Abstract
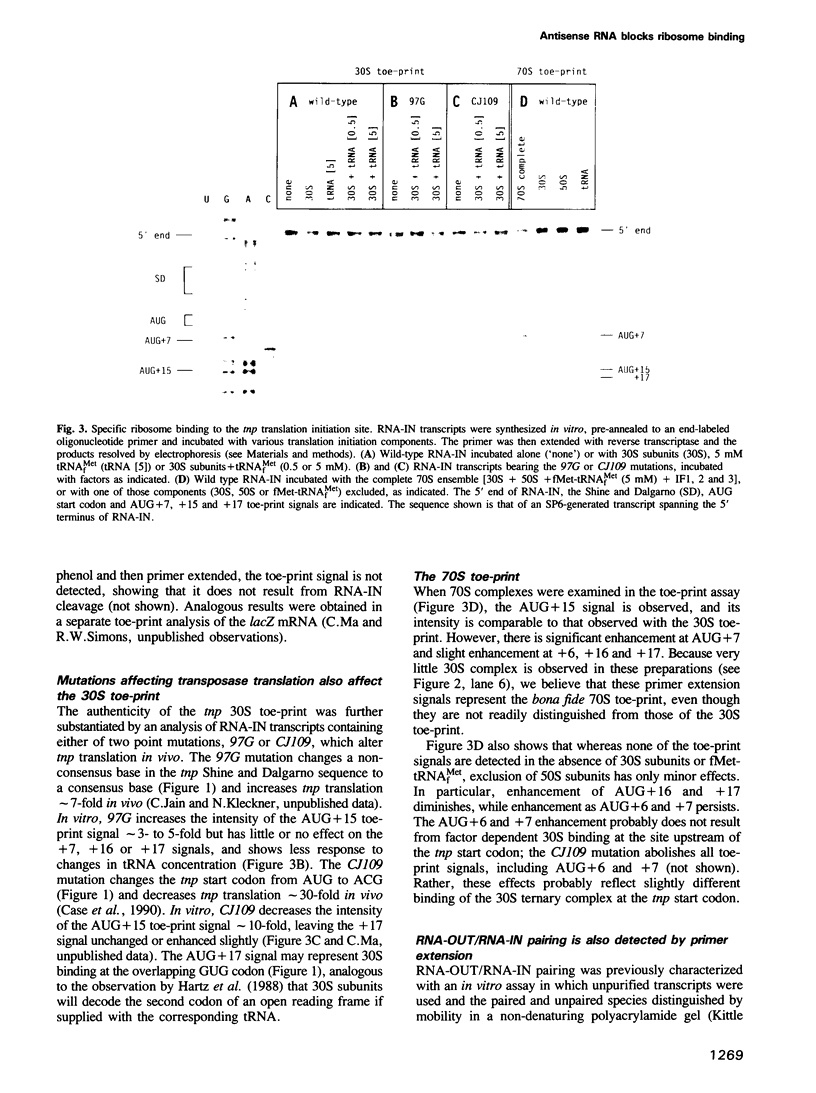
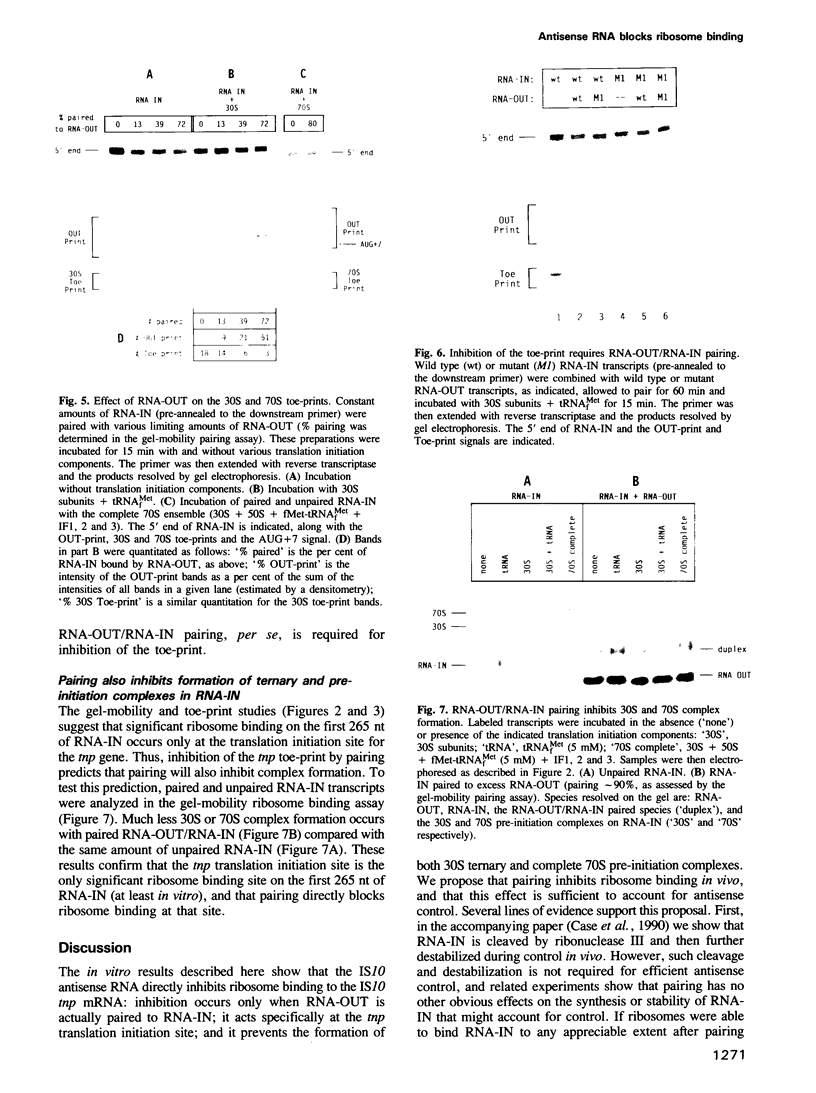
Transposase (tnp) expression from insertion sequence IS10 is controlled, in part, by an antisense RNA, RNA-OUT, which pairs to the translation initiation region of the tnp mRNA, RNA-IN. Genetic experiments suggest that control occurs post-transcriptionally. Here, we present evidence that bears on the control mechanism. Specific ribosome binding at the tnp translation initiation site is demonstrated in vitro. Two mutations that alter tnp translation in vivo are shown to have corresponding effects in vitro. Most importantly, RNA-OUT/RNA-IN pairing is shown to block ribosome binding. In conjunction with the work described in the accompanying paper, we propose that inhibition of ribosome binding also occurs in vivo, and that it is sufficient to account for control. Implications for translational control in analogous systems are discussed.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Backman K., Ptashne M., Gilbert W. Construction of plasmids carrying the cI gene of bacteriophage lambda. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4174–4178. doi: 10.1073/pnas.73.11.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisova G. P., Volkova T. M., Berzin V., Rosenthal G., Gren E. J. The regulatory region of MS2 phage RNA replicase cistron. IV. Functional activity of specific MS2 RNA fragments in formation of the 70 S initiation complex of protein biosynthesis. Nucleic Acids Res. 1979;6(5):1761–1774. doi: 10.1093/nar/6.5.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case C. C., Roels S. M., González J. E., Simons E. L., Simons R. W. Analysis of the promoters and transcripts involved in IS10 anti-sense RNA control. Gene. 1988 Dec 10;72(1-2):219–236. doi: 10.1016/0378-1119(88)90147-3. [DOI] [PubMed] [Google Scholar]
- Case C. C., Roels S. M., Jensen P. D., Lee J., Kleckner N., Simons R. W. The unusual stability of the IS10 anti-sense RNA is critical for its function and is determined by the structure of its stem-domain. EMBO J. 1989 Dec 20;8(13):4297–4305. doi: 10.1002/j.1460-2075.1989.tb08616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case C. C., Simons E. L., Simons R. W. The IS10 transposase mRNA is destabilized during antisense RNA control. EMBO J. 1990 Apr;9(4):1259–1266. doi: 10.1002/j.1460-2075.1990.tb08234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlberg A. E., Dingman C. W., Peacock A. C. Electrophoretic characterization of bacterial polyribosomes in agarose-acrylamide composite gels. J Mol Biol. 1969 Apr 14;41(1):139–147. doi: 10.1016/0022-2836(69)90131-4. [DOI] [PubMed] [Google Scholar]
- Davis M. A., Simons R. W., Kleckner N. Tn10 protects itself at two levels from fortuitous activation by external promoters. Cell. 1985 Nov;43(1):379–387. doi: 10.1016/0092-8674(85)90043-1. [DOI] [PubMed] [Google Scholar]
