Abstract
The proteins of the cellular plasma membrane perform important functions relating to homeostasis and intercellular communication. Due to its overall low cellular abundance, amphipathic character, and low membrane-to-cytoplasm ratio, the plasma membrane proteome has been challenging to isolate and characterize, and is poorly represented in standard liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses. In this study, we employ sucrose gradient ultracentrifugation for the enrichment of the plasma membrane proteome, without chemical labeling and affinity purification, together with GeLCMS and use subsequent bioinformatics tools to select plasma membrane proteins, herein referred to as the surfaceome. Using this methodology, we identify over 1900 cell surface-associated proteins in a human acute myeloid leukemia (AML) cell line. These surface proteins comprise almost 50% of all detected cellular proteins, a number that substantially exceeds the depth of coverage in previously published studies describing the leukemia surfaceome.
Keywords: label-free, proteomics, plasma membrane, GO term, leukemia
Proteins at the cell surface comprise extracellular, transmembrane, and/or intracellular domains that mediate a variety of essential cellular functions. Membrane proteins allow cells to communicate with each other and with the extracellular milieu. They are also critical for the propagation of signaling cascades, interaction with pathogens and responses to environmental changes. Moreover, two thirds of known drug-targets are membrane proteins [1].
Global gene expression studies to date have focused on overall changes at the genomic and transcriptomic levels. However, these have not allowed us to draw reliable conclusions about events at the protein level, since there is a relatively poor correlation between mRNA and protein expression [2–6]. This observation highlights the need for proteomic analyses in any systematic interrogation of cellular phenotype.
To date, in-depth proteomic interrogation of the cell surface landscape has been hampered by the difficulty in extracting highly enriched plasma membrane isolates, low membrane-to-cytoplasm protein abundance ratio, and challenges in resolving and identifying hydrophobic proteins and peptides [7–9]. In an effort to overcome these challenges, a number of strategies have been employed. Chief among these have been chemical tagging methods in conjunction with subcellular fractionation. As an example, biotin labeling of exposed lysine residues of cell surface proteins has been a widely used approach [10–14]. Despite improvements in the purity of the surfaceome achieved with these workflows, the yield in terms of absolute numbers of proteins has remained modest [15]. Enrichments based on exogenous labels depend heavily on labeling efficiency, and bias may be introduced by excluding proteins that do not carry the targeted residue or do not expose the residue to a degree that allows labeling. The biotin labeling efficiency has been reported to range between 20 and 33% [15–17]. A recent study detected 650 plasma membrane proteins from biotinylated cells, representing 50% of all proteins identified and a 33% labeling efficiency [17]. While these findings compare favorably with earlier studies probing the biotin-labeled surfaceome [18–20], the human cell surface proteome is thought to contain around 3700 proteins based on in silico predictions [21]. We hypothesized that some of the limitations of the biotinylation procedure such as low labeling efficiency [15–17], negative impact of derivatization on cell viability, cell permeation of the biotinylation reagent, and imperfect affinity purification could be overcome by a label-free approach. In addition to an improved proteome coverage, it would also greatly simplify the workflow.
The purpose of this study, therefore, was to develop a robust, label-free, non-affinity purified MS-based workflow that would allow a more comprehensive evaluation of the cell surface proteome. To our knowledge, no study until now has captured more than 870 cell surface proteins for a single sample replicate and thus it appears unlikely that a comprehensive proteomic inventory of any particular cell type has as yet been performed.
The surfaceome can in principle be isolated either via chemical labels or extracted from cells based on physicochemical properties. Reports to date have been inconsistent with respect to which of these two approaches results in the highest yield of cell surface proteins [16,22]. Biotin labeling of lysine residues of cell surface exposed proteins has been a popular choice of chemical labeling to identify plasma membrane-associated proteins. A number of recent publications querying the composition of the cell surfaceome have used this approach to specifically enrich for and capture cell surface proteins [13,14,16,18,20,23]. There are however challenges with the biotinylation approach as previously outlined. We therefore developed a robust, label-free, non-affinity-purified workflow for the enrichment and detection of cell surface proteins.
