Abstract
Chinese hamster ovary (CHO)1 cells have been widely used to express heterologous genes and produce therapeutic proteins in biopharmaceutical industry. Different CHO host cells have distinct cell growth rates and protein expression characteristics. In this study, the expression of about 1,307 host proteins in three sublines, i.e. CHO K1, CHO S and CHO/dihydrofolate reductase (dhfr)−, were investigated and compared using proteomic analysis. The proteins involved in cell growth, glycolysis, tricarboxylic acid cycle, transcription, translation and glycosylation were quantitated using Liquid chromatography tandem-mass spectrometry (LC-MS/MS). The key host cell proteins that regulate the kinetics of cell growth and the magnitude of protein expression levels were identified. Furthermore, several rational cell engineering strategies on how to combine the desired features of fast cell growth and efficient production of therapeutic proteins into one new super CHO host cell have been proposed.
Keywords: CHO sublines, comparative proteomics, cell growth, protein expression, regulators
1. Introduction
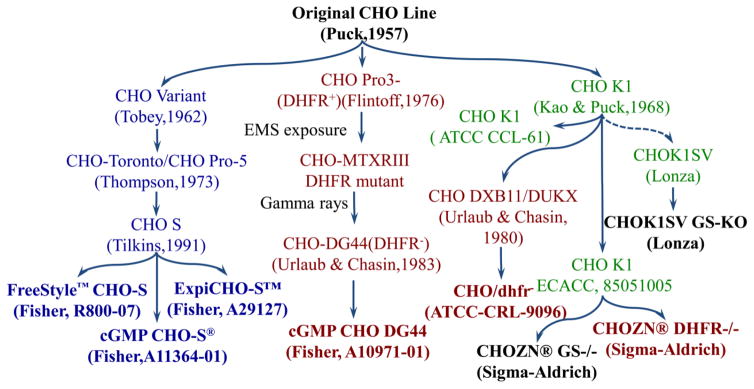
The Chinese hamster ovary (CHO) cells have been widely used to produce protein-based biopharmaceuticals. Compared to other mammalian cells, CHO cells have the unique advantages of robust cell growth, effective post-translational modification, and the well-established standards of good manufacturing practice (GMP). The parental CHO cell line was originally isolated from Chinese hamster by Dr. Theodore T. Puck in 1957 [1], followed by the derivation of multifarious CHO sublines, such as CHO K1, CHO/dhfr−, and CHO S (Fig. 1). The CHO K1 subline was licensed with a glutamine synthetase (GS)-based expression system [2], and a GS negative CHO K1 subline was developed using zinc finger technology [3, 4]. The CHO/dhfr− cells including CHO DXB11 and CHO DG44 sublines were generated using chemical mutagenesis, gamma rays or zinc finger technology to inactivate the enzyme of dihydrofolate reductase (DHFR) [5, 6]. The cGMP bank of another CHO subline, CHO S with characteristics of fast cell growth, was derived from the parental CHO via adaptation [7].
Figure 1.
Cell lineage of CHO cells.
The CHO sublines mentioned above exhibit noteworthy heterogeneity in their phenotypes [8]. For instance, the GS-based gene selection and amplification in CHO K1 enables high protein production, but the application of high concentration of selection reagent methionine sulfoximine MSX in production cell line construction causes unstable protein expression. The selection and amplification of heterologous genes in CHO/dhfr− cells is usually more effective, yet its cell growth is slower than other two sublines. CHO S cell line has relatively higher growth rate or lower doubling time, but it is laborious to develop a high protein producing cell line from this host cell due to the double selection using methotrexate MTX and puromycin. In addition, the clone stability of CHO S-based production cell line is poor, which is caused by the fact that dhfr is an endogenous gene and the gene amplification using high-concentration MTX is necessary. Thus, to improve the production of mmammalian cell-based biopharmaceuticals, it is highly desirable to develop an advanced CHO host cell in which fast cell growth and high protein expression will all be integrated.
The completion of the CHO K1 genome sequencing and the development of proteomics technology have provided both the genetic background and the direct measurement capability to examine the expression levels of the host cell proteins in CHO sublines [9]. Baycin-Hizal et al. have accomplished the first proteomic study of CHO K1 using 120 mass spectrometry analyzes and have identified a total of 6,164 grouped proteins from cellular proteome, secretome and glycoproteome analyzes [10]. A number of other studies have analyzed the extracellular host cell proteins to evaluate the impurities in biopharmaceutical production or optimize cell culture medium [11–14]. In addition, proteomic studies have also been performed to study the effects of cell culture conditions, such as temperature, hyperosmolality, media and feeding strategy, on the expression profile of host cell proteins [15–17].
Cell engineering via gene manipulation could be a powerful tool to construct an innovative host cell. However, the lack of the fundamental understanding of the regulation of cell growth and protein expression has hindered the rational host cell engineering. To our best knowledge, the comparison of the intracellular proteins’ expression among different CHO sublines has not been performed so far. In this study, we aimed to establish a comprehensive understanding of the different phenotypes of three CHO sublines (CHO K1, CHO/dhfr− and CHO S) by comparing their intracellular proteomics profiling. The expression levels of the key enzymes (or proteins) involved in cell growth, glycolysis, tricarboxylic acid (TCA) cycle, transcription, translation and glycosylation were analyzed and compared. The enzymes with different expression levels that correlate to cell growth and protein expression were presented. Finally, the strategies to rationally construct next generation of CHO host cells were also discussed.
2. Materials and methods
2.1 CHO cells and cell culture
Three suspension CHO sublines, including CHO K1, CHO/dhfr− and CHO S, were analyzed in this study. The CHO K1 and CHO S were purchased from Thermo Fisher Scientific (Waltham, MA), and CHO/dhfr− was purchased from ATCC (Manassas, VA). The seed culture of CHO K1, CHO S and CHO/dhfr− were maintained in the three basal media of HyClone CDM4CHO (Hyclone Laboratories, Logan, UT), Gibco CD CHO (Life Technologies, Grand Island, NY) and Sigma EX-CELL CHO DHFR- (Sigma-Aldrich, St. Louis, MO), respectively. All the cell culture media were supplemented with 8 mM L-glutamine (final concentration). The sodium hypoxanthine and thymidine supplements were added to the EX-CELL CHO DHFR- medium. The batch cultures were seeded with viable cell density of 0.3×106 cells/mL. The cells were cultivated with triplication in suspension cultures in 125-mL disposable shaker flasks at 37 °C, 5% CO2 and 120 rpm in a humidified incubator (Caron, Marietta, OH).