- Farabaugh P. J. Sequence of the lacI gene. Nature. 1978 Aug 24;274(5673):765–769. doi: 10.1038/274765a0. [DOI] [PubMed] [Google Scholar]
- Foster T. J., Lundblad V., Hanley-Way S., Halling S. M., Kleckner N. Three Tn10-associated excision events: relationship to transposition and role of direct and inverted repeats. Cell. 1981 Jan;23(1):215–227. doi: 10.1016/0092-8674(81)90286-5. [DOI] [PubMed] [Google Scholar]
- Gerdes K., Helin K., Christensen O. W., Løbner-Olesen A. Translational control and differential RNA decay are key elements regulating postsegregational expression of the killer protein encoded by the parB locus of plasmid R1. J Mol Biol. 1988 Sep 5;203(1):119–129. doi: 10.1016/0022-2836(88)90096-4. [DOI] [PubMed] [Google Scholar]
- Gold L. Posttranscriptional regulatory mechanisms in Escherichia coli. Annu Rev Biochem. 1988;57:199–233. doi: 10.1146/annurev.bi.57.070188.001215. [DOI] [PubMed] [Google Scholar]
- Hall M. N., Gabay J., Débarbouillé M., Schwartz M. A role for mRNA secondary structure in the control of translation initiation. Nature. 1982 Feb 18;295(5850):616–618. doi: 10.1038/295616a0. [DOI] [PubMed] [Google Scholar]
- Halling S. M., Simons R. W., Way J. C., Walsh R. B., Kleckner N. DNA sequence organization of IS10-right of Tn10 and comparison with IS10-left. Proc Natl Acad Sci U S A. 1982 Apr;79(8):2608–2612. doi: 10.1073/pnas.79.8.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartz D., McPheeters D. S., Gold L. Selection of the initiator tRNA by Escherichia coli initiation factors. Genes Dev. 1989 Dec;3(12A):1899–1912. doi: 10.1101/gad.3.12a.1899. [DOI] [PubMed] [Google Scholar]
- Hartz D., McPheeters D. S., Traut R., Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- Kaempfer R., Jay G. Binding of messenger RNA in initiation of prokaryotic translation. Methods Enzymol. 1979;60:332–343. doi: 10.1016/s0076-6879(79)60031-9. [DOI] [PubMed] [Google Scholar]
- Kang C., Wu C. W. Studies on SP6 promoter using a new plasmid vector that allows gene insertion at the transcription initiation site. Nucleic Acids Res. 1987 Mar 11;15(5):2279–2294. doi: 10.1093/nar/15.5.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Meyer R. J. Copy-number of broad host-range plasmid R1162 is regulated by a small RNA. Nucleic Acids Res. 1986 Oct 24;14(20):8027–8046. doi: 10.1093/nar/14.20.8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittle J. D., Simons R. W., Lee J., Kleckner N. Insertion sequence IS10 anti-sense pairing initiates by an interaction between the 5' end of the target RNA and a loop in the anti-sense RNA. J Mol Biol. 1989 Dec 5;210(3):561–572. doi: 10.1016/0022-2836(89)90132-0. [DOI] [PubMed] [Google Scholar]