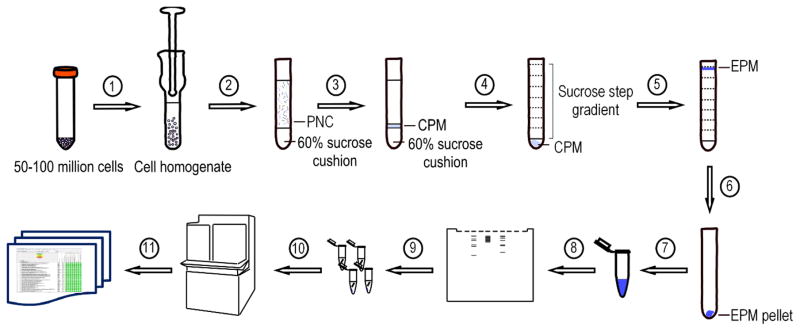
To this end, two density-equilibrium ultracentrifugation cycles were performed [24,25] (Figure 1). We used the human AML cell line MOLM-14 for the isolation and enrichment of the cell surfaceome.
Figure 1.
Experimental workflow for the proposed label-free, non-affinity-enriched surfaceome protocol. (1) Resuspend cells in buffer and homogenize with a Dounce homogenizer. Spin homogenate at 1,000g for 10 minutes at 4°C. (2) Layer the PNC over a 60% sucrose cushion and centrifuge at 100,000g for an hour at 4°C. (3) Recover the CPM fraction from the top of the sucrose cushion and transfer to a centrifuge tube. (4) Layer sucrose step gradient on top of the CPM. Centrifuge at 100,000g overnight at 4°C. (5) Recover the EPM fraction at the 37% sucrose fraction border phase and transfer to a centrifuge tube. (6) Fill the tube with HEPES buffer and centrifuge at 150,000g for an hour 4°C. (7) Dissolve the EPM pellet in 300 μL ammonium bicarbonate buffer. Acetone precipitate all or part of the protein sample. (8) Denature protein sample and run on an SDS-PAGE. (9) Cut the gel in thin slices and perform in-gel trypsin digestion. (10) Extract the tryptic peptides and analyze by LC-MS/MS. (11) Bioinformatic data processing and analysis.
PNC, post-nuclear cytosol; CPM, crude plasma membrane; EPM, enriched plasma membrane
In the first isolation step we centrifuged the post-nuclear lysate over a 60% sucrose cushion to enrich for the crude plasma membrane (CPM) fraction at the border phase. Subsequently, a sucrose step gradient was layered over the CPM for further enrichment of the plasma membrane (EPM) fraction by centrifugation overnight at 100,000g. During this centrifugation the plasma membrane will float to the top of the sucrose gradient, which provides better recovery and resolution than sedimentation (Experimental Procedures_SuppInfo).
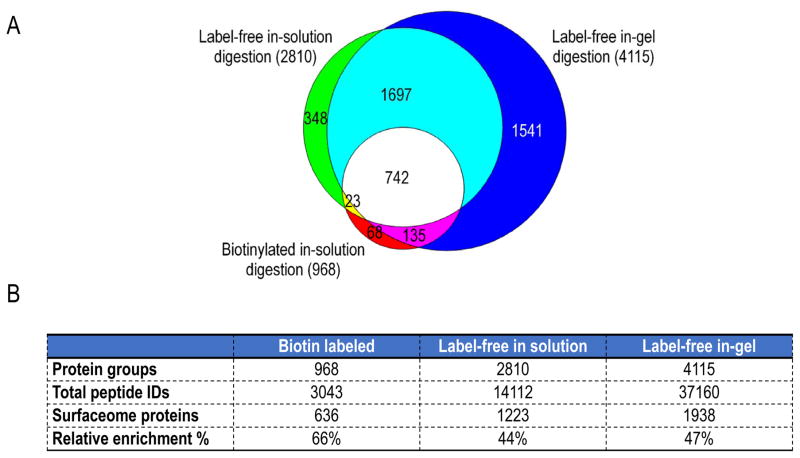
To our knowledge, there has not been a direct comparison between label-free, non-affinity-purified and biotin-labeled surfaceome workflows. Weekes and colleagues compared whole cell lysate and crude membrane preparations, but not enriched plasma membrane preparations, with biotinylated plasma membrane isolates [15]. We therefore first compared biotin-labeled isolation with label-free, non-affinity purified isolation of the cell surfaceome. The cell surface proteome of MOLM-14 cells was captured via lysine biotinylation, following a published protocol [14] and compared to the MOLM-14 surfaceome isolated with our proposed label-free workflow. As shown in Figure 2, we isolated 2810 protein groups with the label-free approach and 968 protein groups via lysine biotinylation, a similar number to published data using biotinylation methods [17,19,20]. Around 80% of the proteins captured via biotinylation were also isolated in the label-free workflow.
Figure 2.