2.2 Extraction and digestion of host proteins
To prepare proteomics samples, the cell cultures were sampled between early and mid-log phases, i.e. day 3 (CHO K1 and CHO S) and day 4 (CHO/dhfr−). At sampling points, the average viable cell densities were 2.2×106 cells/mL and the viabilities were maintained at > 99%. Three flasks of each cell were carried out to collect cell samples for the extraction of host cell proteins. The CHO cells collected from batch cultures were centrifuged at 8,000 rpm for 5 mins at 4 °C, washed for three times using PBS buffer, and stored at −80 °C for further proteomic analysis. All reagents and supplements used in this study were purchased from Thermo Fisher Scientific unless otherwise specified.
The detailed procedure of host cell protein extraction and digestion was described in previous publications [18, 19]. In brief, the host cell proteins were first extracted from cell pellets using M-PER, denatured and run into a 10% SDS Bis-Tris PAGE. Then the sliced protein gel was equilibrated in 100 mM ammonium bicarbonate, reduced, carbidomethylated, dehydrated and digested with Trypsin Gold (Promega, Madison, WI). Finally, the digested peptide was extracted, concentrated and resolubilized in 20 μL of 5% CAN/0.1% formic acid prior to analysis by 1D reverse phase LC-nESI-MS2.
2.3 LC-MS/MS analysis
LC-MS/MS was applied to acquire the high-quality peptide precursor and fragment ion data as described in literature [19]. Each proteomics sample was injected to LC-MS/MS with triplication. A 1260 Infinity nHPLC stack (Agilent, Santa Clara, CA) equipped with a Jupiter C-18 column (300 Å, 5 micron, 75 micron I.D. × 15 cm, Phenomenex) was run to separate the digested peptides. The peptides were eluted using 0%–30% acetonitrile in D.I. H2O containing 0.1% formic acid with a flow rate of 0.3 μL/min. The peptide fractions were sprayed into a hybrid mass spectrometer (MS, Thermo Orbitrap Velos Pro) equipped with a nano-electrospray source to gain proteomics data. All data were collected in collision-induced dissociation mode. The instrument configuration during data collection followed previous publication [18–20].
2.4 Protein identification
The collected XCalibur RAW files were centroided and converted to MzXML format using ReAdW and converted to mgf files using MzXML2Search. The data were searched with SEQUEST against UniProt-derived proteome databases of both mouse and rat. The searching parameters include trypsin digestion, two missed cleavages sites, 20 ppm of precursor mass tolerance, 0.36 Da fragment ion tolerance, variable modification M at 15.9949, and static modification C at 57.0293. The peptides’ searches were performed with a species specific subset of the UniRef100 database. The identified peptides were filtered, grouped, and quantified using Scaffold (Protein Sciences, Portland, OR). The peptide and protein probabilities were set at > 90.0% and > 99.0%, respectively, while the false rate was set at lower than 1.0% to retain protein with high confidence. The identified CHO host cell proteins were described using UniRef100 ID and their expression levels were depicted using the normalized spectra count abundance between samples. The key software tools used in the statistical analysis are SEQUEST and Proteomesoftware. The average spectral count data calculated from the triplicated experiments were presented.
3. Results and discussion
3.1 Comparative proteomics
The CHO K1, CHO/dhfr− and CHO S cells have been widely used to produce cell-based biopharmaceuticals. Nevertheless, these three sublines possess quite different desired features for the production of therapeutic proteins. In this study, we applied comparative proteomic analysis and examined the expression profiling of more than 1,300 intracellular proteins of CHO K1, CHO/dhfr− and CHO S host cells. The key enzymes that regulate cell growth, metabolism of carbon and energy, and protein expression were analyzed and compared. The UniRef ID, function, names, average spectral count, and probability of these enzymes are presented in Tables 1–4. The complete datasets, including raw MS data, search parameters, search results, reference search database and summarized data with statistical analysis, were also deposited in the public repository PeptideAtlas (http://www.peptideatlas.org/, accession no. PASS00963).
Table 1.
Representative proteins regulating cell growth in CHO K1, CHO S, and CHO/dhfr−.
| Protein | Spectral Counta | ||||
|---|---|---|---|---|---|
|
| |||||
| UniRef IDb | Namec | CHO K1 | CHO S | CHO/dhfr− | Probability |
| G3V6P7 | Myosin, heavy polypeptide 9, non-muscle (myh9) | 52±9.9 | 175±7.1 | 36±11 | 1 |
| Q8BMK4 | Cytoskeleton-associated protein 4 (ckap4) | 19.5±0.7 | 25±7.1 | 7.5±0.7 | 1 |
| B2GV99 | Myl6 protein (myl6) | 15.5±3.5 | 23±4.2 | 10±2.8 | 1 |
| P57780 | Alpha-actinin-4 (actn4) | 60±4.2 | 86±8.5 | 41.5±4.9 | 1 |
| Q58E70 | Tpm3 protein (tpm3) | 17.5±3.5 | 36.5±3.5 | 25.5±3.5 | 1 |
| P09495 | Tropomyosin alpha-4 chain (tpm4) | 23±5.6 | 45±5.7 | 35.5±2.1 | 1 |
| P11440 | Cyclin-dependent kinase 1 (cdk1) | 6.5±2.1 | 13±0 | 4±4 | 1 |
| P68373 | Tubulin alpha-1C chain (tuba1c) | 89.5±0.7 | 122±12.7 | 82±4.2 | 1 |
| Q6P9T8 | Tubulin beta-4B chain (tubb4b) | 93±5.6 | 127±26.8 | 87±4.2 | 1 |
| B7FAU9 | Filamin, alpha (flna) | 134±16.9 | 127.5±4.9 | 0±0 | 1 |
| P20152 | Vimentin (vim) | 216.5±30.4 | 3±0 | 55±0.7 | 1 |
| P45592 | Cofilin-1 (cfl1) | 46.5±2.1 | 37±0 | 20.5±0.7 | 1 |
Note:
The average spectral count data with standard deviation that were calculated from triplicated experiments were presented.
The annotation was performed using Mouse UniRef100 database. The identified proteins were presented with UniRef100 ID.
All raw MS data, search parameters, search database and summarized data with statistic analysis were deposited to a public repository PeptideAtlas (http://www.peptideatlas.org/, accession no. PASS00963).
Table 4.