- Krinke L., Wulff D. L. OOP RNA, produced from multicopy plasmids, inhibits lambda cII gene expression through an RNase III-dependent mechanism. Genes Dev. 1987 Nov;1(9):1005–1013. doi: 10.1101/gad.1.9.1005. [DOI] [PubMed] [Google Scholar]
- Kröger M., Hobom G. Structural analysis of insertion sequence IS5. Nature. 1982 May 13;297(5862):159–162. doi: 10.1038/297159a0. [DOI] [PubMed] [Google Scholar]
- Lee Y., Schmidt F. J. Characterization of the in vivo RNA product of the pOUT promoter of IS10R. J Bacteriol. 1985 Nov;164(2):556–562. doi: 10.1128/jb.164.2.556-562.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S. M., Wu T. H., Chiang C. H., Susskind M. M., McClure W. R. Control of gene expression in bacteriophage P22 by a small antisense RNA. I. Characterization in vitro of the Psar promoter and the sar RNA transcript. Genes Dev. 1987 Apr;1(2):197–203. doi: 10.1101/gad.1.2.197. [DOI] [PubMed] [Google Scholar]
- Lodish H. F. Thermal melting of bacteriophage f2 RNA and initiation of synthesis of the maturation protein. J Mol Biol. 1971 Mar 28;56(3):627–632. doi: 10.1016/0022-2836(71)90406-2. [DOI] [PubMed] [Google Scholar]
- Macdonald P. M., Kutter E., Mosig G. Regulation of a bacteriophage T4 late gene, soc, which maps in an early region. Genetics. 1984 Jan;106(1):17–27. doi: 10.1093/genetics/106.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPheeters D. S., Stormo G. D., Gold L. Autogenous regulatory site on the bacteriophage T4 gene 32 messenger RNA. J Mol Biol. 1988 Jun 5;201(3):517–535. doi: 10.1016/0022-2836(88)90634-1. [DOI] [PubMed] [Google Scholar]
- Merril C. R., Gottesman M. E., Adhya S. L. Escherichia coli gal operon proteins made after prophage lambda induction. J Bacteriol. 1981 Sep;147(3):875–887. doi: 10.1128/jb.147.3.875-887.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T., Chou M. Y., Inouye M. A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript (micRNA). Proc Natl Acad Sci U S A. 1984 Apr;81(7):1966–1970. doi: 10.1073/pnas.81.7.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson L. M., Stormo G. D., Niece R. L., Reznikoff W. S. lacZ translation initiation mutations. J Mol Biol. 1984 Aug 25;177(4):663–683. doi: 10.1016/0022-2836(84)90043-3. [DOI] [PubMed] [Google Scholar]
- Nomura M., Gourse R., Baughman G. Regulation of the synthesis of ribosomes and ribosomal components. Annu Rev Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- Raleigh E. A., Kleckner N. Quantitation of insertion sequence IS10 transposase gene expression by a method generally applicable to any rarely expressed gene. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1787–1791. doi: 10.1073/pnas.83.6.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D., Hoopes B. C., McClure W. R., Kleckner N. IS10 transposition is regulated by DNA adenine methylation. Cell. 1985 Nov;43(1):117–130. doi: 10.1016/0092-8674(85)90017-0. [DOI] [PubMed] [Google Scholar]
