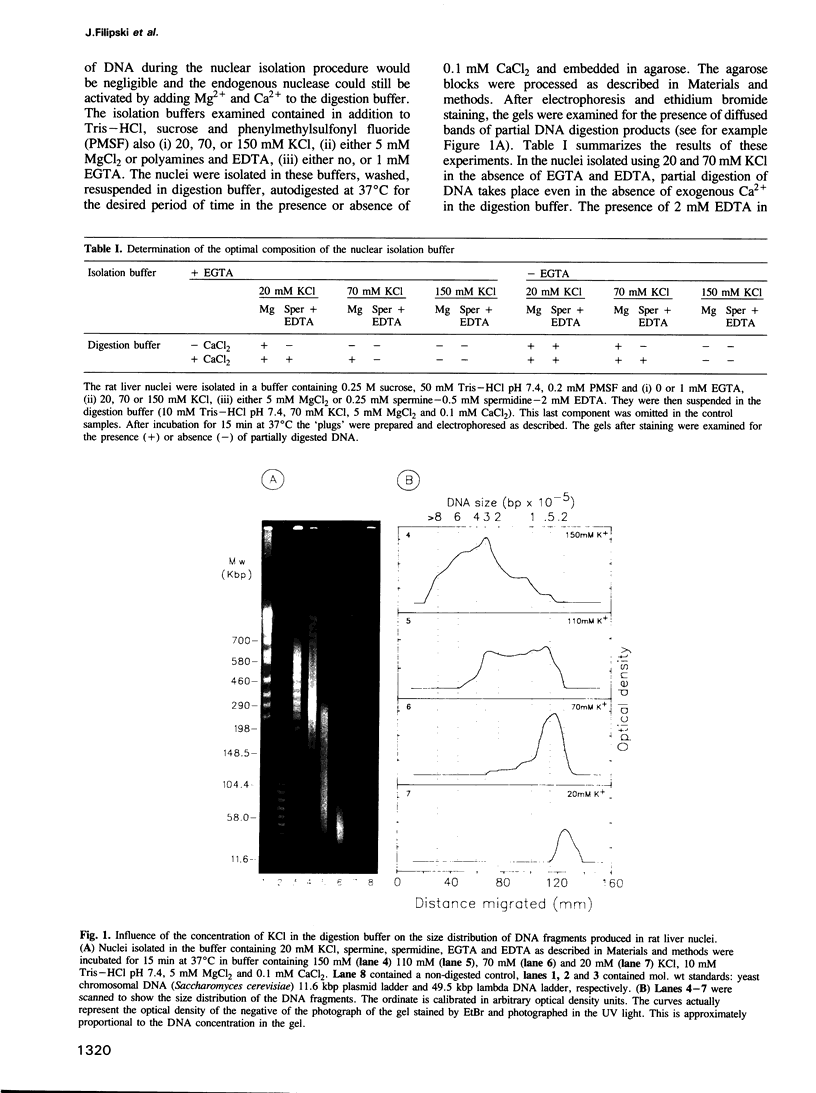
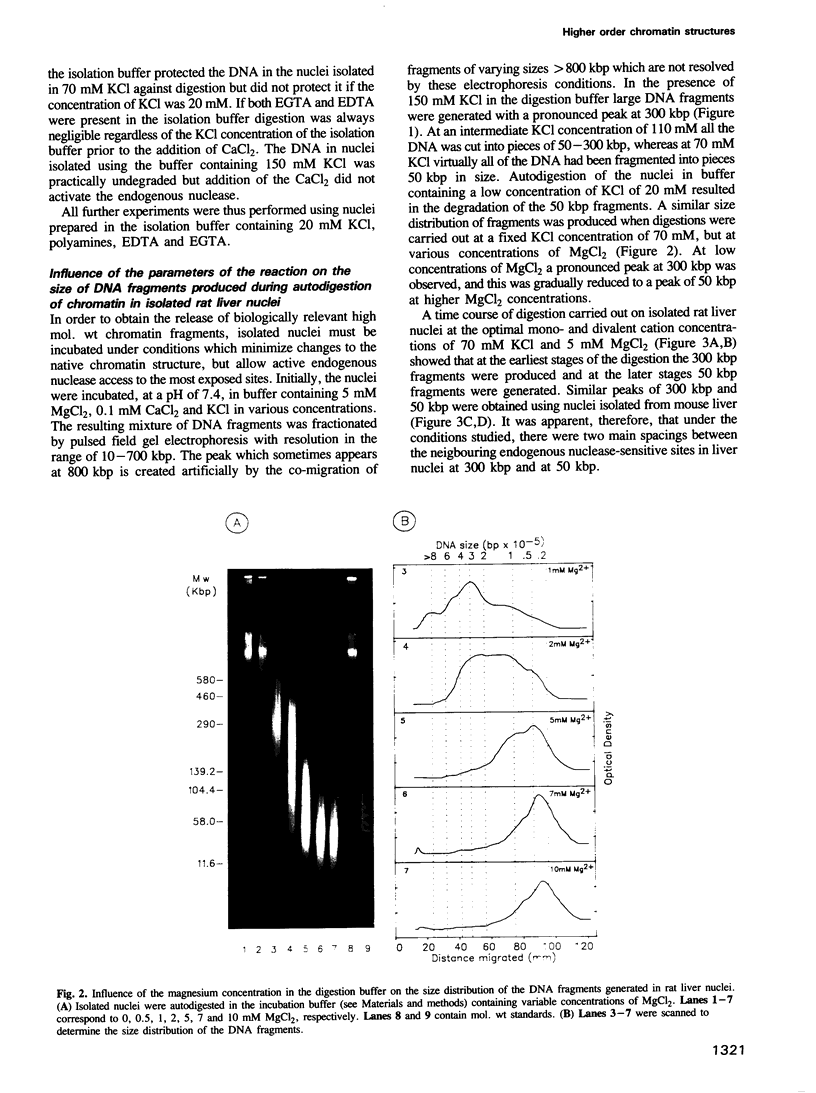
Abstract
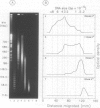
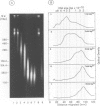
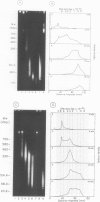
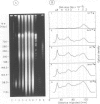
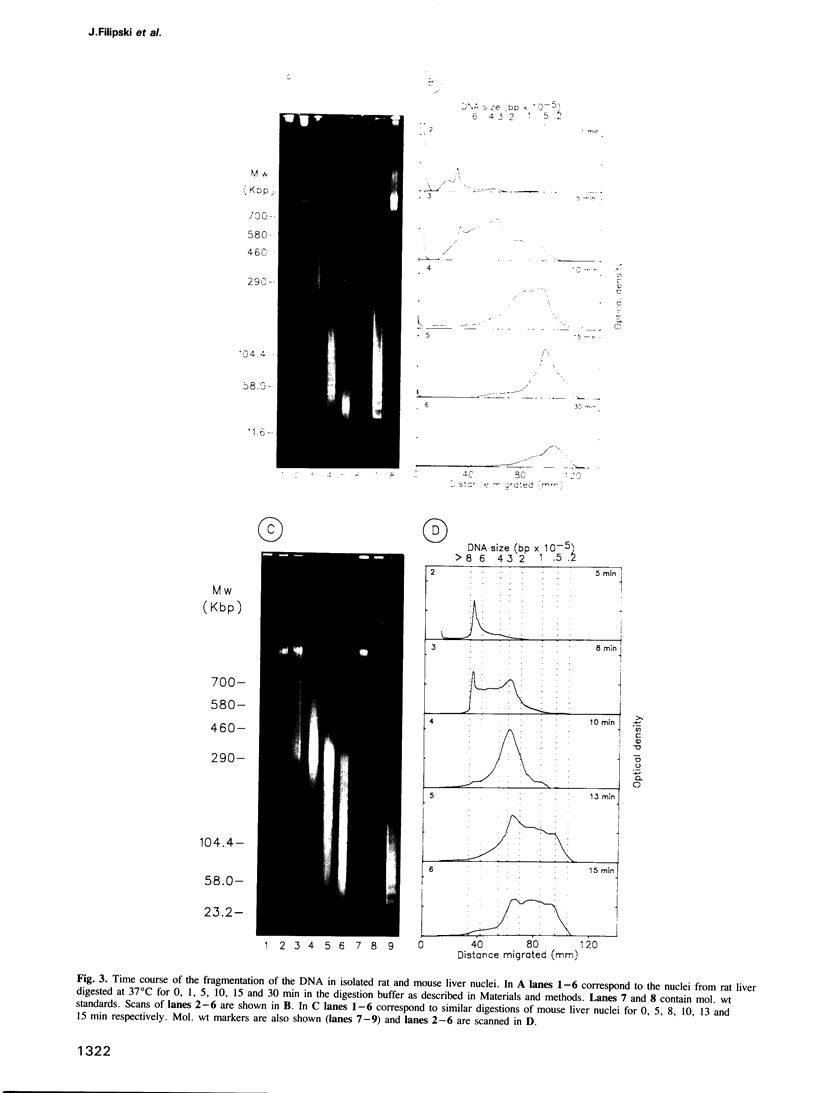
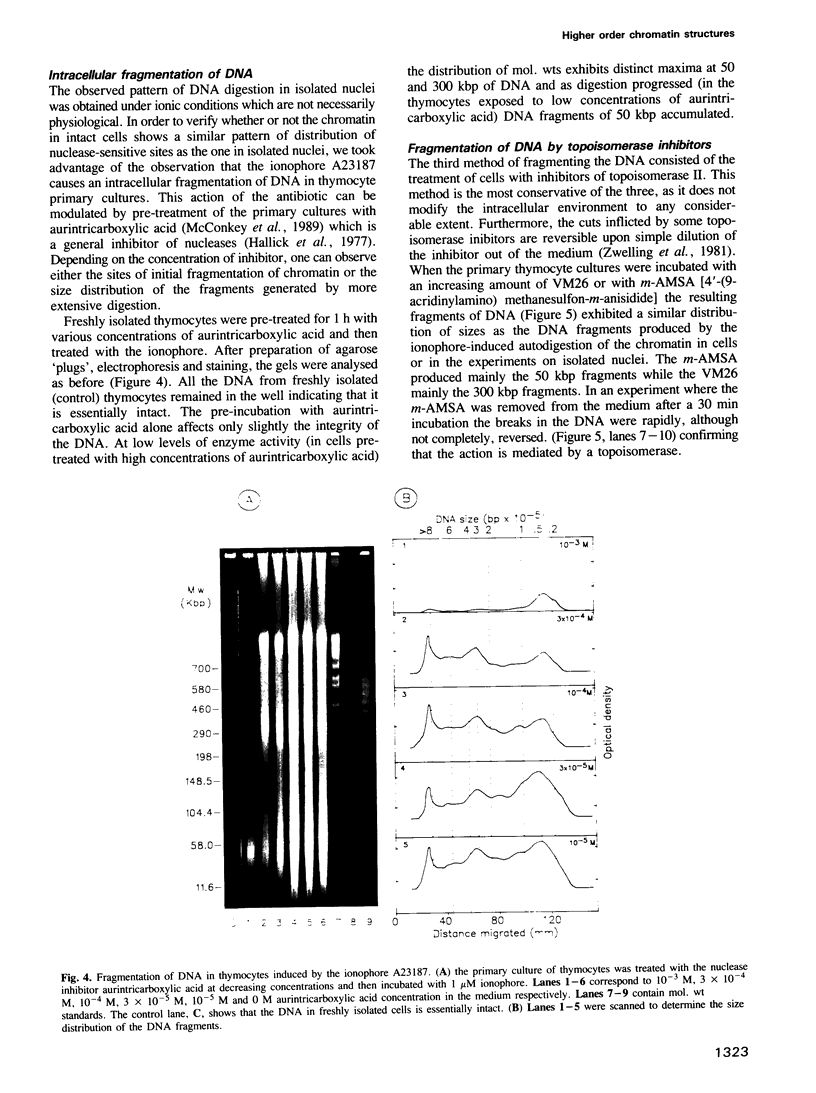
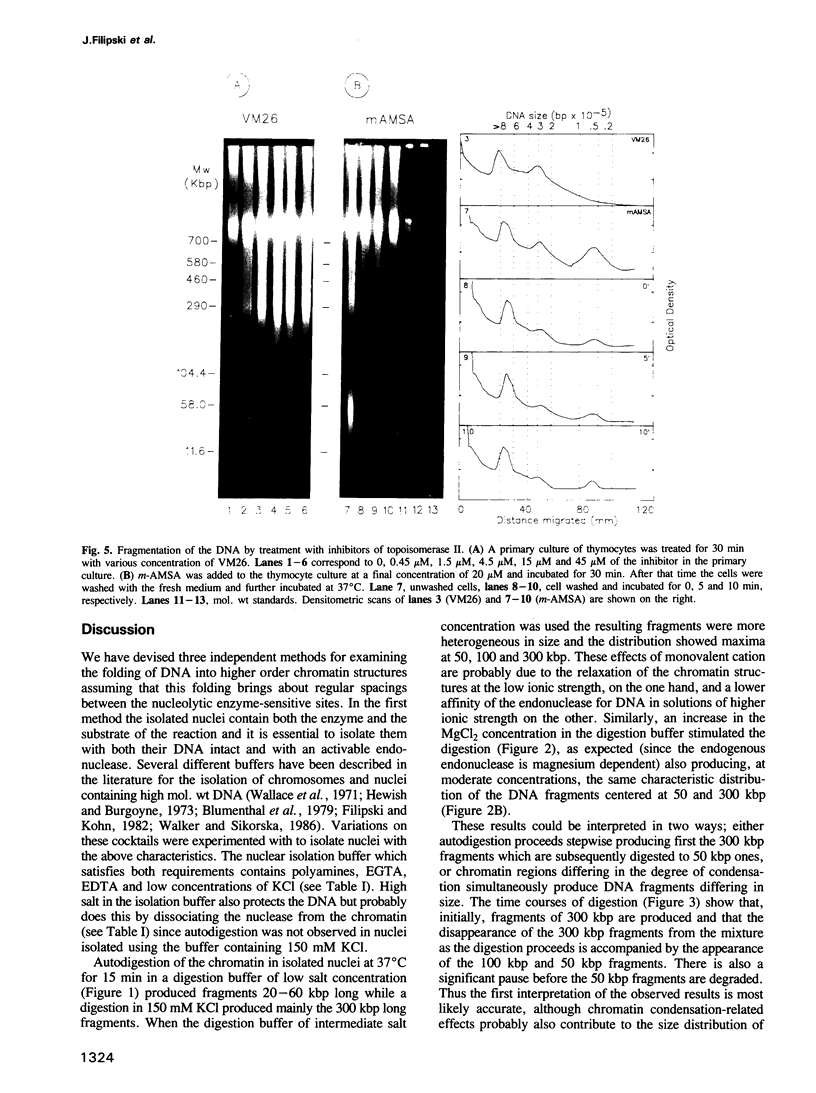
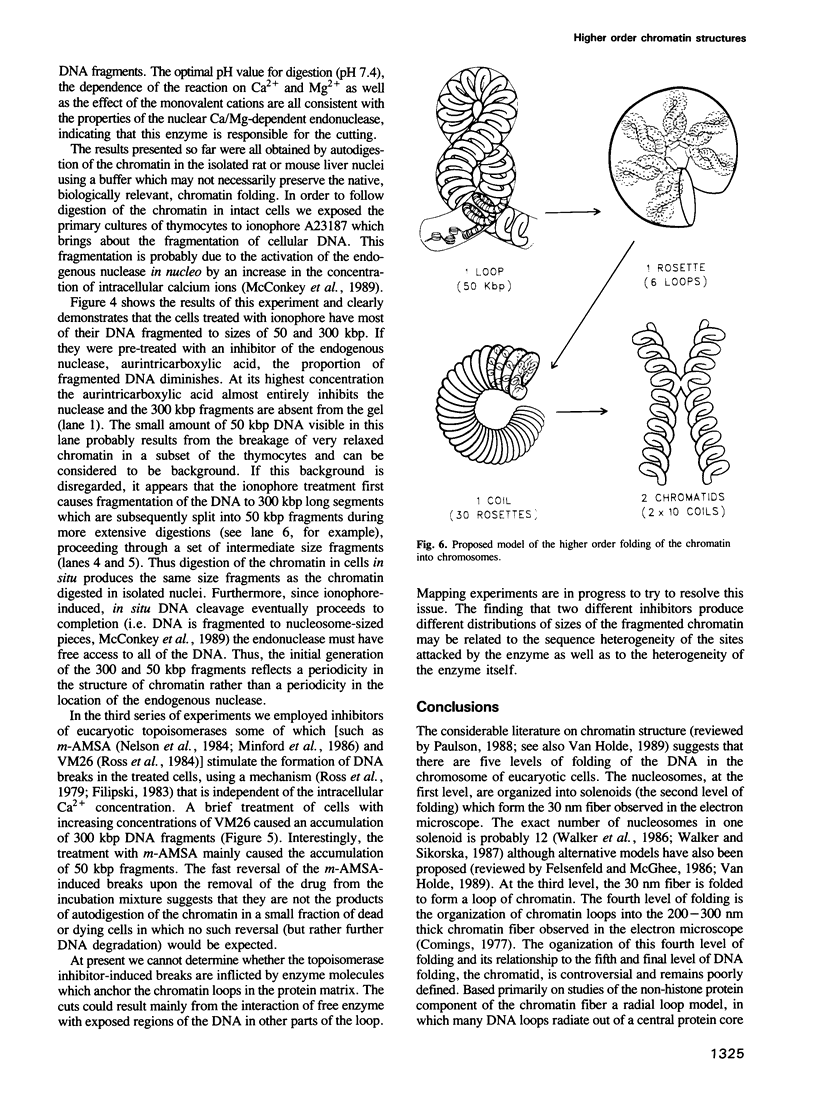
Each level of DNA folding in cells corresponds to a distinct chromatin structure. The basic chromatin units, nucleosomes, are arranged into solenoids which form chromatin loops. To characterize better the loop organization of chromatin we have assumed that the accessibility of DNA inside these structures is lower than on the outside and examined the size distribution of high mol. wt DNA fragments obtained from cells and isolated nuclei after digestion with endogenous nuclease or topoisomerase II. The largest discrete fragments obtained contain 300 kbp of DNA. Their further degradation proceeds through another discrete size step of 50 kbp. This suggests that chromatin loops contain approximately 50 kbp of DNA and that they are grouped into hexameric rosettes at the next higher level of chromatin structure. Based upon these observations a model by which the 30 nm chromatin fibre can be folded up into compact metaphase chromosomes is also described.