Post-translation modification proteins with different expressions in CHO K1, CHO S, and CHO/dhfr−.
| Protein | Spectral Count | ||||
|---|---|---|---|---|---|
|
| |||||
| UniRef ID | Name | CHO K1 | CHO S | CHO/dhfr− | Probability |
| P46978 | Glycosyltransferase subunit STT3A (stt3a) | 13±8.4 | 6±0 | 2±0 | 1 |
| Q3TDQ1 | Glycosyltransferase subunit STT3B (stt3b) | 4±0 | 2.5±0.7 | 0.5±0.7 | 0.99 |
| O54734 | Glycosyltransferase 48 kDa subunit (ddost) | 13±0 | 13.5±2.1 | 6.5±3.5 | 1 |
| Q91YQ5 | Glycosyltransferase subunit 1 (rpn1) | 26.5±2.1 | 26±5.6 | 24±0 | 0.99 |
| P06761 | 78 kDa glucose-regulated protein (hspa5) | 113.5±6.3 | 126.5±5.6 | 190.5±47.3 | 1 |
| D3ZAN3 | Alpha glucosidase 2 alpha neutral subunit (ganab) | 23±2.8 | 33±0 | 17±7.0 | 0.99 |
| Q9JLA3 | UDP-glucose:glycoprotein glucosyltransferase 1 (uggt1) | 19.5±0.7 | 22.5±0.7 | 13±4.2 | 1 |
| P18418 | Calreticulin (calr) | 30.5±7.7 | 51.5±7.7 | 27.5±2.1 | 1 |
| P35564 | Calnexin (canx) | 11±1.4 | 11.5±6.3 | 8.5±3.5 | 0.99 |
| P08113 | Endoplasmin (hsp90b1) | 129.5±9.1 | 170±24.1 | 142.5±28.9 | 1 |
| P27773 | Protein disulfide-isomerase A3 (pdia3) | 75±2.8 | 101.5±7.7 | 117.5±6.3 | 1 |
| P24368 | Peptidyl-prolyl cis-trans isomerase B (ppib) | 9±1.4 | 20±1.4 | 24.5±2.1 | 1 |
3.2 Cell growth
Our experiments on cell growth showed that the cell doubling time for CHO K1, CHO/dhfr− and CHO S cells was 24±2 h, 27±2 h, and 18±2 h, respectively. It is obvious that the CHO S cells grew the fastest and the CHO/dhfr− cells grew the slowest, as previously reported [21]. The cell growth related host cell proteins that showed > 50% change of expression levels between CHO S and the other two sublines are summarized in Table 1.
As shown in Table 1, nine proteins had obviously higher expression levels in CHO S. These proteins include myosin (myh9), cytoskeleton-associated protein 4 (ckap4), myl6 protein (myl6), alpha-actinin-4 (actn4), tropomyosin alpha-3 (tpm3), tropomyosin alpha-4 (tpm4), cyclin-dependent kinase (cdk), tubulin alpha (tuba), and tubulin beta (tubb4b). Both myosin and cyclin-dependent kinase were expressed at > 2-fold higher levels in CHO S than in CHO K1 or CHO/dhfr− cells. Previous studies have shown that the non-muscle myosin has multiple functions, e.g., cytokinesis, cell division, cellular movement and maintenance of cell shape, via diverse isoforms, phosphorylation and/or protein binding patterns [22]. Cyclin-dependent kinase plays an important role in regulating cell cycle, transcription and mRNA processing. Our results suggest that the higher expression levels of myosin and cyclin-dependent kinase are likely positively correlated with the faster cell growth rate of CHO S cells. In contrast, seven of these nine proteins except Tpm3 and Tpm4 had the lowest expression levels in CHO/dhfr− cells (Table 1), suggesting that these proteins may account for the slow growth of CHO/dhfr− cells.
Interestingly, our data show that the expression levels of filamin (flna), vimentin (vim) and cofilin-1 (cfl1) in CHO K1 cells were significantly higher than CHO S and/or CHO/dhfr− (Table 1). Studies have shown that filamin α regulates cell shape and migration by remodeling the cytoskeleton [23] and enhances the surface expression of glycoprotein [24]. Protein vimentin has been reported to support the anchoring of organelles to nucleus, endoplasmic reticulum, and mitochondria [25]. The binding of filamin α to vimentin has been considered as a pivotal factor that controls cell adhesion and spreading [26]. Protein cofilin functions as a homeostatic regulator in cell biology through regulating the actin-filament dynamics [27, 28]. The higher expression indicated that these three proteins could play important role in regulating the cell growth of CHO K1.
3.3 Carbon and energy metabolism
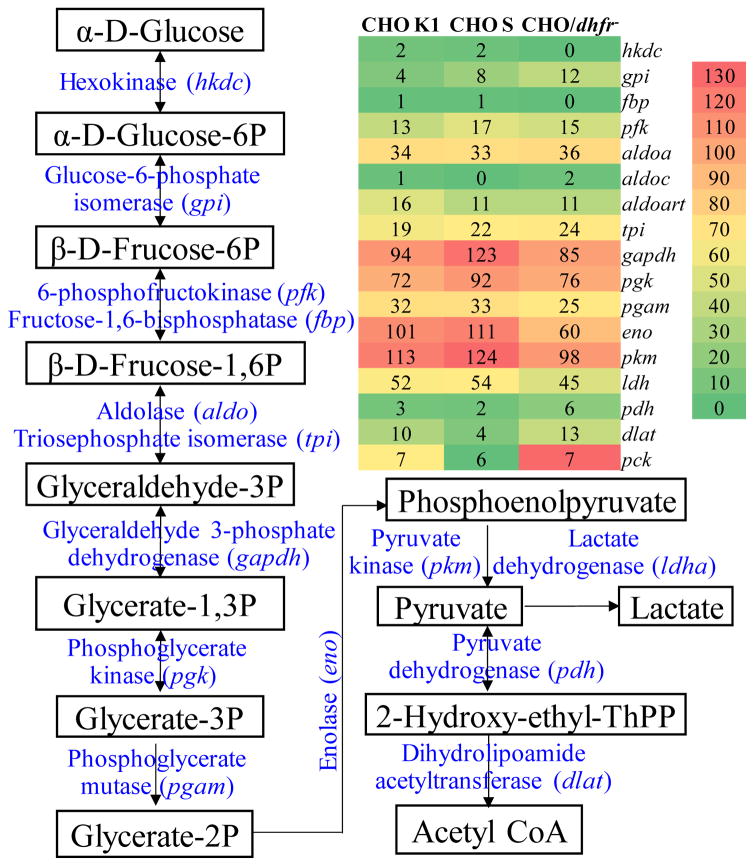
The metabolism analysis of glucose and lactate in host cell cultures were analyzed. The results showed that the glucose specific consumption rate was 297.4 pg/(cell•day), 243.7 pg/(cell•day) and 365.1 pg/(cell•day), and the lactate specific accumulation rate was 172.3 pg/(cell•day), 147.2 pg/(cell•day), and 233.5 pg/(cell•day), for the CHO K1, CHO/dhfr− and CHO S cells, respectively. It is clear that the CHO S has the highest glucose catabolism and the CHO/dhfr− has the lowest glucose catabolism. Intracellular metabolism of carbon and energy is crucial to cell growth, heterologous protein expression and other cellular activities. Therefore, we investigated the expression levels of the proteins involved in glycolysis and TCA cycle.
