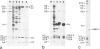Abstract
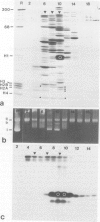
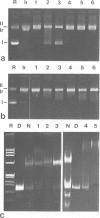
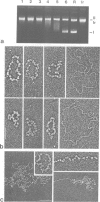
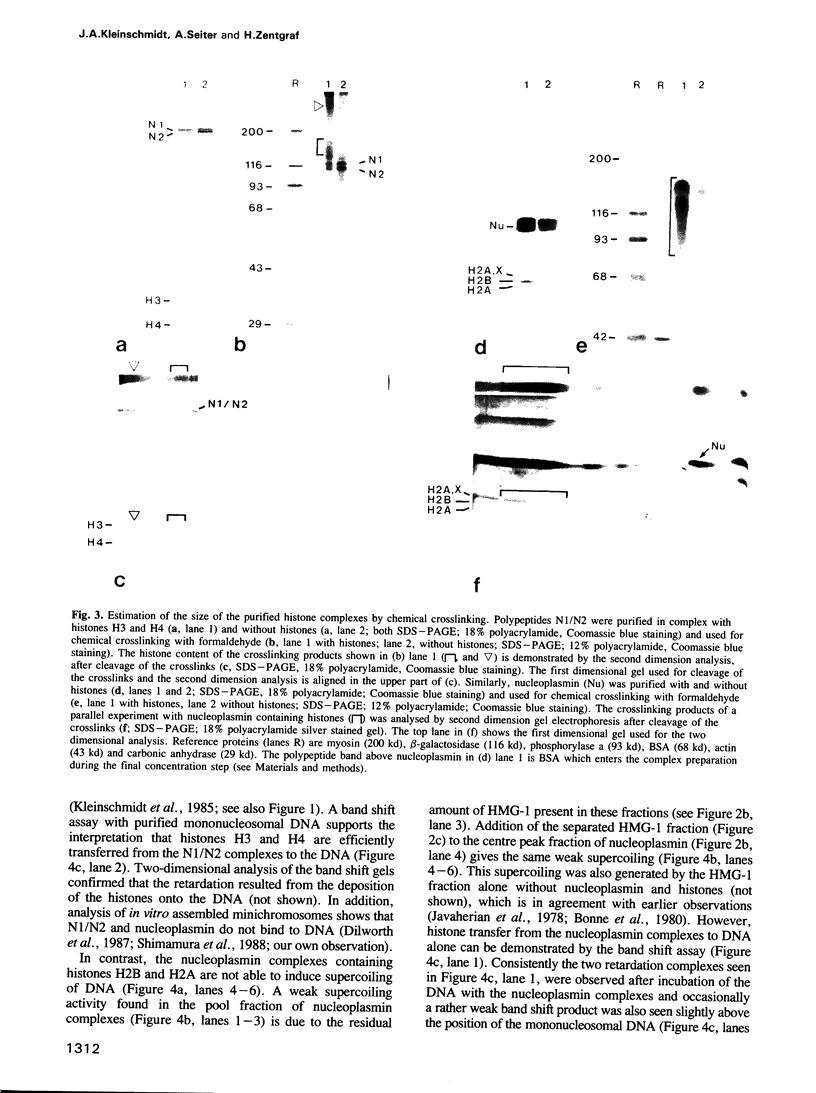
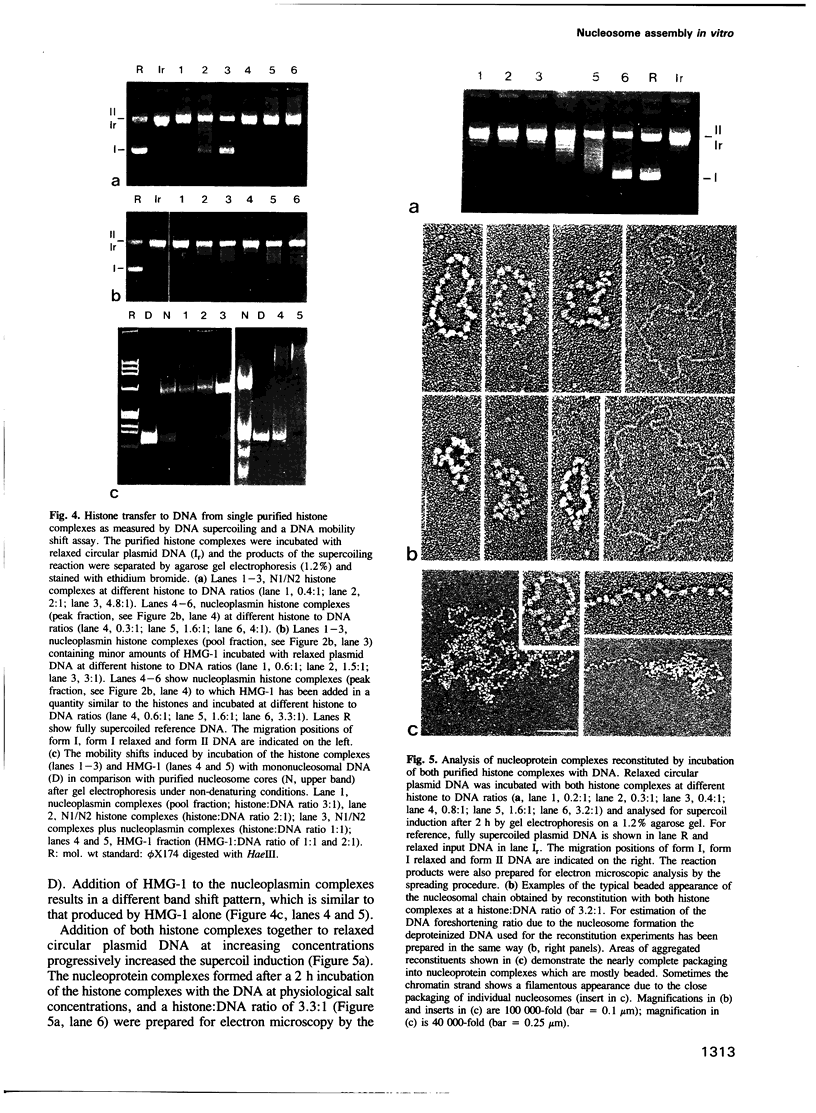
High speed supernatants of Xenopus laevis oocyte nuclei efficiently assemble DNA into nucleosomes in vitro under physiological salt conditions. The assembly activity cofractionates with two histone complexes composed of the acidic protein N1/N2 in complex with histones H3 and H4, and nucleoplasmin in complex with histones H2B and H2A. Both histone complexes have been purified and their nucleosome assembly activities have been analysed separately and in combination. While the histones from the N1/N2 complexes are efficiently transferred to DNA and induce supercoils into relaxed circular plasmid DNA, the nucleoplasmin complexes show no supercoil induction, but can also transfer their histones to DNA. In combination, the complexes act synergistically in supercoil induction thereby increasing the velocity and the number of supercoils induced. Electron microscopic analysis of the reaction products shows fully packaged nucleoprotein structures with the typical nucleosomal appearance resulting in a compaction ratio of 2.8 under low ionic strength conditions. The high mobility group protein HMG-1, which is also present in the soluble nuclear homogenate from X. laevis oocytes, is not required for nucleosome core assembly. Fractionation experiments show that the synergistic effect in the supercoiling reaction can be exerted by histones H3 and H4 bound to DNA and the nucleoplasmin complexes alone. This indicates that it is not the synchronous action of both complexes which is required for nucleosome assembly, but that their cooperative action can be resolved into two steps: deposition of H3 and H4 from the N1/N2 complexes onto the DNA and completion of nucleosome core formation by addition of H2B and H2A from the nucleoplasmin complexes.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adamson E. D., Woodland H. R. Histone synthesis in early amphibian development: histone and DNA syntheses are not co-ordinated. J Mol Biol. 1974 Sep 15;88(2):263–285. doi: 10.1016/0022-2836(74)90481-1. [DOI] [PubMed] [Google Scholar]
- Almouzni G., Méchali M. Assembly of spaced chromatin promoted by DNA synthesis in extracts from Xenopus eggs. EMBO J. 1988 Mar;7(3):665–672. doi: 10.1002/j.1460-2075.1988.tb02861.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragay A. M., Diaz P., Daban J. R. Association of nucleosome core particle DNA with different histone oligomers. Transfer of histones between DNA-(H2A,H2B) and DNA-(H3,H4) complexes. J Mol Biol. 1988 Nov 5;204(1):141–154. doi: 10.1016/0022-2836(88)90605-5. [DOI] [PubMed] [Google Scholar]