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berrios M., Osheroff N., Fisher P. A. In situ localization of DNA topoisomerase II, a major polypeptide component of the Drosophila nuclear matrix fraction. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4142–4146. doi: 10.1073/pnas.82.12.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal A. B., Dieden J. D., Kapp L. N., Sedat J. W. Rapid isolation of metaphase chromosomes containing high molecular weight DNA. J Cell Biol. 1979 Apr;81(1):255–259. doi: 10.1083/jcb.81.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boy de la Tour E., Laemmli U. K. The metaphase scaffold is helically folded: sister chromatids have predominantly opposite helical handedness. Cell. 1988 Dec 23;55(6):937–944. doi: 10.1016/0092-8674(88)90239-5. [DOI] [PubMed] [Google Scholar]
- Cook P. R., Brazell I. A. Supercoils in human DNA. J Cell Sci. 1975 Nov;19(2):261–279. doi: 10.1242/jcs.19.2.261. [DOI] [PubMed] [Google Scholar]
- DiNardo S., Voelkel K., Sternglanz R. DNA topoisomerase II mutant of Saccharomyces cerevisiae: topoisomerase II is required for segregation of daughter molecules at the termination of DNA replication. Proc Natl Acad Sci U S A. 1984 May;81(9):2616–2620. doi: 10.1073/pnas.81.9.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W. C., Heck M. M. Localization of topoisomerase II in mitotic chromosomes. J Cell Biol. 1985 May;100(5):1716–1725. doi: 10.1083/jcb.100.5.