As presented in the glycolysis pathway in Fig. 2, the ATP-dependent hexokinase (hkdc, UniRef100_Q91W97) showed low expression in all three CHO sublines. Since the reaction of “glucose → glucose-6P” catalyzed by hexokinase is the first step in glycolysis, the up-regulation of hexokinase could improve the efficiency of glucose consumption and catabolism. Five enzymes in the glycolysis pathway of CHO cells, namely glyceraldehyde 3-phosphate dehydrogenase (gapdh, UniRef100_P04797), ADP-dependent phosphoglycerate kinase (pgk, UniRef100_P09411), enolase (eno, UniRef100_P04764), ADP-dependent pyruvate kinase (pkm, UniRef100_Q6P7S0), and lactate dehydrogenase (ldh, UniRef100_P04642), had relatively higher expression levels (Fig. 2). The up-regulation of Pgk and Pkm could enhance glycolysis by providing more ATP through “1,3-biphosphoglycerate → glyceraldehyde 3-phosphate” and “phosphoenolpyruvate → pyruvate”. Pyruvate is a key metabolic intermediate subsequently entering into the TCA cycle within mitochondria or converting into lactate within cytosol, so the higher levels of Pkm provide TCA cycle raw materials effectively for metabolism of carbon and energy in these cells.
Figure 2.
Heat map of protein expression involved in glycolysis. Red: high expression, Yellow: medium expression, and Green: low expression.
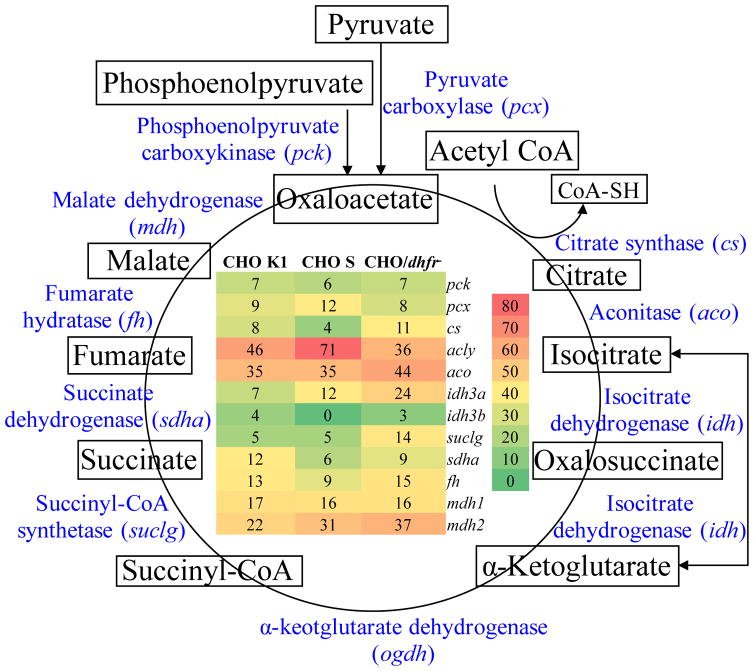
As shown in Fig. 3, almost all the enzymes in TCA cycle except ketoglutarate dehydrogenase (ogdh) had been detected and analyzed. The heat map showed that the ATP-dependent citrate lyase (acly, UniRef100_Q3V117) that catalyzes the formation of oxaloacetate and acetyl-CoA had significantly high expression in CHO S with high spectra count of 71, indicating a good target to up-regulate in cell engineering. In addition, six proteins showed relatively higher expression levels and played important roles in TCA cycle in CHO/dhfr−, including citrate synthase (cs, UniRef100_G3V936), aconitate hydratase (aco2, UniRef100_Q9ER34), isocitrate dehydrogenase (idh3a, UniRef100_F1LNF7), succinyl-CoA synthetase (suclg1, UniRef100_P13086), malate dehydrogenase 2 (mdh2, UniRef100_P04636), and fumarate hydratase (fh, UniRef100_P14408).
Figure 3.
Heat map of protein expression involved in TCA cycle. Red: high expression, Yellow: medium expression, and Green: low expression.
3.4 Protein expression: transcription and translation
Heterologous protein expression is a complex process including transcription, translation, post translational modifications and folding, which can significantly affect the cellular dynamics of host cells. Proteome of CHO cells reflecting the host cellular machinery will facilitate our understanding of the protein production bottlenecks.
The proteins associated with transcription that had obviously different expression levels among the three CHO sublines are summarized in Table 2. The UniRef ID, protein name, gene name, MS counts and probability were reported. Seven of the transcription correlated proteins showed higher expression levels in CHO K1 than in CHO S or CHO/dhfr−. These seven proteins include ribonucleoside-diphosphate reductase (rrm1), protein Ascc3l1 (snrnp200), nucleolin (ncl), DEAH (Asp-Glu-Ala-His) box polypeptide 15 (dhx15), nucleolin-related protein NRP (nrp), pre-mRNA-processing-splicing factor 8 (prpf8), and nuclear ribonucleoprotein F (hnrnpf). CHO K1 subline is capable to produce heterologous proteins at commercial scale, and the reported high titer (e.g. > 5 g/L) of therapeutic proteins was produced by using CHO K1 cells [29]. These seven host cell proteins in transcription could contribute to the high production of biopharmaceuticals by CHO K1 cells.
Table 2.
