostelium discoideum tRNA genes can be expressed efficiently in vivo in yeast, and transcription products are processed to mature tRNAs. However, primary transcripts of a variant tRNA(Val)(UAC) gene are processing deficient under standard growth conditions (30 degrees C), due to a slightly altered 5' flanking region. A stable extended amino acid acceptor stem, which seems to be required to compensate a G5-G68 mismatch, cannot form. This mismatch destabilizes secondary and probably tertiary structures to such an extent that recognition of processing enzyme(s) under normal conditions (30 degrees C) is impaired. Growing yeast cells at reduced temperature (22 degrees C) can phenotypically complement the processing defect. This observation provides a new concept for the temperature dependent expression of protein coding genes which carry a nonsense codon. Translation of corresponding messages can be controlled by products of a temperature sensitive su-tRNA gene. We successfully tested this concept with two amber suppressors derived from a tRNA(Glu)(UUC) gene from D. discoideum. One of the variant tRNA genes codes for a product with a destabilized amino acid acceptor stem. Primary transcripts of this particular su-tRNA(Glu)(CUA) gene are processed only at reduced growth temperatures and consequently function as temperature sensitive suppressors only under these conditions.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abelson J. RNA processing and the intervening sequence problem. Annu Rev Biochem. 1979;48:1035–1069. doi: 10.1146/annurev.bi.48.070179.005131. [DOI] [PubMed] [Google Scholar]
- Allison D. S., Hall B. D. Effects of alterations in the 3' flanking sequence on in vivo and in vitro expression of the yeast SUP4-o tRNATyr gene. EMBO J. 1985 Oct;4(10):2657–2664. doi: 10.1002/j.1460-2075.1985.tb03984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold G. J., Gross H. J. Unrelated leader sequences can efficiently promote human tRNA gene transcription. Gene. 1987;51(2-3):237–246. doi: 10.1016/0378-1119(87)90312-x. [DOI] [PubMed] [Google Scholar]
- Baldi M. I., Mattoccia E., Tocchini-Valentini G. P. Role of RNA structure in splicing: excision of the intervening sequence in yeast tRNA3leu is dependent on the formation of a D stem. Cell. 1983 Nov;35(1):109–115. doi: 10.1016/0092-8674(83)90213-1. [DOI] [PubMed] [Google Scholar]
- Björk G. R., Ericson J. U., Gustafsson C. E., Hagervall T. G., Jönsson Y. H., Wikström P. M. Transfer RNA modification. Annu Rev Biochem. 1987;56:263–287. doi: 10.1146/annurev.bi.56.070187.001403. [DOI] [PubMed] [Google Scholar]
- Castagnoli L., Ciliberto G., Cortese R. Processing of eukaryotic tRNA precursors: secondary structure of the precursor specific sequences affects the rate but not the accuracy of processing reactions. Nucleic Acids Res. 1982 Jul 24;10(14):4135–4145. doi: 10.1093/nar/10.14.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaño J. G., Tobian J. A., Zasloff M. Purification and characterization of an endonuclease from Xenopus laevis ovaries which accurately processes the 3' terminus of human pre-tRNA-Met(i) (3' pre-tRNase). J Biol Chem. 1985 Jul 25;260(15):9002–9008. [PubMed] [Google Scholar]
- Choffat Y., Suter B., Behra R., Kubli E. Pseudouridine modification in the tRNA(Tyr) anticodon is dependent on the presence, but independent of the size and sequence, of the intron in eucaryotic tRNA(Tyr) genes. Mol Cell Biol. 1988 Aug;8(8):3332–3337. doi: 10.1128/mcb.8.8.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFranco D., Sharp S., Söll D. Identification of regulatory sequences contained in the 5'-flanking region of Drosophila lysine tRNA2 genes. J Biol Chem. 1981 Dec 10;256(23):12424–12429. [PubMed] [Google Scholar]
- Deutscher M. P. Processing of tRNA in prokaryotes and eukaryotes. CRC Crit Rev Biochem. 1984;17(1):45–71. doi: 10.3109/10409238409110269. [DOI] [PubMed] [Google Scholar]
- Dingermann T., Amon-Böhm E., Bertling W., Marschalek R., Nerke K. A family of non-allelic tRNA(ValGUU) genes from the cellular slime mold Dictyostelium discoideum. Gene. 1988 Dec 20;73(2):373–384. doi: 10.1016/0378-1119(88)90502-1. [DOI] [PubMed] [Google Scholar]