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld G., McGhee J. D. Structure of the 30 nm chromatin fiber. Cell. 1986 Feb 14;44(3):375–377. doi: 10.1016/0092-8674(86)90456-3. [DOI] [PubMed] [Google Scholar]
- Filipski J. Competitive inhibition of nicking--closing enzymes may explain some biological effects of DNA intercalators. FEBS Lett. 1983 Aug 8;159(1-2):6–12. doi: 10.1016/0014-5793(83)80406-2. [DOI] [PubMed] [Google Scholar]
- Filipski J., Kohn K. W. Ellipticine-induced protein-associated DNA breaks in isolated L1210 nuclei. Biochim Biophys Acta. 1982 Sep 27;698(3):280–286. doi: 10.1016/0167-4781(82)90158-0. [DOI] [PubMed] [Google Scholar]
- Filipski J., Yin J., Kohn K. W. Reconstitution of intercalator-induced DNA scission by an active component from nuclear extracts. Biochim Biophys Acta. 1983 Oct 13;741(1):116–122. doi: 10.1016/0167-4781(83)90017-9. [DOI] [PubMed] [Google Scholar]
- Haapala O., Nokkala S. Structure of human metaphase chromosomes. Hereditas. 1982;96(2):215–228. doi: 10.1111/j.1601-5223.1982.tb00852.x. [DOI] [PubMed] [Google Scholar]
- Hallick R. B., Chelm B. K., Gray P. W., Orozco E. M., Jr Use of aurintricarboxylic acid as an inhibitor of nucleases during nucleic acid isolation. Nucleic Acids Res. 1977 Sep;4(9):3055–3064. doi: 10.1093/nar/4.9.