Key enzymes involved in transcription that showed different expression in CHO K1, CHO S, and CHO/dhfr−.
| Protein | Spectral Count | ||||
|---|---|---|---|---|---|
|
| |||||
| UniRef ID | Name | CHO K1 | CHO S | CHO/dhfr− | Probability |
| P07742 | Ribonucleoside-diphosphate reductase (rrm1) | 41.5±0.7 | 8±0 | 1±1.4 | 1 |
| F1LNJ2 | Protein Ascc3l1 (snrnp200) | 35.5±6.3 | 2±1.4 | 23±2.8 | 1 |
| P13383 | Nucleolin (ncl) | 72.5±6.3 | 21±1.4 | 39.5±2.1 | 1 |
| D3ZD97 | DEAH (Asp-Glu-Ala-His) box polypeptide 15 (dhx15) | 21.5±6.3 | 8±7.0 | 18±1.4 | 1 |
| Q9QZX1 | Nucleolin-related protein NRP (nrp) | 48.5±6.3 | 19±1.4 | 34±0 | 1 |
| Q99PV0 | Pre-mRNA-processing-splicing factor 8 (prpf8) | 30±7.0 | 0±0 | 17.5±6.3 | 1 |
| Q9Z2X1 | Nuclear ribonucleoprotein F (hnrnpf) | 22.5±4.9 | 21.5±0.7 | 8.5±2.1 | 1 |
| Q4JG03 | Mcl-1 ubiquitin ligase (huwe1) | 4±1.4 | 21.5±9.1 | 11.5±2.1 | 1 |
| B7ZWF1 | Ddx3x protein (ddx3x) | 22.5±3.5 | 36.5±2.1 | 10±2.8 | 1 |
| O08629 | Transcription intermediary factor 1-beta (trim28) | 16.5±0.7 | 21±0 | 10±1.4 | 1 |
| A1L333 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 (ddx5) | 21±1.4 | 24.5±12.0 | 12±2.8 | 1 |
| P63017 | Heat shock cognate 71 kDa protein (hspa8) | 145±24.0 | 135.5±31.8 | 123.5±4.9 | 1 |
The proteins in translation process that showed distinct expression levels among three CHO sublines are listed in Table 3. The six enzymes that correlate with high capability of heterologous protein expression in CHO K1 cells, including serine/arginine-rich-splicing factor 1 (srsf1), elongation factor 1-gamma (eef1g), 40S ribosomal protein S12 (rps12), 40S ribosomal protein S4, X isoform (rps4x), 40S ribosomal protein S9 (rps9), and 60S ribosomal protein L3 (rpl3), were identified (Table 3). Another six proteins, namely translation initiation factor 3 (eif3l), heat shock protein HSP 90-alpha (hsp90aa1), rps16 protein (rps16), 60S ribosomal protein L18 (rpl18), 60S ribosomal protein L9 (rpl9), and regulator of nonsense transcripts 1 (upf1), were found to be expressed at the highest levels in CHO S cells (Table 3). All three CHO sublines expressed significantly high levels of elongation factor 2 (eef2) with MS counts of 118–130 and elongation factor 1-alpha 1 (eef1a1) with MS counts of 65–74. Elongation factors have been cloned to optimize expression vector and improved biopharmaceutical production [30]. The high levels of elongation factors detected in this study also confirmed that these three CHO sublines are good host cells to produce heterologous proteins.
Table 3.
Host cell proteins involved in translation regulation in CHO K1, CHO S, and CHO/dhfr−.
| Protein | Spectral Count | ||||
|---|---|---|---|---|---|
|
| |||||
| UniRef ID | Name | CHO K1 | CHO S | CHO/dhfr− | Probability |
| H7BX95 | Serine/arginine-rich-splicing factor 1 (srsf1) | 21.5±0.7 | 14±1.4 | 8±1.4 | 1 |
| Q68FR6 | Elongation factor 1-gamma (eef1g) | 59±2.8 | 40.5±6.3 | 27±0 | 0.99 |
| P63323 | 40S ribosomal protein S12 (rps12) | 28±2.8 | 21.5±2.1 | 14±1.4 | 0.93 |
| Q545F8 | 40S ribosomal protein S4, X isoform (rps4x) | 28.5±0.7 | 26±1.4 | 14.5±2.1 | 1 |
| D3YWH9 | 40S ribosomal protein S9 (rps9) | 27.5±12.0 | 20.5±7.7 | 13.5±0.7 | 1 |
| P21531 | 60S ribosomal protein L3 (rpl3) | 24±1.4 | 16.5±2.1 | 12±2.8 | 0.99 |
| G3V7G9 | Translation initiation factor 3, subunit L (eif3l) | 16±2.8 | 23.5±4.9 | 6.5±2.1 | 1 |
| P82995 | Heat shock protein HSP 90-alpha (hsp90aa1) | 95.5±2.1 | 166±22.6 | 48.5±3.5 | 1 |
| Q5CZY9 | Rps16 protein (rps16) | 24±7.0 | 31±4.2 | 14.5±2.1 | 1 |
| P35980 | 60S ribosomal protein L18 (rpl18) | 20±2.8 | 23.5±3.5 | 11±0 | 1 |
| P17077 | 60S ribosomal protein L9 (rpl9) | 27.5±4.9 | 28.5±3.5 | 13.5±3.5 | 1 |
| Q9EPU0 | Regulator of nonsense transcripts 1 (upf1) | 17.5±4.9 | 24±4.2 | 4±1.4 | 1 |
| F1M9V7 | Protein Npepps (npepps) | 8.5±0.7 | 14.5±0.7 | 24±2.8 | 1 |
| P50475 | Alanine--tRNA ligase, cytoplasmic (aars) | 28.5±6.3 | 15.5±0.7 | 32.5±17.6 | 0.99 |
| P05197 | Elongation factor 2 (eef2) | 126±8.4 | 130±14.1 | 118±0 | 1 |
| P10126 | Elongation factor 1-alpha 1 (eef1a1) | 74±9.9 | 71±11.3 | 65±4.2 | 1 |
3.5 Protein expression: post-translational modification and refolding
CHO cells enable glycosylation and sialylation of polypeptides in endoplasmic reticulum and Golgi, which is important for the refolding, stability and bioactivity of synthesized proteins. N-glycosylation begins with the addition of core oligosaccharide (Glc3Man9GlcNAC2) to the specific asparagine residues of the nascent polypeptides, which is catalyzed by oligosaccharyltransferases [31]. We found that all four glycosyltransferase subunits, i.e. subunit 1 (rpn1), STT3A (stt3a), STT3B (stt3b) and 48 kDa subunit (ddost), were expressed at low levels in CHO/dhfr− cells (Table 4), suggesting that the glycosylation of heterologous therapeutic proteins could be improved by up-regulating the expression of glycosyltransferase in CHO/dhfr− cells. Multiple enzymes are involved in the reactions post glycosylation, such as the glucosidase (ganab) that trims glucose, the lectin chaperones (calr, canx) that recognize and bind to monoglucosylate glycoforms (Glc1Man9GlcNAC2 protein) as a folding acceleration signal, and the UDP-glucose:glycoprotein glucosyltransferase (uggt1) that senses misfolded glycoprotein and properly folded glycoprotein [32]. All these enzymes were expressed at high levels in CHO S cells (Table 4), led us to speculate that the glycoprotein folding cycle, also called calnexin/calreticulin cycle, is very efficient in the CHO S subline.