Comparison of three different surfaceome analysis workflows. (A) Area proportional Venn diagram depicting the overlap of all protein groups identified in the surfaceome isolated with three different methods: biotinylated in-solution digestion, label-free in-solution digestion and label-free in-gel digestion. (B) Absolute and relative numbers of cell surface annotated proteins identified and total numbers of peptides identified with the three different methods compared.
In the studies described above, proteins were hydrolyzed in solution with trypsin. Earlier work had suggested that in particular for plasma membrane proteins, in-gel trypsin digestion outperforms in-solution digestion [26]. The work by Choksawangkarn and colleagues showed not only an increase in the total number of cell surface proteins identified, but also a larger representation of hydrophobic proteins with the in-gel digestion protocol [26]. Therefore, we also compared in-solution and in-gel trypsin digestion in the current label-free workflow. Not surprisingly, we identified the greatest number of protein groups (4115) with the in-gel digestion workflow, in part due to the protein-level fractionation of the in-gel technique, which is absent in the in-solution digestion protocol (Figure 2A). When comparing the three workflows, the label-free in-gel digestion workflow identified the most cell surface proteins per the GO term selection criteria used (Figure 2B and Table 1_SuppInfo).
These studies demonstrate that our label-free strategy can bypass some of the inherent limitations of the biotinylation procedure, such as low labeling efficiency, cell permeation of the biotin reagent, negative effect of derivatization on cell viability, and difficulty in detection of biotinylated peptides originating from labeling at the protein level [27]. Moreover, the protein-level fractionation incurred by the in-gel trypsin digestion most likely results in more low-abundance proteins identified compared to in-solution techniques. A comparison of spectral counts between the techniques has suggested that some protein may be lost during resolubilization in the in-solution procedure contributing to the identification of fewer plasma membrane proteins [26].
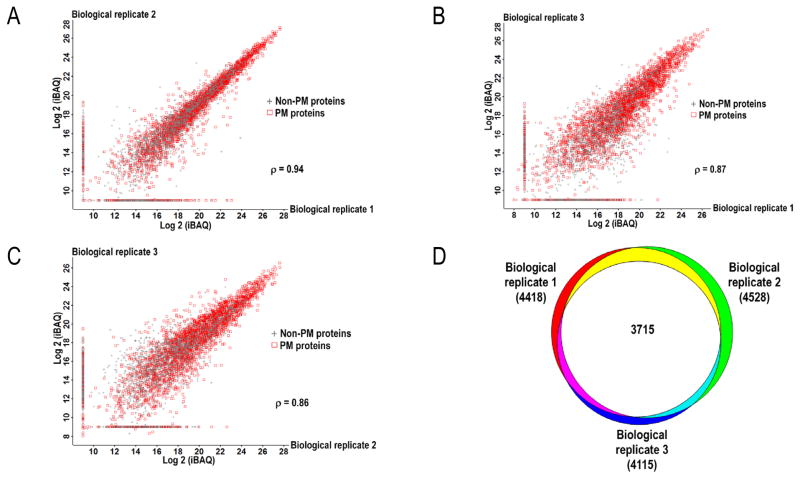
Repeatability and reproducibility are key aspects of any scientific protocol and indispensable measures of a method’s general performance. To these ends, we performed a series of experiments to determine the technical repeatability and biological reproducibility of our new optimized workflow. The technical repeatability of our MS analysis is demonstrated by >90% overlap for proteins identified in three technical repeats, thus demonstrating high test-retest reliability (Figure 1_SuppInfo). The MaxQuant iBAQ values (Experimental Procedures_SuppInfo and Table 2_SuppInfo) for the technical repeats were plotted in pairwise comparisons for each protein, and the Pearson correlation coefficient (ρ) was calculated for each comparison. All comparisons yielded Pearson coefficients close to 1, indicating a near perfect 1:1 relationship between repeats.
Equal comparisons were done with the surface proteomes identified from three different batches of MOLM-14 cells processed at three distinct times. As presented in Figure 3, the surfaceome showed approximately 85% biological reproducibility. The Pearson correlation coefficients ranged between 0.86 and 0.94, confirming a strong linear relationship between the biological replicates. These numbers highlight the robustness of this workflow. As these experiments were performed on an immortalized cell line, it was not difficult to obtain large numbers of cells for analyses. However, in future work on primary cells, cell number could be limiting. Therefore, we wanted to investigate whether similar surfaceome coverage could be obtained with fewer than 100 million cells. We extracted EPM fractions in parallel from 50 and 100 million MOLM-14 cells for comparison of identified proteins. An equal amount of protein from the two EPM fractions were in-gel digested and the tryptic peptides were analyzed by LC-MS/MS. We identified a similar number of protein groups in both samples (4712 versus 4640) with 4248 protein groups in common (Table 3_SuppInfo), an average 91% overlap that is similar to the overlap previously observed among biological replicates (Figure 2). We therefore concluded that use of 50 million cells was sufficient for this protocol and did not compromise depth of protein coverage. Equivalent experiments using 25 million cells indicated similar proteome coverage (Figure 2_SuppInfo).