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. The calcium dependent endonuclease activity of isolated nuclear preparations. Relationships between its occurrence and the occurrence of other classes of enzymes found in nuclear preparations. Biochem Biophys Res Commun. 1973 May 15;52(2):475–481. doi: 10.1016/0006-291x(73)90736-5. [DOI] [PubMed] [Google Scholar]
- Igó-Kemenes T., Zachau H. G. Domains in chromatin structure. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):109–118. doi: 10.1101/sqb.1978.042.01.012. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K., Cheng S. M., Adolph K. W., Paulson J. R., Brown J. A., Baumbach W. R. Metaphase chromosome structure: the role of nonhistone proteins. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):351–360. doi: 10.1101/sqb.1978.042.01.036. [DOI] [PubMed] [Google Scholar]
- McConkey D. J., Hartzell P., Nicotera P., Orrenius S. Calcium-activated DNA fragmentation kills immature thymocytes. FASEB J. 1989 May;3(7):1843–1849. doi: 10.1096/fasebj.3.7.2497041. [DOI] [PubMed] [Google Scholar]
- Minford J., Pommier Y., Filipski J., Kohn K. W., Kerrigan D., Mattern M., Michaels S., Schwartz R., Zwelling L. A. Isolation of intercalator-dependent protein-linked DNA strand cleavage activity from cell nuclei and identification as topoisomerase II. Biochemistry. 1986 Jan 14;25(1):9–16. doi: 10.1021/bi00349a002. [DOI] [PubMed] [Google Scholar]
