Abstract
Androgen insensitivity syndrome (AIS) is the most common cause of disorders of sex development usually caused by mutations in the androgen receptor (AR) gene. AIS is characterized by a poor genotype–phenotype correlation, and many patients with clinically presumed AIS do not seem to have mutations in the AR gene. We therefore aimed at identifying a biomarker enabling the assessment of the cellular function of the AR as a transcriptional activator. In the first step, we used complementary DNA (cDNA) microarrays for a genome-wide screen for androgen-regulated genes in two normal male primary scrotal skin fibroblast strains compared to two labia majora fibroblast strains from 46,XY females with complete AIS (CAIS). Apolipoprotein D (APOD) and two further transcripts were significantly upregulated by dihydrotestosterone (DHT) in scrotum fibroblasts, while CAIS labia majora cells were unresponsive. Microarray data were well correlated with quantitative real-time polymerase chain reaction (qRT-PCR; R=0.93). Subsequently, we used qRT-PCR in independent new cell cultures and confirmed the significant DHT-dependent upregulation of APOD in five normal scrotum strains [13.5±8.2 (SD)-fold] compared with three CAIS strains (1.2±0.7-fold, p= 0.028; t test) and six partial androgen insensitivity syndrome strains (2±1.3-fold, p=0.034; t test). Moreover, two different 17β-hydroxysteroid dehydrogenase III deficiency labia majora strains showed APOD induction in the range of normal scrotum (9.96±1.4-fold), supporting AR specificity. Therefore, qRT-PCR of APOD messenger RNA transcription in primary cultures of labioscrotal skin fibroblasts is a promising tool for assessing AR function, potentially allowing a function-based diagnostic evaluation of AIS in the future.
Keywords: Androgen insensitivity syndrome, Disorders of sex development, Apolipoprotein D, Genital fibroblasts
Introduction
Normal male external genital differentiation is dependent on normal androgen action via the androgen receptor (AR) in the external genital target tissues. The AR is an androgen-activated transcription factor controlling transcription rates of androgen-regulated target genes. Mutations in the X-chromosomal AR gene inhibit AR function and in turn lead to the androgen insensitivity syndrome (AIS) in 46,XY individuals [1, 2]. The clinical spectrum of AIS ranges from complete AIS (CAIS) with normal female external genitalia due to complete inactivation of the AR to partial AIS (PAIS) with varying degrees of genital ambiguity, including mild forms with only slight defects of virilization or isolated infertility caused by partial insufficiency of AR function [1, 2]. On the hormonal level, AIS is characterized by normal androgen biosynthesis and normal or elevated gonadotrophins [1].
Intriguingly, many patients with clinically evident AIS do not have documented mutations in the AR gene [2]. These individuals might have defects in yet uncharacterized co-regulators of AR signaling. For instance, there is evidence that co-regulators such as transcriptional intermediary factor 2 (TIF2) are involved in the pathogenesis of AIS [3–5]. An androgen receptor co-activator defect has been published to be the sole cause of AIS in the absence of an AR gene mutation [6]. In some patients, reduced AR transcription due to unknown mechanisms may also underlie AIS [7]. These considerations hamper clinical management due to the lack of a defined molecular diagnosis and clinical prognosis in a significant subset of patients. Moreover, in AIS patients with an established AR gene mutation, the correlation between genotype and phenotype is often poor, even within the same family [8–10]. Therefore, the discovery of androgen-regulated target genes reflecting AR signaling in patient-derived cellular material could provide opportunities for developing a bioassay for the diagnosis of AIS based on functional criteria.