We also investigated the enzymes facilitating protein folding. As shown in Table 4, we found that both CHO S and CHO/dhfr− cells had high expression levels of the 78 kDa glucose-regulated protein (hspa5, a.k.a. immunoglobulin heavy chain binding proteins/“BiP/Grp78”) that binds the nascent nonglycosylated proteins and/or supports protein refolding [33], and the protein disulphide isomerases (pdia3) and the peptidyl-prolyl cis-trans isomerase (ppib) that directly catalyze the rate-limiting steps in protein folding [34, 35].
3.6 Strategies to engineer host cell
In this study, the intracellular proteins involved in the regulation of cell growth, glycolysis, TCA cycle, transcription, translation and glycosylation were studied and compared among three different CHO hosts. The proteomic analysis indicated some possible strategies of rational host cell engineering for fast cell growth, high protein expression and preferable protein quality, which warrants further evaluation and studies.
Cell growth regulation
Over-expression of anti-apoptotic genes, e.g., bcl-2 or bcl-xL, has been tried to increase the viability of CHO cells, thereby the protein production [36, 37]. Meanwhile, gene regulation and culture condition optimization has also been used to induce G1 phase arrest during cell cycle to improve cellular metabolism [38, 39]. Different from those previous studies, the comparative proteomics in our study has identified some cytoskeletal proteins (i.e. filamin and vimentin) with significantly different expression levels among the three CHO sublines tested. These cytoskeletal proteins can maintain cell shape, keep intracellular transport and affect the formation of mitotic spindles for cell division in addition to the regulation of protein translation process [40]. Thus, we suggest that by manipulating the expression levels of these cytoskeleton-associated proteins, the cell growth and protein production in CHO cells may be improved.
Protein expression regulation
Several strategies, including host cell engineering, expression vector optimization, high producing cell line development, and production process parameters optimization, can be used to improve protein production. Cell engineering is a powerful tool to enhance protein production but gene manipulation requires a comprehensive understanding of host cell regulation of transcription, translation and post-translational modification. Studies have shown that the introduction of an artificial zinc finger transcription factor (ZFP-TF) in CHO cells has improved antibody production by 10-fold [41], and the overexpression of E2F-1 transcription factor has led to 20% increase of cell viability [42]. In our current study, we have identified a few transcription regulators in CHO S (snrnp200 and prpf8) and CHO/dhfr− (rrm1) cells, of which the up-regulation could be used to improve transcription efficiency. Translation elongation factor has been used in vector optimization, but the translation initiation is a key rate-limiting step that is more desirable to control the translation [43]. Our proteomics results suggest that the translation initiation factors, 40S ribosomal protein and 60S ribosomal protein, can be used to enhance the translation in CHO/dhfr− cells. We have also found that the elongation factors are expressed at high levels in all three CHO sublines, so to further up-regulate the expression of translation elongation factors is not a good choice for host cell engineering.
Protein quality regulation
Reinhart et al. reported that the choice of CHO subline would affect the glycan structure of the antibodies produced besides cell culture conditions [44]. O’Callaghan et al. also reported that CHO clonal hosts had different glycosylation capability [45]. Therefore, the genetic engineering of the post-translational modification of CHO host cells can be an effective approach to enhance clinical efficacy of therapeutic proteins [46]. The proteomic analysis in our study suggests that the overexpression of the glycosyltransferase may be able to improve protein quality in CHO/dhfr− cells. And at the same time, the post-glycosylation modification could also be improved by up-regulating the expression of the correlated enzymes. Since the enzymes that catalyze protein refolding have high expression levels in all three CHO hosts, the results suggest that it may not be practical to manipulate the refolding process.
4. Conclusions
In this study, comparative proteomics was applied to systematically characterize and compare the expression levels of intracellular proteins in three different CHO host cell sublines (CHO K1, CHO S, and CHO/dhfr−). The study provides new insight into the host cell regulation of cell growth, metabolism as well as protein production and protein quality, and the potentially associated host regulators were analyzed. Finally, the strategies to engineer different CHO host cells were also proposed, which may also provide a guideline for the design of a novel CHO host. The work could be considered as a reference for CHO subline selection, rational cell engineering and even bioprocess optimization to effectively produce biopharmaceuticals with high titer and better quality.
Highlights.
Comparative proteomic analysis of three CHO sublines was performed
Host cell proteins regulating cell growth and protein expression were identified
Rational cell engineering strategies were proposed
Acknowledgments
This study was supported by the National Institute of Health (Grant Number R21 HL 127599A1, 2016) and the start-up fund from the University of Alabama at Birmingham (UAB). The proteomic analysis was conducted at the UAB Comprehensive Cancer Center Mass Spectrometry/Proteomics (MSP) Shared Facility.