- Axel R., Melchior W., Jr, Sollner-Webb B., Felsenfeld G. Specific sites of interaction between histones and DNA in chromatin. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4101–4105. doi: 10.1073/pnas.71.10.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonne-Andrea C., Harper F., Sobczak J., De Recondo A. M. Rat liver HMG1: a physiological nucleosome assembly factor. EMBO J. 1984 May;3(5):1193–1199. doi: 10.1002/j.1460-2075.1984.tb01950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonne C., Duguet M., de Recondo A. M. Single-strand DNA binding protein from rat liver: interactions with supercoiled DNA. Nucleic Acids Res. 1980 Nov 11;8(21):4955–4968. doi: 10.1093/nar/8.21.4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürglin T. R., Mattaj I. W., Newmeyer D. D., Zeller R., De Robertis E. M. Cloning of nucleoplasmin from Xenopus laevis oocytes and analysis of its developmental expression. Genes Dev. 1987 Mar;1(1):97–107. doi: 10.1101/gad.1.1.97. [DOI] [PubMed] [Google Scholar]
- Cereghini S., Yaniv M. Assembly of transfected DNA into chromatin: structural changes in the origin-promoter-enhancer region upon replication. EMBO J. 1984 Jun;3(6):1243–1253. doi: 10.1002/j.1460-2075.1984.tb01959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotten M., Chalkley R. Purification of a novel, nucleoplasmin-like protein from somatic nuclei. EMBO J. 1987 Dec 20;6(13):3945–3954. doi: 10.1002/j.1460-2075.1987.tb02736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremisi C., Pignatti P. F., Croissant O., Yaniv M. Chromatin-like structures in polyoma virus and simian virus 10 lytic cycle. J Virol. 1975 Jan;17(1):204–211. doi: 10.1128/jvi.17.1.204-211.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crémisi C., Yaniv M. Sequential assembly of newly synthesized histones on replicating SV40 DNA. Biochem Biophys Res Commun. 1980 Feb 27;92(4):1117–1123. doi: 10.1016/0006-291x(80)90402-7. [DOI] [PubMed] [Google Scholar]
- Dilworth S. M., Black S. J., Laskey R. A. Two complexes that contain histones are required for nucleosome assembly in vitro: role of nucleoplasmin and N1 in Xenopus egg extracts. Cell. 1987 Dec 24;51(6):1009–1018. doi: 10.1016/0092-8674(87)90587-3. [DOI] [PubMed] [Google Scholar]
- Dilworth S. M., Dingwall C. Chromatin assembly in vitro and in vivo. Bioessays. 1988 Aug-Sep;9(2-3):44–49. doi: 10.1002/bies.950090203. [DOI] [PubMed] [Google Scholar]
- Dingwall C., Dilworth S. M., Black S. J., Kearsey S. E., Cox L. S., Laskey R. A. Nucleoplasmin cDNA sequence reveals polyglutamic acid tracts and a cluster of sequences homologous to putative nuclear localization signals. EMBO J. 1987 Jan;6(1):69–74. doi: 10.1002/j.1460-2075.1987.tb04720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glikin G. C., Ruberti I., Worcel A. Chromatin assembly in Xenopus oocytes: in vitro studies. Cell. 1984 May;37(1):33–41. doi: 10.1016/0092-8674(84)90298-8. [DOI] [PubMed] [Google Scholar]
- Griffith J. D. Chromatin structure: deduced from a minichromosome. Science. 1975 Mar 28;187(4182):1202–1203. doi: 10.1126/science.187.4182.1202. [DOI] [PubMed] [Google Scholar]