Historically, the clinical diagnosis of AIS was confirmed by reduced affinity of binding of androgen in cultured genital skin fibroblasts. However, this method is restricted to patients having molecular defects in the ligand-binding domain of the AR [11]. The degree of AR dysfunction in AIS was also used to be assessed by the degree of decrease of steroid hormone binding globulin in response to stanozolol [12, 13]. However, it is unclear whether this test reflects the competence of the AR as a transcriptional regulator, making test results difficult to interpret. Attempts to identify androgen-regulated genes in AR-expressing male foreskin-derived fibroblasts—a commonly used “normal control” in human genital differentiation research—either failed or showed inconsistent results [14–16], including our own genome-wide studies [17, 18]. In part, these inconsistencies may be explained by the fact that postnatal genital fibroblasts represent end-stage differentiated cells that have become unresponsive to androgen signaling since genital morphogenesis has been completed [17, 18]. We have recently shown that skin fibroblasts derived from the scrotum, which is the male homologue of the labia majora in females, display significant differences of gene expression patterns compared with foreskin fibroblasts, indicating that these two cell types are very different entities [19]. Although differentiation of cells is usually accompanied by specific epigenomic modifications [20, 21], we speculated that scrotum fibroblasts, in contrast to foreskin fibroblasts, could have retained AR activity. We therefore analyzed for the first time dihydrotestosterone (DHT)-induced gene transcription in scrotum fibroblasts in the presence or absence of the demethylating agent 5-aza-deoxy-cytidine (AZA), first by complementary DNA (cDNA) microarrays and subsequently by quantitative real-time polymerase chain reaction (qRT-PCR). We found significant upregulation of apolipoprotein D (APOD) in scrotum fibroblasts from normal males, while those from individuals with PAIS and CAIS showed severely reduced or absent response. AZA did neither affect APOD-expression nor unveil further AR targets.
Materials and methods
Experimental subjects
The study was approved by the ethical committee of the University of Lübeck, Germany. A written informed consent was obtained from normal subjects, patients, and their parents.
Fibroblast strains, cell culture, and treatment conditions
Scrotal skin fibroblasts were obtained from eight males with phenotypically normal male external genitalia. 46,XY labia majora and labioscrotal-derived fibroblasts were obtained from six PAIS and five CAIS patients with proven inactivating mutations of the AR gene. In addition, two labia-majora-derived fibroblast cultures with 17β-HSDIII deficiency due to documented inactivating mutations were analyzed (Table 1). Fibroblasts were grown at 37°C with 5% CO2 in phenol red free Dulbecco’s modified Eagle’s medium (Gibco BRL, Eggenstein, Germany) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin/streptomycin, 2 mM L-glutamine (Sigma, St. Louis, MO, USA), and 20 mM HEPES buffer (Gibco BRL). All hormone treatments were carried out with these medium constituents, but with charcoal-treated 10% FBS.
Table 1.
Fibroblast strains
| Cell strain/passage number | Anatomic origin/embryonal anlagen | Anatomic phenotype of external genitalia/AIS stage | Age at biopsy (years; months) | Selected clinical background and functional data (androgen binding, AR genotype, specific remarks) |
|---|---|---|---|---|
| S 3 | Scrotum, right side | Normal male | 12;7 | Torsion of left testis 4 weeks ago, clinical indication for surgery: prophylactic orchidopexy on the right side, no androgen binding data |
| 3 | Labioscrotal swellings | |||
| S 4 | Scrotum, left side | Normal male | 1;3 | Minor deviation of penile shaft but no hypospadias, maldescensus testis on both sides, indication for surgery: orchidopexy; Kd 0.12 nM, Bmax 44 fmol/mg protein |
| 6 | Labioscrotal swellings | |||
| S 5 | Scrotum, right side | Normal male | 1;7 | Suspected inguinal testis on the right side. Clinical indication for surgery: orchidopexy, upon surgery no testicular tissue present on the right, Kd 0.09 nM, Bmax 28 fmol/mg protein |
| 8 | Labioscrotal swellings | |||
| S 8 | Scrotum, raphe | Normal male | 31;0 | Infertility, azoospermia, clinical indication for surgery: testicular sperm extraction, hormones before biopsy: LH=3.5 U/L, FSH=2.7 U/L; testosterone=276.31 mg/dl; fibroblast genomic DNA sequence analysis upon knowledge of APOD RT-PCR revealed I841S point mutation within the ligand binding domain of AR |