- Nelson E. M., Tewey K. M., Liu L. F. Mechanism of antitumor drug action: poisoning of mammalian DNA topoisomerase II on DNA by 4'-(9-acridinylamino)-methanesulfon-m-anisidide. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1361–1365. doi: 10.1073/pnas.81.5.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnuki Y. Structure of chromosomes. I. Morphological studies of the spiral structure of human somatic chromosomes. Chromosoma. 1968;25(4):402–428. doi: 10.1007/BF02327721. [DOI] [PubMed] [Google Scholar]
- Paulson J. R., Laemmli U. K. The structure of histone-depleted metaphase chromosomes. Cell. 1977 Nov;12(3):817–828. doi: 10.1016/0092-8674(77)90280-x. [DOI] [PubMed] [Google Scholar]
- Pienta K. J., Coffey D. S. A structural analysis of the role of the nuclear matrix and DNA loops in the organization of the nucleus and chromosome. J Cell Sci Suppl. 1984;1:123–135. doi: 10.1242/jcs.1984.supplement_1.9. [DOI] [PubMed] [Google Scholar]
- Rattner J. B., Lin C. C. Radial loops and helical coils coexist in metaphase chromosomes. Cell. 1985 Aug;42(1):291–296. doi: 10.1016/s0092-8674(85)80124-0. [DOI] [PubMed] [Google Scholar]
- Ross W. E., Glaubiger D., Kohn K. W. Qualitative and quantitative aspects of intercalator-induced DNA strand breaks. Biochim Biophys Acta. 1979 Mar 28;562(1):41–50. doi: 10.1016/0005-2787(79)90124-2. [DOI] [PubMed] [Google Scholar]