Figure 3.
(A–C) Biological reproducibility assessed by the calculated Pearson correlation coefficient (ρ) for each pairwise comparison between three biological replicates prepared and analyzed on different days from different batches of cells. MaxQuant Log2 (iBAQ) values present in one but absent in another pairwise comparison were given an arbitrary value of 9 for visualization purposes. (D) Venn diagram depicts the overlap in detected proteins between the biological replicates. PM, plasma membrane
It remains generally unknown in all cell types which proteins are present at the cell surface and how their expression changes in response to physiological and non-physiological cues. In this study, we sought to capture not only integral membrane proteins, but also peripherally associated membrane proteins. We thus used GO annotation terminology to define the surfaceome, including cell surface (GO:0009986), plasma membrane (GO:0005886), extracellular space (GO:0005615), extracellular region (GO:0005576), and integral component of plasma membrane (GO:0005887) [16,17,22]. For a more comprehensive cell surfaceome coverage, we expanded this core list with additional GO terms including: cell outer membrane (GO:0009279), external side of plasma membrane (GO:0009897), extracellular exosome (GO:0070062) and membrane raft (GO:0045121) (Experimental Procedures_SuppInfo). Hörmann and colleagues recently reported that the majority of non-plasma membrane annotated proteins are in fact co-purified due to interaction with plasma membrane annotated proteins. They noted that out of 201 proteins that were not annotated as “plasma membrane”, “cell surface”, “cell membrane” or “extracellular”, 140 were found to interact with at least one plasma membrane protein [17]. This strongly argues for broader GO term inclusion when defining the surfaceome in order not to exclude legitimate cell surface proteins or to limit the potential of this approach.
In our label-free, in-gel digested surfaceome we identified 4115 protein groups, out of which 1938 protein groups (47%) were plasma membrane-associated according to our definition. As shown in Figure 2B, the corresponding numbers for the biotinylated surfaceome were 968 and 636 protein groups (66%), respectively. While these numbers suggest a higher relative enrichment of cell surface proteins with lysine biotinylation, corroborating data reported in earlier studies [13], the total number of proteins and the number of plasma membrane-associated proteins are dramatically improved with the label-free isolation in these studies (Figure 2 and Table 1_SuppInfo). In addition, we queried the proteins identified in the label-free workflow for known AML cell surface antigens. We found a high representation of previously published AML cell surface proteins among the captured surface antigens (Table 4_SuppInfo), which provides additional validation of this method.
To date, few groups have attempted to characterize the plasma membrane proteome of myeloid leukemia cells. Compared to previous publications on the leukemia cell surfaceome [18,20,22], we identified on average at least twice more cell surface-associated proteins in a leukemia cell line (Figure 3A_SuppInfo). Two earlier published studies relied on biotin labeling to capture the cell surfaceome in cultured leukemia cells [18,20] and identified a total of 897 and 823 proteins, respectively. In one of the studies, Strassberger and colleagues compared the biotinylated surfaceome of human myeloid leukemia cell lines and normal human granulocytes and identified 320 differentially expressed cell surface proteins [20]. We compared our current data and noted that we had also identified 57% of the proteins reported in the Strassberger study. Reciprocally, 9% of the proteins identified in our workflow were detected in the Strassberger study (Figure 3A_SuppInfo). The additional earlier study by Hofmann and colleagues identified 538 cell surface proteins in two acute promyelocytic leukemia cell lines, a distinct subtype of AML, combining different biotinylation protocols [18]. Fifty-seven percent of the described proteins were also present in our dataset (Figure 3A_SuppInfo). In these two reports the percentage of putative plasma membrane proteins ranged from 44 to 60%. In a third study, Bonardi et al. analyzed the membrane preparations of two non-biotin-labeled leukemia patient samples and identified 867 and 610 plasma membrane-associated proteins in the CD34+-sorted population from each specimen, corresponding to less than 30% of all proteins detected. The improved depth of analysis reported here was gained not only from our combination of material handling procedures (plasma membrane enrichment, SDS-PAGE) and LC separation (90 minute UPLC gradient) but also from the use of accurate mass, high resolution, mass spectrometry instrumentation used here. With our protocol, we identified on average >1900 cell surface-associated and >4000 total protein groups in a single sample replicate, thereby demonstrating the higher discovery potential of this improved workflow.