- Ishimi Y., Hirosumi J., Sato W., Sugasawa K., Yokota S., Hanaoka F., Yamada M. Purification and initial characterization of a protein which facilitates assembly of nucleosome-like structure from mammalian cells. Eur J Biochem. 1984 Aug 1;142(3):431–439. doi: 10.1111/j.1432-1033.1984.tb08305.x. [DOI] [PubMed] [Google Scholar]
- Ishimi Y., Kojima M., Yamada M., Hanaoka F. Binding mode of nucleosome-assembly protein (AP-I) and histones. Eur J Biochem. 1987 Jan 2;162(1):19–24. doi: 10.1111/j.1432-1033.1987.tb10535.x. [DOI] [PubMed] [Google Scholar]
- Ishimi Y., Yasuda H., Hirosumi J., Hanaoka F., Yamada M. A protein which facilitates assembly of nucleosome-like structures in vitro in mammalian cells. J Biochem. 1983 Sep;94(3):735–744. doi: 10.1093/oxfordjournals.jbchem.a134414. [DOI] [PubMed] [Google Scholar]
- Jackson V., Chalkley R. A new method for the isolation of replicative chromatin: selective deposition of histone on both new and old DNA. Cell. 1981 Jan;23(1):121–134. doi: 10.1016/0092-8674(81)90277-4. [DOI] [PubMed] [Google Scholar]
- Jackson V. Studies on histone organization in the nucleosome using formaldehyde as a reversible cross-linking agent. Cell. 1978 Nov;15(3):945–954. doi: 10.1016/0092-8674(78)90278-7. [DOI] [PubMed] [Google Scholar]
- Javaherian K., Liu J. F., Wang J. C. Nonhistone proteins HMG1 and HMG2 change the DNA helical structure. Science. 1978 Mar 24;199(4335):1345–1346. doi: 10.1126/science.628842. [DOI] [PubMed] [Google Scholar]
- Kleinschmidt J. A., Dingwall C., Maier G., Franke W. W. Molecular characterization of a karyophilic, histone-binding protein: cDNA cloning, amino acid sequence and expression of nuclear protein N1/N2 of Xenopus laevis. EMBO J. 1986 Dec 20;5(13):3547–3552. doi: 10.1002/j.1460-2075.1986.tb04681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschmidt J. A., Fortkamp E., Krohne G., Zentgraf H., Franke W. W. Co-existence of two different types of soluble histone complexes in nuclei of Xenopus laevis oocytes. J Biol Chem. 1985 Jan 25;260(2):1166–1176. [PubMed] [Google Scholar]
- Kleinschmidt J. A., Franke W. W. Soluble acidic complexes containing histones H3 and H4 in nuclei of Xenopus laevis oocytes. Cell. 1982 Jul;29(3):799–809. doi: 10.1016/0092-8674(82)90442-1. [DOI] [PubMed] [Google Scholar]
- Kleinschmidt J. A., Scheer U., Dabauvalle M. C., Bustin M., Franke W. W. High mobility group proteins of amphibian oocytes: a large storage pool of a soluble high mobility group-1-like protein and involvement in transcriptional events. J Cell Biol. 1983 Sep;97(3):838–848. doi: 10.1083/jcb.97.3.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschmidt J. A., Seiter A. Identification of domains involved in nuclear uptake and histone binding of protein N1 of Xenopus laevis. EMBO J. 1988 Jun;7(6):1605–1614. doi: 10.1002/j.1460-2075.1988.tb02986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskey R. A., Earnshaw W. C. Nucleosome assembly. Nature. 1980 Aug 21;286(5775):763–767. doi: 10.1038/286763a0. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Honda B. M., Mills A. D., Finch J. T. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature. 