- Ross W., Rowe T., Glisson B., Yalowich J., Liu L. Role of topoisomerase II in mediating epipodophyllotoxin-induced DNA cleavage. Cancer Res. 1984 Dec;44(12 Pt 1):5857–5860. [PubMed] [Google Scholar]
- Sikorska M., MacManus J. P., Walker P. R., Whitfield J. F. The protein kinases of rat liver nuclei. Biochem Biophys Res Commun. 1980 Apr 29;93(4):1196–1203. doi: 10.1016/0006-291x(80)90616-6. [DOI] [PubMed] [Google Scholar]
- Smith C. L., Cantor C. R. Purification, specific fragmentation, and separation of large DNA molecules. Methods Enzymol. 1987;155:449–467. doi: 10.1016/0076-6879(87)55030-3. [DOI] [PubMed] [Google Scholar]
- Strätling W. H., Grade C., Hörz W. Ca/Mg-dependent endonuclease from porcine liver. Purification, properties, and sequence specificity. J Biol Chem. 1984 May 10;259(9):5893–5898. [PubMed] [Google Scholar]
- Vogelstein B., Pardoll D. M., Coffey D. S. Supercoiled loops and eucaryotic DNA replicaton. Cell. 1980 Nov;22(1 Pt 1):79–85. doi: 10.1016/0092-8674(80)90156-7. [DOI] [PubMed] [Google Scholar]
- Walker P. R., Sikorska M. Chromatin structure. Evidence that the 30-nm fiber is a helical coil with 12 nucleosomes/turn. J Biol Chem. 1987 Sep 5;262(25):12223–12227. [PubMed] [Google Scholar]
- Walker P. R., Sikorska M. Modulation of the sensitivity of chromatin to exogenous nucleases: implications for the apparent increased sensitivity of transcriptionally active genes. Biochemistry. 1986 Jul 1;25(13):3839–3845. doi: 10.1021/bi00361a015. [DOI] [PubMed] [Google Scholar]