- Dingermann T., Brechner T., Marschalek R., Amon-Böhm E., Welker D. L. tRNAGlu(GAA) genes from the cellular slime mold Dictyostelium discoideum. DNA. 1989 Apr;8(3):193–204. doi: 10.1089/dna.1.1989.8.193. [DOI] [PubMed] [Google Scholar]
- Dingermann T., Burke D. J., Sharp S., Schaack J., Söll D. The 5- flanking sequences of Drosophila tRNAArg genes control their in vitro transcription in a Drosophila cell extract. J Biol Chem. 1982 Dec 25;257(24):14738–14744. [PubMed] [Google Scholar]
- Dingermann T., Nerke K., Blöcker H., Frank R. Structural requirements for the synthesis of tRNATrp from Dictyostelium discoideum in yeast. Biochimie. 1988 Jun;70(6):711–719. doi: 10.1016/0300-9084(88)90099-5. [DOI] [PubMed] [Google Scholar]
- Dingermann T., Nerke K., Marschalek R. Influence of different 5'-flanking sequences of tRNA genes on their in vivo transcription efficiencies in Saccharomyces cerevisiae. Eur J Biochem. 1987 Dec 30;170(1-2):217–224. doi: 10.1111/j.1432-1033.1987.tb13689.x. [DOI] [PubMed] [Google Scholar]
- Frendewey D., Dingermann T., Cooley L., Söll D. Processing of precursor tRNAs in Drosophila. Processing of the 3' end involves an endonucleolytic cleavage and occurs after 5' end maturation. J Biol Chem. 1985 Jan 10;260(1):449–454. [PubMed] [Google Scholar]
- Garber R. L., Altman S. In vitro processing of B. mori transfer RNA precursor molecules. Cell. 1979 Jun;17(2):389–397. doi: 10.1016/0092-8674(79)90165-x. [DOI] [PubMed] [Google Scholar]
- Garber R. L., Gage L. P. Transcription of a cloned Bombyx mori tRNA2Ala gene: nucleotide sequence of the tRNA precursor and its processing in vitro. Cell. 1979 Nov;18(3):817–828. doi: 10.1016/0092-8674(79)90134-x. [DOI] [PubMed] [Google Scholar]
- Geiduschek E. P., Tocchini-Valentini G. P. Transcription by RNA polymerase III. Annu Rev Biochem. 1988;57:873–914. doi: 10.1146/annurev.bi.57.070188.004301. [DOI] [PubMed] [Google Scholar]
- Hagenbüchle O., Larson D., Hall G. I., Sprague K. U. The primary transcription product of a silkworm alanine tRNA gene: identification of in vitro sites of initiation, termination and processing. Cell. 1979 Dec;18(4):1217–1229. doi: 10.1016/0092-8674(79)90234-4. [DOI] [PubMed] [Google Scholar]
- Hipskind R. A., Clarkson S. G. 5'-flanking sequences that inhibit in vitro transcription of a xenopus laevis tRNA gene. Cell. 1983 Oct;34(3):881–890. doi: 10.1016/0092-8674(83)90545-7. [DOI] [PubMed] [Google Scholar]
- Hottinger H., Pearson D., Yamao F., Gamulin V., Cooley L., Cooper T., Söll D. Nonsense suppression in Schizosaccharomyces pombe: the S. pombe Sup3-e tRNASerUGA gene is active in S. cerevisiae. Mol Gen Genet. 1982;188(2):219–224. doi: 10.1007/BF00332678. [DOI] [PubMed] [Google Scholar]
- Hudziak R. M., Laski F. A., RajBhandary U. L., Sharp P. A., Capecchi M. R. Establishment of mammalian cell lines containing multiple nonsense mutations and functional suppressor tRNA genes. Cell. 1982 Nov;31(1):137–146. doi: 10.1016/0092-8674(82)90413-5. [DOI] [PubMed] [Google Scholar]
- Kuchino Y., Beier H., Akita N., Nishimura S. Natural UAG suppressor glutamine tRNA is elevated in mouse cells infected with Moloney murine leukemia virus. Proc Natl Acad Sci U S A. 1987 May;84(9):2668–2672. doi: 10.1073/pnas.84.9.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laski F. A., Belagaje R., Hudziak R. M., Capecchi M. R., Norton G. P., Palese P., RajBhandary U. L., Sharp P. A. Synthesis of an ochre suppressor tRNA gene and expression in mammalian cells. EMBO J. 1984 Nov;3(11):2445–2452. doi: 10.1002/j.1460-2075.1984.tb02154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lofquist A., Sharp S. The 5'-flanking sequences of Drosophila melanogaster tRNA5Asn genes differentially arrest RNA polymerase III. J Biol Chem. 1986 Nov 5;261(31):14600–14606. [PubMed] [Google Scholar]
- Mao J., Schmidt O., Söll D. Dimeric transfer RNA precursors in S. pombe. Cell. 1980 Sep;21(2):509–516. doi: 10.1016/0092-8674(80)90488-2. [DOI] [PubMed] [Google Scholar]