1978 Oct 5;275(5679):416–420. doi: 10.1038/275416a0. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Honda B. M., Mills A. D., Morris N. R., Wyllie A. H., Mertz J. E., De Roberts E. M., Gurdon J. B. Chromatin assembly and transcription in eggs and oocytes of Xenopus laevis. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):171–178. doi: 10.1101/sqb.1978.042.01.019. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D., Morris N. R. Assembly of SV40 chromatin in a cell-free system from Xenopus eggs. Cell. 1977 Feb;10(2):237–243. doi: 10.1016/0092-8674(77)90217-3. [DOI] [PubMed] [Google Scholar]
- Müller U., Zentgraf H., Eicken I., Keller W. Higher order structure of simian virus 40 chromatin. Science. 1978 Aug 4;201(4354):406–415. doi: 10.1126/science.208155. [DOI] [PubMed] [Google Scholar]
- Nelson T., Hsieh T. S., Brutlag D. Extracts of Drosophila embryos mediate chromatin assembly in vitro. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5510–5514. doi: 10.1073/pnas.76.11.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson T., Wiegand R., Brutlag D. Ribonucleic acid and other polyanions facilitate chromatin assembly in vitro. Biochemistry. 1981 Apr 28;20(9):2594–2601. doi: 10.1021/bi00512a035. [DOI] [PubMed] [Google Scholar]
- Oudet P., Gross-Bellard M., Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975 Apr;4(4):281–300. doi: 10.1016/0092-8674(75)90149-x. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Campos A., Shimamura A., Worcel A. Assembly and properties of chromatin containing histone H1. J Mol Biol. 1989 Sep 5;209(1):135–150. doi: 10.1016/0022-2836(89)90177-0. [DOI] [PubMed] [Google Scholar]
- Ruiz-Carrillo A., Jorcano J. L., Eder G., Lurz R. In vitro core particle and nucleosome assembly at physiological ionic strength. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3284–3288. doi: 10.1073/pnas.76.7.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U., Zentgraf H. Nucleosomal and supranucleosomal organization of transcriptionally inactive rDNA circles in Dytiscus oocytes. Chromosoma. 1978 Nov 22;69(2):243–254. doi: 10.1007/BF00329922. [DOI] [PubMed] [Google Scholar]
- Shimamura A., Tremethick D., Worcel A. Characterization of the repressed 5S DNA minichromosomes assembled in vitro with a high-speed supernatant of Xenopus laevis oocytes. Mol Cell Biol. 1988 Oct;8(10):4257–4269. doi: 10.1128/mcb.8.10.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. C., Dworkin-Rastl E., Dworkin M. B. Expression of a histone H1-like protein is restricted to early Xenopus development. Genes Dev. 1988 Oct;2(10):1284–1295. doi: 10.1101/gad.2.10.1284. [DOI] [PubMed] [Google Scholar]