- Walker P. R., Sikorska M., Whitfield J. F. Chromatin structure. Nuclease digestion profiles reflect intermediate stages in the folding of the 30-nm fiber rather than the existence of subunit beads. J Biol Chem. 1986 May 25;261(15):7044–7051. [PubMed] [Google Scholar]
- Wallace P. G., Hewish D. R., Venning M. M., Burgoyne L. A. Multiple forms of mammalian deoxyribonucleic acid polymerase. An attempt to relate their interactions with nuclei and free deoxyribonucleic acid in vitro with their possible functions in vivo. Biochem J. 1971 Nov;125(1):47–54. doi: 10.1042/bj1250047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield J. F., Perris A. D., Youdale T. Destruction of the nuclear morphology of thymic lymphocytes by the corticosteroid cortisol. Exp Cell Res. 1968 Oct;52(2):349–362. doi: 10.1016/0014-4827(68)90476-x. [DOI] [PubMed] [Google Scholar]
- Zwelling L. A., Michaels S., Erickson L. C., Ungerleider R. S., Nichols M., Kohn K. W. Protein-associated deoxyribonucleic acid strand breaks in L1210 cells treated with the deoxyribonucleic acid intercalating agents 4'-(9-acridinylamino) methanesulfon-m-anisidide and adriamycin. Biochemistry. 1981 Nov 10;20(23):6553–6563. doi: 10.1021/bi00526a006. [DOI] [PubMed] [Google Scholar]