| 9 | Labioscrotal swellings | |||
| S 9 | Scrotum, raphe | Normal male | 34;0 | Infertility, azoospermia, clinical indication for surgery: testicular sperm extraction; Kd 0.08 nM, Bmax 55 fmol/mg protein |
| 7 | Labioscrotal swellings | |||
| S 12 | Scrotum, left side | Normal male | 9;2 | Maldescensus testis on the left side, clinical indication for surgery: orchidopexy on the left, Kd 0.07 nM, Bmax 37.09 fmol/mg protein |
| 9 | Labioscrotal swellings | |||
| S 13 | Scrotum, raphe | Normal male | 58;1 | Infertility, azoospermia; clinical indication for surgery: testicular sperm extraction; no androgen binding data |
| 5 | Labioscrotal swellings | |||
| S 15 | Scrotum, raphe | Normal male | 5;2 | Phimosis; clinical indication for surgery: circumcision; no androgen binding data |
| 7 | Labioscrotal swellings | |||
| ARD 446 | Scrotum | Predominantly male (AIS2) | 7;8 | Silent mutation GGG>GGA in codon 795 of the AR-gene; Kd 0.07 nM, Bmax 26 fmol/mg protein |
| 6 | Labioscrotal swellings | |||
| ARD 001 | Labia majora/scrotum | Ambiguous (AIS3) | 0;9 | Exonic splice site mutation AGC>AGT in codon 888 leading to aberrant splicing |
| 3 | Labioscrotal swellings | |||
| ARD 084 | Labia majora/scrotum | Ambiguous (AIS3) | 0;8 | No mutation in the whole coding region of the AR gene, reduced AR-mRNA, reduced AR-protein and reduced ligand binding (Kd 0.05 nM, Bmax 6 fmol/mg protein) |
| 9 | Labioscrotal swellings | |||
| ARD 534 | Labia majora/scrotum | Ambiguous (AIS3) | 2;4 | Arg608Lys |
| 10 | Labioscrotal swellings | |||
| ARD 377 | Labia majora/scrotum | Predominantly female (AIS4) | 1;2 | Ile841Ser; Kd 0.55 nM, Bmax 17 fmol/mg protein |
| 4 | Labioscrotal swellings | |||
| ARD 659 | Labia majora | Predominantly female (AIS4) | 3;10 | Ala870Gly |
| 4 | Labioscrotal swellings | |||
| ARD 402 | Labia majora | Normal female (CAIS) | 1;0 | 2-bp deletion in exon1, frameshift, premature stop codon, negative androgen binding, very low AR-mRNA transcription, no AR-protein in Western immunoblot |
| Labioscrotal swellings | ||||
| ARD 411 | Labia majora | Normal female (CAIS) | 0;4 | Arg855Cys, negative androgen binding |
| 7 | Labioscrotal swellings | |||
| ARD 682 | Labia majora | Normal female (CAIS) | 14;10 | Gln59stop, negative androgen binding |
| 8 | Labioscrotal swellings | No AR-protein in Western blotting | ||
| ARD 1097 | Labia majora | Normal female (CAIS) | 1;3 | Pro390Ser+Arg855Gly; negative androgen binding |
| 4 | Labioscrotal swellings | |||
| ARD 1144 | Labia majora | Normal female (CAIS) | 4;3 | Pathological androgen binding, Kd 1.59 nM, Bmax 14 fmol/mg protein, no mutation detected |
| 4 | Labioscrotal swellings | |||
| ARD 111 | Labia majora | Normal female | 10;9 | Homozygous disruptive mutation 325+4A/T of the 17β-HSDIII gene |
| 7 | Labioscrotal swellings | |||
| ARD 1373 | Labia majora | Normal female | 13;4 | Homozygous disruptive mutation 325+4A/T of the 17β-HSDIII gene |
| 9 | Labioscrotal swellings |
Fibroblast strains; ARD patient strain-ID, S normal male scrotal fibroblasts. Normal ranges for androgen (methyltrienolone) binding: Bmax (binding capacity), 13–116 fmol/mg protein; Kd (dissociation constant), 0.03–0.13 nM
In the first set of experiments (experimental set 1), two different scrotum fibroblasts strains (S4, S15) and two different 46,XY CAIS labia majora fibroblasts strains (ARD411, ARD1097) were selected (Table 1) for a genome-wide screening by microarray and subsequent independent validation by qRT-PCR. They were treated with either (1) 10 nM DHT (in ethanol) plus 2 μg/ml AZA (in acetic acid); (2) 10 nM DHT (in ethanol) plus acetic acid; (3) ethanol plus AZA (in acetic acid); and (4) ethanol plus acetic acid. DHT (Merck, Darmstadt, Germany) was dissolved in ethanol (final dilution in culture media=1:10,000). AZA (Sigma, Taufkirchen, Germany) was dissolved in acetic acid (final dilution in culture media=0.5%). Passage numbers were between 3 and 9 and 1 × 106 cells were seeded per culture (T-175 flasks; day 1). DHT was introduced 24 h after seeding the cells (day 2) and was repeated once every 48 h for a total of six times (last treatment at day 14). After the final hormone treatment, cells remained in culture for 96 h prior to harvest (day 18). By this method, fibroblasts become confluent (G0), and we have demonstrated previously that these conditions lead to highly reproducible and stable gene transcription profiles as needed for genome-wide microarray experiments with non-immortalized, primary cultures of fibroblasts [17].