Footnotes
CHO: Chinese hamster ovary, DHFR: dihydrofolate reductase, LC-MS/MS: Liquid chromatography tandem-mass spectrometry
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Puck TT. The genetics of somatic mammalian cells. Adv Biol Med Phys. 1957;5:75–101. doi: 10.1016/b978-1-4832-3111-2.50006-7. [DOI] [PubMed] [Google Scholar]
- 2.Kao FT, Puck TT. Genetics of somatic mammalian cells, VII. Induction and isolation of nutritional mutants in Chinese hamster cells. Proc Natl Acad Sci U S A. 1968;60:1275–1281. doi: 10.1073/pnas.60.4.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fan L, Kadura I, Krebs LE, Hatfield CC, Shaw MM, Frye CC. Improving the efficiency of CHO cell line generation using glutamine synthetase gene knockout cells. Biotechnol Bioeng. 2012;109:1007–1015. doi: 10.1002/bit.24365. [DOI] [PubMed] [Google Scholar]
- 4.Liu PQ, Chan EM, Cost GJ, Zhang L, Wang J, Miller JC, Guschin DY, Reik A, Holmes MC, Mott JE, Collingwood TN, Gregory PD. Generation of a triple-gene knockout mammalian cell line using engineered zinc-finger nucleases. Biotechnol Bioeng. 2010;106:97–105. doi: 10.1002/bit.22654. [DOI] [PubMed] [Google Scholar]
- 5.Urlaub G, Chasin LA. Isolation of Chinese hamster cell mutants deficient in dihydrofolate reductase activity. Proc Natl Acad Sci U S A. 1980;77:4216–4220. doi: 10.1073/pnas.77.7.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santiago Y, Chan E, Liu PQ, Orlando S, Zhang L, Urnov FD, Holmes MC, Guschin D, Waite A, Miller JC, Rebar EJ, Gregory PD, Klug A, Collingwood TN. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc Natl Acad Sci U S A. 2008;105:5809–5814. doi: 10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson LH, Baker RM. Isolation of mutants of cultured mammalian cells. Methods Cell Biol. 1973;6:209–281. doi: 10.1016/s0091-679x(08)60052-7. [DOI] [PubMed] [Google Scholar]
- 8.Wurm FM. CHO Quasispecies—Implications for Manufacturing Processes. Processes. 2013;1:296–311. [Google Scholar]
- 9.Xu X, Nagarajan H, Lewis NE, Pan S, Cai Z, Liu X, Chen W, Xie M, Wang W, Hammond S, Andersen MR, Neff N, Passarelli B, Koh W, Fan HC, Wang J, Gui Y, Lee KH, Betenbaugh MJ, Quake SR, Famili I, Palsson BO, Wang J. The genomic sequence of the Chinese hamster ovary (CHO)-K1 cell line. Nat Biotechnol. 2011;29:735–741. doi: 10.1038/nbt.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baycin-Hizal D, Tabb DL, Chaerkady R, Chen L, Lewis NE, Nagarajan H, Sarkaria V, Kumar A, Wolozny D, Colao J, Jacobson E, Tian Y, O’Meally RN, Krag SS, Cole RN, Palsson BO, Zhang H, Betenbaugh M. Proteomic analysis of Chinese hamster ovary cells. J Proteome Res. 2012;11:5265–5276. doi: 10.1021/pr300476w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim UM, Yap MG, Lim YP, Goh LT, Ng SK. Identification of autocrine growth factors secreted by CHO cells for applications in single-cell cloning media. J Proteome Res. 2013;12:3496–3510. doi: 10.1021/pr400352n. [DOI] [PubMed] [Google Scholar]
- 12.Valente KN, Schaefer AK, Kempton HR, Lenhoff AM, Lee KH. Recovery of Chinese hamster ovary host cell proteins for proteomic analysis. Biotechnol J. 2014;9:87–99. doi: 10.1002/biot.201300190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bracewell DG, Francis R, Smales CM. The future of host cell protein (HCP) identification during process development and manufacturing linked to a risk-based management for their control. Biotechnol Bioeng. 2015;112:1727–1737. doi: 10.1002/bit.25628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hogwood CE, Tait AS, Koloteva-Levine N, Bracewell DG, Smales CM. The dynamics of the CHO host cell protein profile during clarification and protein A capture in a platform antibody purification process. Biotechnol Bioeng. 2013;110:240–251. doi: 10.1002/bit.24607. [DOI] [PubMed] [Google Scholar]
- 15.Kantardjieff A, Jacob NM, Yee JC, Epstein E, Kok YJ, Philp R, Betenbaugh M, Hu WS. Transcriptome and proteome analysis of Chinese hamster ovary cells under low temperature and butyrate treatment. J Biotechnol. 2010;145:143–159. doi: 10.1016/j.jbiotec.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 16.Lee MS, Kim KW, Kim YH, Lee GM. Proteome analysis of antibody-expressing CHO cells in response to hyperosmotic pressure. Biotechnol Prog. 2003;19:1734–1741. doi: 10.1021/bp034093a. [DOI] [PubMed] [Google Scholar]
- 17.Liu Z, Dai S, Bones J, Ray S, Cha S, Karger BL, Li JJ, Wilson L, Hinckle G, Rossomando A. A quantitative proteomic analysis of cellular responses to high glucose media in Chinese hamster ovary cells. Biotechnol Prog. 2015;31:1026–1038. doi: 10.1002/btpr.2090. [DOI] [PubMed] [Google Scholar]
- 18.Ma C, Kojima K, Xu N, Mobley J, Zhou L, Yang ST, Liu XM. Comparative proteomics analysis of high n-butanol producing metabolically engineered Clostridium tyrobutyricum. J Biotechnol. 2015;193:108–119. doi: 10.1016/j.jbiotec.2014.10.036. [DOI] [PubMed] [Google Scholar]
- 19.Riby J, Mobley J, Zhang J, Bracci PM, Skibola CF. Serum protein profiling in diffuse large B-cell lymphoma. Proteomics Clin Appl. 2016;10:1113–1121. doi: 10.1002/prca.201600074. [DOI] [PubMed] [Google Scholar]
- 20.Ludwig MR, Kojima K, Bowersock GJ, Chen D, Jhala NC, Buchsbaum DJ, Grizzle WE, Klug CA, Mobley JA. Surveying the serologic proteome in a tissue-specific kras(G12D) knockin mouse model of pancreatic cancer. Proteomics. 2016;16:516–531. doi: 10.1002/pmic.201500133. [DOI] [PubMed] [Google Scholar]
- 21.Wolfgang Sommeregger AG, Sterovsky Thomas, Casanova Emilio, Kunert Renate. Powerful expression in Chinese Hamster Ovary cells using bacterial artificial chromosomes: Parameters influencing productivity. BMC Proceedings. 2013;7:25–26. [Google Scholar]
- 22.Takubo T, Wakui S, Daigo K, Kurokata K, Ohashi T, Katayama K, Hino M. Expression of non-muscle type myosin heavy polypeptide 9 (MYH9) in mammalian cells. Eur J Histochem. 2003;47:345–352. [PubMed] [Google Scholar]