To our knowledge, no study to date has reported more than 870 plasma membrane-associated proteins in a single sample replicate analysis [16,18,20,22,23,28], which corresponds to approximately 25% of the predicted cell surface protein landscape [21]. In summary, we demonstrate that this new proteomic workflow, which combines improvements in overall plasma membrane enrichment, GeLC-MS/MS and bioinformatic analyses, provides superior depth of cell surface coverage compared to previous studies that may facilitate identification of new protein targets for subsequent experimental studies. This process, although labor intensive, requires fewer preparative steps than most biotinylation protocols, and allows for comprehensive surfaceome profiling with limited starting material. Thus this protocol may have general applicability in the interrogation of cancer cell surfaceomes.
Supplementary Material
Acknowledgments
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [29] partner repository with the dataset identifier PXD004801.
We gratefully thank Dr. Claudio Giraudo for assistance with experimental equipment and for helpful discussions. We appreciate the expert instrument service provided by field service engineers, Dan French and Dennis Heim. These studies were supported by a Leukemia & Lymphoma Society Specialized Center of Research Grant (SG), the National Cancer Institute (K08CA184418 to SKT), a Hyundai Quantum Award (RA), the National Institutes of Health (U54HD086984 to SHS) and the Kick Leuk Foundation.
The authors have declared no conflict of interest.
Abbreviations
- PM
plasma membrane
- PNC
post-nuclear cytosol
- EPM
enriched plasma membrane
- CPM
crude plasma membrane
- iBAQ
intensity-based absolute quantification
References
- 1.Yıldırım MA, Goh KI, Cusick ME, Barabási AL, Vidal M. Drug—target network. Nat Biotechnol. 2007;25:1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- 2.Schwanhäusser B, Busse D, Li N, Dittmar G, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 3.Ghazalpour A, Bennett B, Petyuk VA, Orozco L, et al. Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genet. 2011;7:e1001393. doi: 10.1371/journal.pgen.1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang B, Wang J, Wang X, Zhu J, et al. Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513:382–387. doi: 10.1038/nature13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Battle A, Khan Z, Wang SH, Mitrano A, et al. Genomic variation. Impact of regulatory variation from RNA to protein. Science. 2015;347:664–667. doi: 10.1126/science.1260793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jovanovic M, Rooney MS, Mertins P, Przybylski D, et al. Immunogenetics. Dynamic profiling of the protein life cycle in response to pathogens. Science. 2015;347:1259038. doi: 10.1126/science.1259038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Washburn MP, Wolters D, Yates JR. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 8.Wu CC, Yates JR. The application of mass spectrometry to membrane proteomics. Nat Biotechnol. 2003;21:262–267. doi: 10.1038/nbt0303-262. [DOI] [PubMed] [Google Scholar]
- 9.Eichacker LA, Granvogl B, Mirus O, Muller BC, et al. Hiding behind Hydrophobicity: TRANSMEMBRANE SEGMENTS IN MASS SPECTROMETRY. J Biol Chem. 2004;279:50915–50922. doi: 10.1074/jbc.M405875200. [DOI] [PubMed] [Google Scholar]
- 10.Nunomura K. Cell Surface Labeling and Mass Spectrometry Reveal Diversity of Cell Surface Markers and Signaling Molecules Expressed in Undifferentiated Mouse Embryonic Stem Cells. Mol Cell Proteomics. 2005;4:1968–1976. doi: 10.1074/mcp.M500216-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Rybak JN, Ettorre A, Kaissling B, Giavazzi R, et al. In vivo protein biotinylation for identification of organ-specific antigens accessible from the vasculature. Nat Methods. 2005;2:291–298. doi: 10.1038/nmeth745. [DOI] [PubMed] [Google Scholar]
- 12.Zhang W, Zhou G, Zhao Y, White MA, Zhao Y. Affinity enrichment of plasma membrane for proteomics analysis. ELECTROPHORESIS. 2003;24:2855–2863. doi: 10.1002/elps.200305569. [DOI] [PubMed] [Google Scholar]