- Simons R. W., Hoopes B. C., McClure W. R., Kleckner N. Three promoters near the termini of IS10: pIN, pOUT, and pIII. Cell. 1983 Sep;34(2):673–682. doi: 10.1016/0092-8674(83)90400-2. [DOI] [PubMed] [Google Scholar]
- Simons R. W., Houman F., Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53(1):85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Simons R. W., Kleckner N. Biological regulation by antisense RNA in prokaryotes. Annu Rev Genet. 1988;22:567–600. doi: 10.1146/annurev.ge.22.120188.003031. [DOI] [PubMed] [Google Scholar]
- Simons R. W., Kleckner N. Translational control of IS10 transposition. Cell. 1983 Sep;34(2):683–691. doi: 10.1016/0092-8674(83)90401-4. [DOI] [PubMed] [Google Scholar]
- Simons R. W. Naturally occurring antisense RNA control--a brief review. Gene. 1988 Dec 10;72(1-2):35–44. doi: 10.1016/0378-1119(88)90125-4. [DOI] [PubMed] [Google Scholar]
- Tai P. C., Wallace B. J., Davis B. D. Actions of aurintricarboxylate, kasugamycin, and pactamycin on Escherichia coli polysomes. Biochemistry. 1973 Feb;12(4):616–620. doi: 10.1021/bi00728a008. [DOI] [PubMed] [Google Scholar]
- Timmerman K. P., Tu C. P. Complete sequence of IS3. Nucleic Acids Res. 1985 Mar 25;13(6):2127–2139. doi: 10.1093/nar/13.6.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duin J., Overbeek G. P., Backendorf C. Functional recognition of phage RNA by 30-S ribosomal subunits in the absence of initiator tRNA. Eur J Biochem. 1980 Sep;110(2):593–597. doi: 10.1111/j.1432-1033.1980.tb04903.x. [DOI] [PubMed] [Google Scholar]
- Way J. C., Kleckner N. Essential sites at transposon Tn 10 termini. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3452–3456. doi: 10.1073/pnas.81.11.3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter R. B., Morrissey L., Gauss P., Gold L., Hsu T., Karam J. Bacteriophage T4 regA protein binds to mRNAs and prevents translation initiation. Proc Natl Acad Sci U S A. 1987 Nov;84(22):7822–7826. doi: 10.1073/pnas.84.22.7822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. H., Liao S. M., McClure W. R., Susskind M. M. Control of gene expression in bacteriophage P22 by a small antisense RNA. II. Characterization of mutants defective in repression. Genes Dev. 1987 Apr;1(2):204–212. doi: 10.1101/gad.1.2.204. [DOI] [PubMed] [Google Scholar]
- Wulff D. L., Mahoney M., Shatzman A., Rosenberg M. Mutational analysis of a regulatory region in bacteriophage lambda that has overlapping signals for the initiation of transcription and translation. Proc Natl Acad Sci U S A. 1984 Jan;81(2):555–559. doi: 10.1073/pnas.81.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]