- 23.Calderwood DA, Huttenlocher A, Kiosses WB, Rose DM, Woodside DG, Schwartz MA, Ginsberg MH. Increased filamin binding to beta-integrin cytoplasmic domains inhibits cell migration. Nat Cell Biol. 2001;3:1060–1068. doi: 10.1038/ncb1201-1060. [DOI] [PubMed] [Google Scholar]
- 24.Feng S, Lu X, Kroll MH. Filamin A binding stabilizes nascent glycoprotein Ibalpha trafficking and thereby enhances its surface expression. J Biol Chem. 2005;280:6709–6715. doi: 10.1074/jbc.M413590200. [DOI] [PubMed] [Google Scholar]
- 25.Ivaska J, Pallari HM, Nevo J, Eriksson JE. Novel functions of vimentin in cell adhesion, migration, and signaling. Exp Cell Res. 2007;313:2050–2062. doi: 10.1016/j.yexcr.2007.03.040. [DOI] [PubMed] [Google Scholar]
- 26.Ben-Ze’ev A. Cell configuration-related control of vimentin biosynthesis and phosphorylation in cultured mammalian cells. J Cell Biol. 1983;97:858–865. doi: 10.1083/jcb.97.3.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammond S, Lee KH. RNA interference of cofilin in Chinese hamster ovary cells improves recombinant protein productivity. Biotechnol Bioeng. 2012;109:528–535. doi: 10.1002/bit.23322. [DOI] [PubMed] [Google Scholar]
- 28.Bernstein BW, Bamburg JR. ADF/cofilin: a functional node in cell biology. Trends Cell Biol. 2010;20:187–195. doi: 10.1016/j.tcb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuystermans D, Al-Rubeai M. Mammalian cell line selection strategies for high-producer. In: Al-Rubeai M, editor. Animal Cell Culture. Springer International Publishing; Switzerland: 2015. pp. 327–372. [Google Scholar]
- 30.Edamatsu H, Kaziro Y, Itoh H. Inducible high-level expression vector for mammalian cells, pEF-LAC carrying human elongation factor 1alpha promoter and lac operator. Gene. 1997;187:289–294. doi: 10.1016/s0378-1119(96)00768-8. [DOI] [PubMed] [Google Scholar]
- 31.Baiet B, Burel C, Saint-Jean B, Louvet R, Menu-Bouaouiche L, Kiefer-Meyer MC, Mathieu-Rivet E, Lefebvre T, Castel H, Carlier A, Cadoret JP, Lerouge P, Bardor M. N-glycans of Phaeodactylum tricornutum diatom and functional characterization of its N-acetylglucosaminyltransferase I enzyme. J Biol Chem. 2011;286:6152–6164. doi: 10.1074/jbc.M110.175711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ito Y, Takeda Y, Seko A, Izumi M, Kajihara Y. Functional analysis of endoplasmic reticulum glucosyltransferase (UGGT): Synthetic chemistry’s initiative in glycobiology. Semin Cell Dev Biol. 2015;41:90–98. doi: 10.1016/j.semcdb.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 33.Benyair R, Ron E, Lederkremer GZ. Protein quality control, retention, and degradation at the endoplasmic reticulum. Int Rev Cell Mol Biol. 2011;292:197–280. doi: 10.1016/B978-0-12-386033-0.00005-0. [DOI] [PubMed] [Google Scholar]
- 34.Marzec M, Eletto D, Argon Y. GRP94: An HSP90-like protein specialized for protein folding and quality control in the endoplasmic reticulum. Biochim Biophys Acta. 2012;1823:774–787. doi: 10.1016/j.bbamcr.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schiene-Fischer C, Yu C. Receptor accessory folding helper enzymes: the functional role of peptidyl prolyl cis/trans isomerases. FEBS Lett. 2001;495:1–6. doi: 10.1016/s0014-5793(01)02326-2. [DOI] [PubMed] [Google Scholar]
- 36.Fussenegger M, Fassnacht D, Schwartz R, Zanghi JA, Graf M, Bailey JE, Portner R. Regulated overexpression of the survival factor bcl-2 in CHO cells increases viable cell density in batch culture and decreases DNA release in extended fixed-bed cultivation. Cytotechnology. 2000;32:45–61. doi: 10.1023/A:1008168522385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Majors BS, Betenbaugh MJ, Pederson NE, Chiang GG. Enhancement of transient gene expression and culture viability using Chinese hamster ovary cells overexpressing Bcl-x(L) Biotechnol Bioeng. 2008;101:567–578. doi: 10.1002/bit.21917. [DOI] [PubMed] [Google Scholar]
- 38.Kumar N, Gammell P, Clynes M. Proliferation control strategies to improve productivity and survival during CHO based production culture: A summary of recent methods employed and the effects of proliferation control in product secreting CHO cell lines. Cytotechnology. 2007;53:33–46. doi: 10.1007/s10616-007-9047-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park JH, Noh SM, Woo JR, Kim JW, Lee GM. Valeric acid induces cell cycle arrest at G1 phase in CHO cell cultures and improves recombinant antibody productivity. Biotechnol J. 2016;11:487–496. doi: 10.1002/biot.201500327. [DOI] [PubMed] [Google Scholar]
- 40.Kim S, Coulombe PA. Emerging role for the cytoskeleton as an organizer and regulator of translation. Nat Rev Mol Cell Biol. 2010;11:75–81. doi: 10.1038/nrm2818. [DOI] [PubMed] [Google Scholar]
- 41.Kwon RJ, Kim SK, Lee SI, Hwang SJ, Lee GM, Kim JS, Seol W. Artificial transcription factors increase production of recombinant antibodies in Chinese hamster ovary cells. Biotechnol Lett. 2006;28:9–15. doi: 10.1007/s10529-005-4680-7. [DOI] [PubMed] [Google Scholar]
- 42.Majors BS, Arden N, Oyler GA, Chiang GG, Pederson NE, Betenbaugh MJ. E2F-1 overexpression increases viable cell density in batch cultures of Chinese hamster ovary cells. J Biotechnol. 2008;138:103–106. doi: 10.1016/j.jbiotec.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 43.Merrick WC. Initiation of protein biosynthesis in eukaryotes. Biochemistry and Molecular Biology Education. 2003;31:378–385. [Google Scholar]
- 44.David Reinhart LD, Sommeregger Wolfgang, Gili Andreas, Schafellner Stanislaus, Castan Andreas, Kaisermayer Christian, Kunert Renate. Influence of cell culture media and feed supplements on cell metabolism and quality of IgG produced in CHO-K1, CHO-S, and CHO-DG44. BMC Proceedings. 2015;9:1–3. [Google Scholar]
- 45.O’Callaghan PM, Berthelot ME, Young RJ, Graham JW, Racher AJ, Aldana D. Diversity in host clone performance within a Chinese hamster ovary cell line. Biotechnol Prog. 2015;31:1187–1200. doi: 10.1002/btpr.2097. [DOI] [PubMed] [Google Scholar]
- 46.Xu N, Ma C, Sun W, Wu Y, Liu XM. Achievements and perspectives in Chinese hamster ovary host cell engineering. Pharmaceutical Bioprocessing. 2015;3:285–292. [Google Scholar]