- 13.Wollscheid B, Bausch-Fluck D, Henderson C, O’Brien R, et al. Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins. Nat Biotechnol. 2009;27:378–386. doi: 10.1038/nbt.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bausch-Fluck D, Hofmann A, Wollscheid B. In: Liver Proteomics. Josic D, Hixson DC, editors. Humana Press; Totowa, NJ: 2012. pp. 1–16. [Google Scholar]
- 15.Weekes MP, Antrobus R, Lill RJ, Duncan LM, et al. Comparative analysis of techniques to purify plasma membrane proteins. J Biomol Tech. 2010:21. [PMC free article] [PubMed] [Google Scholar]
- 16.Ozlu N, Qureshi MH, Toyoda Y, Renard BY, et al. Quantitative comparison of a human cancer cell surface proteome between interphase and mitosis. EMBO J. 2015;34:251–265. doi: 10.15252/embj.201385162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hörmann K, Stukalov A, Müller AC, Heinz LX, et al. A Surface Biotinylation Strategy for Reproducible Plasma Membrane Protein Purification and Tracking of Genetic and Drug-Induced Alterations. J Proteome Res. 2016;15:647–658. doi: 10.1021/acs.jproteome.5b01066. [DOI] [PubMed] [Google Scholar]
- 18.Hofmann A, Gerrits B, Schmidt A, Bock T, et al. Proteomic cell surface phenotyping of differentiating acute myeloid leukemia cells. Blood. 2010;116:e26–e34. doi: 10.1182/blood-2010-02-271270. [DOI] [PubMed] [Google Scholar]
- 19.Mirkowska P, Hofmann A, Sedek L, Slamova L, et al. Leukemia surfaceome analysis reveals new disease-associated features. Blood. 2013;121:e149–e159. doi: 10.1182/blood-2012-11-468702. [DOI] [PubMed] [Google Scholar]
- 20.Strassberger V, Gutbrodt KL, Krall N, Roesli C, et al. A comprehensive surface proteome analysis of myeloid leukemia cell lines for therapeutic antibody development. J Proteomics. 2014;99:138–151. doi: 10.1016/j.jprot.2014.01.022. [DOI] [PubMed] [Google Scholar]
- 21.Da Cunha JPC, Galante PAF, De Souza JE, De Souza RF, et al. Bioinformatics construction of the human cell surfaceome. Proc Natl Acad Sci. 2009;106:16752–16757. doi: 10.1073/pnas.0907939106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonardi F, Fusetti F, Deelen P, van Gosliga D, et al. A Proteomics and Transcriptomics Approach to Identify Leukemic Stem Cell (LSC) Markers. Mol Cell Proteomics. 2013;12:626–637. doi: 10.1074/mcp.M112.021931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hofmann A, Thiesler T, Gerrits B, Behnke S, et al. Surfaceome of classical Hodgkin and non-Hodgkin lymphoma. Proteomics Clin Appl. 2015;9:661–670. doi: 10.1002/prca.201400146. [DOI] [PubMed] [Google Scholar]
- 24.Zhang LJ, Wang XE, Peng X, Wei YJ, et al. Proteomic analysis of low-abundant integral plasma membrane proteins based on gels. Cell Mol Life Sci CMLS. 2006;63:1790–1804. doi: 10.1007/s00018-006-6126-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyd RS, Dyer MJS, Cain K. Proteomic analysis of cell surface membrane proteins in leukemic cells. Methods Mol Biol Clifton NJ. 2007;370:135–146. doi: 10.1007/978-1-59745-353-0_11. [DOI] [PubMed] [Google Scholar]
- 26.Choksawangkarn W, Edwards N, Wang Y, Gutierrez P, Fenselau C. Comparative Study of Workflows Optimized for In-gel, In-solution, and On-filter Proteolysis in the Analysis of Plasma Membrane Proteins. J Proteome Res. 2012;11:3030–3034. doi: 10.1021/pr300188b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schiapparelli LM, McClatchy DB, Liu HH, Sharma P, et al. Direct Detection of Biotinylated Proteins by Mass Spectrometry. J Proteome Res. 2014;13:3966–3978. doi: 10.1021/pr5002862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gomes-Alves P, Serra M, Brito C, R-Borlado L, et al. Exploring analytical proteomics platforms toward the definition of human cardiac stem cells receptome. Proteomics. 2015;15:1332–1337. doi: 10.1002/pmic.201400318. [DOI] [PubMed] [Google Scholar]
- 29.Vizcaíno JA, Csordas A, del-Toro N, Dianes JA, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.