In the second set of experiments (experimental set 2), a completely independent new set of fibroblast cultures derived from different patients and normal controls (phenotypic normal males: S3, S5, S8, S9, S12, S13; CAIS: ARD682, ARD1144, ARD402, PAIS: ARD446, ARD534, ARD001, ARD084, ARD377, ARD659; 17β-HSDIII deficiency: ARD111, ARD1373) received either 10 nM DHT plus ethanol or ethanol alone under the same treatment regimen as described above, and RNA was subsequently subjected to qRT-PCR.
RNA isolation, amplification, and reference RNA
Total RNA was isolated from fibroblasts using Trizol (Invitrogen, Paisley, UK) followed by RNeasy mini kit protocol (Qiagen, Hilden, Germany). DNA contamination was removed using the DNA free kit (Ambion, Austin, USA). Total RNA and universal human reference RNA (Stratagene Europe, Amsterdam, The Netherlands) were amplified according to the MessageAmp II aRNA kit protocol (Ambion). Total RNA and the amplified antisense RNA (aRNA) were quantified using the 2100 Bioanalyzer (Agilent, Palo Alto, USA).
Microarray hybridization
Spotted cDNA microarrays (Stanford Functional Genomics Facility, Stanford, CA, USA) were used for gene expression studies containing approximately 43,000 features representing 32,968 unique human genes. Labeling of aRNA and subsequent cDNA hybridizations were performed according to published protocols (http://cmgm.stanford.edu/pbrown/protocols/index.html). While 3 μg of each fibroblast aRNA was labeled with Cy5 (pseudocolor red), 2 μg of a universal human reference RNA was labeled with Cy3 (pseudocolor green; Amersham Pharmacia Biotech Europe, Freiburg, Germany). Competitive hybridizations of Cy5- and Cy3-labeled probes on a total of 16 microarrays were performed for 14–18 h over night at 65°C in a water bath. Hybridized and washed microarrays were scanned using a Genepix 4000B microarray scanner (Axon, Foster City, USA). Images were analyzed with GenePix Pro 4.0 software (Axon Instruments, Foster City, USA).
Microarray data analysis
Only spots with fluorescence signals of ≥1.5-fold over array background in either the experimental (Cy5) or the reference (Cy3) channel were considered based on the 16 microarray experiments representing the four experimental conditions corresponding to the four used cell strains in the first set of treatments (experimental set 1). Cy5/Cy3 fluorescence ratios for all genes were normalized to obtain an average absolute log2 red/green ratio of 0 in each microarray. Fluorescence ratios for each gene were centered (by mean) and genes with at least 80% interpretable data in the microarrays were used. We further considered only those transcripts whose log2 red (Cy5)/green (Cy3) ratio differed from the mean expression level across all 16 experiments by at least 1.0 (corresponding to twofold upregulation or twofold downregulation) in at least four of 16 experiments. In the first, we performed unsupervised hierarchical clustering analysis (Pearson correlation metrics, average linkage clustering) [22]. Data were displayed by TreeView [22]. Secondly, we applied the significance analysis of microarrays (SAM) procedure [23], a well-established and stringent biomathematical method to identify transcripts with statistically significant differences in expression levels on microarrays, and we compared all DHT-treated samples with all untreated samples in the normal scrotum fibroblasts independent from the presence or absence of AZA. Moreover, we have compared all AZA-treated samples with all non-AZA samples independent from DHT treatment or AR status by SAM.