- Marschalek R., Dingermann T. Identification of a protein factor binding to the 5'-flanking region of a tRNA gene and being involved in modulation of tRNA gene transcription in vivo in Saccharomyces cerevisiae. Nucleic Acids Res. 1988 Jul 25;16(14B):6737–6752. doi: 10.1093/nar/16.14.6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson J. M., Meuris P., Grunstein M., Abelson J., Miller J. H. Expression of a set of synthetic suppressor tRNA(Phe) genes in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6815–6819. doi: 10.1073/pnas.84.19.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattoccia E., Baldi M. I., Pande G., Ogden R., Tocchini-Valentini G. P. Mutation in the a block of the yeast tRNAleu3 gene that allows transcription but abolishes splicing and 5'-end maturation. Cell. 1983 Jan;32(1):67–76. doi: 10.1016/0092-8674(83)90497-x. [DOI] [PubMed] [Google Scholar]
- Melton D. A., De Robertis E. M., Cortese R. Order and intracellular location of the events involved in the maturation of a spliced tRNA. Nature. 1980 Mar 13;284(5752):143–148. doi: 10.1038/284143a0. [DOI] [PubMed] [Google Scholar]
- Nichols M., Söll D., Willis I. Yeast RNase P: catalytic activity and substrate binding are separate functions. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1379–1383. doi: 10.1073/pnas.85.5.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K., Kurjan J., Hall B. D., De Robertis E. M. Genetic analysis of the processing of a spliced tRNA. EMBO J. 1982;1(2):263–268. doi: 10.1002/j.1460-2075.1982.tb01157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasse-Messenguy F., Fink G. R. Temperature-sensitive nonsense suppressors in yeast. Genetics. 1973 Nov;75(3):459–464. doi: 10.1093/genetics/75.3.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond K. C., Raymond G. J., Johnson J. D. In vivo modulation of yeast tRNA gene expression by 5'-flanking sequences. EMBO J. 1985 Oct;4(10):2649–2656. doi: 10.1002/j.1460-2075.1985.tb03983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salser W. Globin mRNA sequences: analysis of base pairing and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):985–1002. doi: 10.1101/sqb.1978.042.01.099. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedivy J. M., Capone J. P., RajBhandary U. L., Sharp P. A. An inducible mammalian amber suppressor: propagation of a poliovirus mutant. Cell. 1987 Jul 31;50(3):379–389. doi: 10.1016/0092-8674(87)90492-2. [DOI] [PubMed] [Google Scholar]
- Sharp S. J., Schaack J., Cooley L., Burke D. J., Söll D. Structure and transcription of eukaryotic tRNA genes. CRC Crit Rev Biochem. 1985;19(2):107–144. doi: 10.3109/10409238509082541. [DOI] [PubMed] [Google Scholar]
- Shaw K. J., Olson M. V. Effects of altered 5'-flanking sequences on the in vivo expression of a Saccharomyces cerevisiae tRNATyr gene. Mol Cell Biol. 1984 Apr;4(4):657–665. doi: 10.1128/mcb.4.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague K. U., Hagenbüchle O., Zuniga M. C. The nucleotide sequence of two silk gland alanine tRNAs: implications for fibroin synthesis and for initiator tRNA structure. Cell. 1977 Jul;11(3):561–570. doi: 10.1016/0092-8674(77)90074-5. [DOI] [PubMed] [Google Scholar]
- Sprague K. U., Larson D., Morton D. 5' flanking sequence signals are required for activity of silkworm alanine tRNA genes in homologous in vitro transcription systems. Cell. 1980 Nov;22(1 Pt 1):171–178. doi: 10.1016/0092-8674(80)90165-8. [DOI] [PubMed] [Google Scholar]
- Sprinzl M., Hartmann T., Weber J., Blank J., Zeidler R. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1989;17 (Suppl):r1–172. doi: 10.1093/nar/17.suppl.r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple G. F., Dozy A. M., Roy K. L., Kan Y. W. Construction of a functional human suppressor tRNA gene: an approach to gene therapy for beta-thalassaemia. Nature. 1982 Apr 8;296(5857):537–540. doi: 10.1038/296537a0. [DOI] [PubMed] [Google Scholar]
- Traboni C., Ciliberto G., Cortese R. Mutations in Box B of the promoter of a eucaryotic tRNAPro gene affect rate of transcription, processing, and stability of the transcripts. Cell. 1984 Jan;36(1):179–187. doi: 10.1016/0092-8674(84)90087-4. [DOI] [PubMed] [Google Scholar]
- Willis I., Nichols M., Chisholm V., Söll D., Heyer W. D., Szankasi P., Amstutz H., Munz P., Kohli J. Functional complementation between mutations in a yeast suppressor tRNA gene reveals potential for evolution of tRNA sequences. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7860–7864. doi: 10.1073/pnas.83.20.7860. [DOI] [PMC free article] [PubMed] [Google Scholar]