- Smith S., Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989 Jul 14;58(1):15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- Stein A., Whitlock J. P., Jr, Bina M. Acidic polypeptides can assemble both histones and chromatin in vitro at physiological ionic strength. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5000–5004. doi: 10.1073/pnas.76.10.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. O., Kornberg R. D. An octamer of histones in chromatin and free in solution. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2626–2630. doi: 10.1073/pnas.72.7.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waithe W. I., Renaud J., Nadeau P., Pallotta D. Histone synthesis by lymphocytes in G0 and G1. Biochemistry. 1983 Apr 12;22(8):1778–1783. doi: 10.1021/bi00277a006. [DOI] [PubMed] [Google Scholar]
- Walker J. M., Gooderham K., Hastings J. R., Mayes E., Johns E. W. The primary structures of non-histone chromosomal proteins HMG 1 and 2. FEBS Lett. 1980 Dec 29;122(2):264–270. doi: 10.1016/0014-5793(80)80453-4. [DOI] [PubMed] [Google Scholar]
- Wedlich D., Dreyer C., Hausen P. Occurrence of a species-specific nuclear antigen in the germ line of Xenopus and its expression from paternal genes in hybrid frogs. Dev Biol. 1985 Mar;108(1):220–234. doi: 10.1016/0012-1606(85)90025-9. [DOI] [PubMed] [Google Scholar]
- West M. H., Bonner W. M. Histone 2A, a heteromorphous family of eight protein species. Biochemistry. 1980 Jul 8;19(14):3238–3245. doi: 10.1021/bi00555a022. [DOI] [PubMed] [Google Scholar]
- Woodland H. R. Histone synthesis during the development of Xenopus. FEBS Lett. 1980 Nov 17;121(1):1–10. doi: 10.1016/0014-5793(80)81252-x. [DOI] [PubMed] [Google Scholar]
- Worcel A., Han S., Wong M. L. Assembly of newly replicated chromatin. Cell. 1978 Nov;15(3):969–977. doi: 10.1016/0092-8674(78)90280-5. [DOI] [PubMed] [Google Scholar]
- Wu R. S., Bonner W. M. Separation of basal histone synthesis from S-phase histone synthesis in dividing cells. Cell. 1981 Dec;27(2 Pt 1):321–330. doi: 10.1016/0092-8674(81)90415-3. [DOI] [PubMed] [Google Scholar]
- Wu R. S., Tsai S., Bonner W. M. Patterns of histone variant synthesis can distinguish G0 from G1 cells. Cell. 1982 Dec;31(2 Pt 1):367–374. doi: 10.1016/0092-8674(82)90130-1. [DOI] [PubMed] [Google Scholar]
- Wyllie A. H., Laskey R. A., Finch J., Gurdon J. B. Selective DNA conservation and chromatin assembly after injection of SV40 DNA into Xenopus oocytes. Dev Biol. 1978 May;64(1):178–188. doi: 10.1016/0012-1606(78)90069-6. [DOI] [PubMed] [Google Scholar]
- Zentgraf H., Franke W. W. Differences of supranucleosomal organization in different kinds of chromatin: cell type-specific globular subunits containing different numbers of nucleosomes. J Cell Biol. 1984 Jul;99(1 Pt 1):272–286. doi: 10.1083/jcb.99.1.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentgraf H., Trendelenburg M. F., Spring H., Scheer U., Franke W. W., Müller U., Drury K. C., Rungger D. Mitochondrial DNA arranged into chromatin-like structures after injection into amphibian oocyte nuclei. Exp Cell Res. 1979 Sep;122(2):363–375. doi: 10.1016/0014-4827(79)90312-4. [DOI] [PubMed] [Google Scholar]
- van Dongen W. M., Moorman A. F., Destrée O. H. The accumulation of the maternal pool of histone H1A during oogenesis in Xenopus laevis. Cell Differ. 1983 May;12(5):257–264. doi: 10.1016/0045-6039(83)90021-0. [DOI] [PubMed] [Google Scholar]