Total RNA amplification, qRT-PCR of APOD
Total RNA was amplified according to the manufacturer’s instructions using the First-strand cDNA synthesis kit (Fermentas, Hannover, Germany). qRT-PCR was applied to a total of eight scrotum fibroblasts strains [two validation experiments on messenger RNA (mRNA) used for microarray hybridizations in the first set of experiments (experimental set 1) plus six independent experiments in the second qRT-PCR-only set (experimental set 2)], five CAIS labia majora fibroblasts strains [two validation experiments on mRNA used for microarray hybridizations in the first set of experiments (experimental set 1) plus three independent experiments in the second qRT-PCR-only set (experimental set 2)], six PAIS fibroblasts strains, and two 17β-HSDIII fibroblast strains [only as part of the second set of qRT-PCR experiments (experimental set 2)] (Table 1). qRT-PCR was performed using the QuantiTect SYBR Green PCR kit (Qiagen). The primers used for PCR (TIB MOLBIOL, Berlin, Germany) had the following sequences: APOD forward primer 5′-CCACCCCAgTTAACCTCACA-3′ and APOD reverse primer 5′-gTgCCgATggCATAAACC-3′. Succinate dehydrogenase subunit A (SDHA) was used as housekeeping gene for normalization. SDHA forward primer was 5′-ACCAggTCACACACTgTTgC-3′ and SDHA reverse primer was 5′-CgTAgAAATgCCACCTCCA-3′. PCR was carried out on the LightCycler (Roche Diagnostics, Mannheim, Germany). In total, 18 μl of PCR master mix including primers and 2 μl of cDNA (approximately 6.25 ng/μl) were pipetted into each capillary. The PCR conditions were 95°C for 15 min, 45 cycles of 94°C for 15 s, 52°C for 25 s, and 72°C for 25 s. Standard curves for APOD and SDHA were obtained by running the PCR with different dilutions of reference cDNA. Standard curves were used to assess the efficiency correction for the experimental probes. Melting curves were analyzed to make sure that the amplified product was specific for APOD and SDHA and devoid of primer dimers.
Results
Microarray data
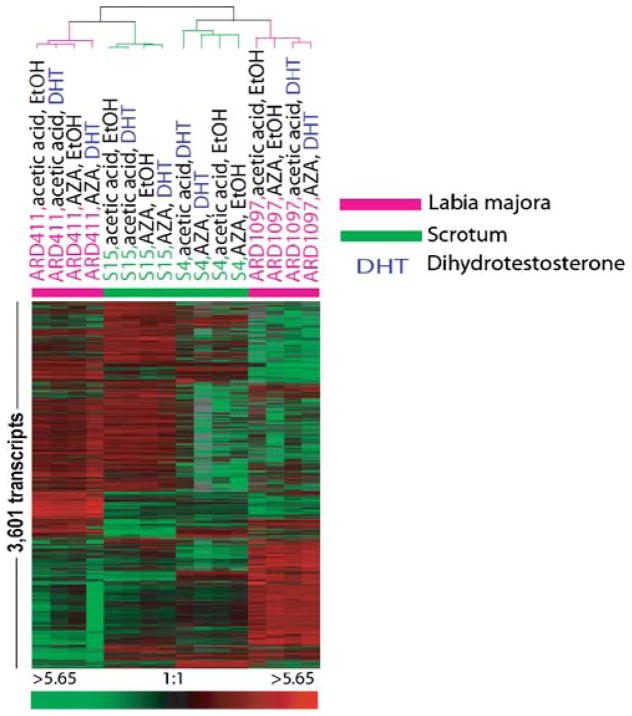
Filtering of all genes of all the 16 microarrays (experimental set 1) as described above resulted in 3,601 transcripts. Unsupervised hierarchical clustering of these transcripts across the 16 microarray experiments demonstrated distinct, reproducible patterns of gene expression (Fig. 1). Clustering sorted all microarray experiments according to the cell strain, indicating the existence of highly strain-specific, stable baseline gene transcription signatures independent from the treatment of the cells. Therefore, strain identity was the overriding biological pattern causing gene expression differences in the 16 microarrays dataset. From the methodological perspective, this observation confirms the high reproducibility of our cell culture strategy in combination with the microarray procedures (Fig. 1), as also previously published [19]. However, our unsupervised analysis did neither expose obvious influences of DHT treatment nor of AZA treatment, suggesting that the cells responded only little to either of these two treatments. Therefore, at this stage of analyses, our finding on scrotal fibroblasts seemed to agree with our previous work showing a lack of response of foreskin fibroblasts to many different protocols of DHT treatment [17, 18].
Fig. 1.
Unsupervised hierarchical cluster analysis of scrotum and 46, XY labia majora fibroblasts (experimental set 1). The heatmap represents hierarchical clustering of 3,601 transcripts which had at least 80% interpretable data across the 16 microarrays and whose expression levels were at least twofold different from the mean expression level across all samples in at least four experiments. Transcripts are represented in rows and experiments in columns. Expression values per gene are centered by the mean log2 Cy5/Cy3 normalized ration. Increasing red intensity corresponds to higher relative transcript levels compared to the mean expression level across all experiments. Increasing green intensity corresponds to relatively decreased transcript levels compared to the mean. The cluster dendrogram demonstrates the degree of relatedness (Pearson correlation) between the expression patterns of the 3,601 genes in the 16 fibroblast samples. The length of the arms of the dendogram reflects the degree of correlation between the samples normalized ratio.
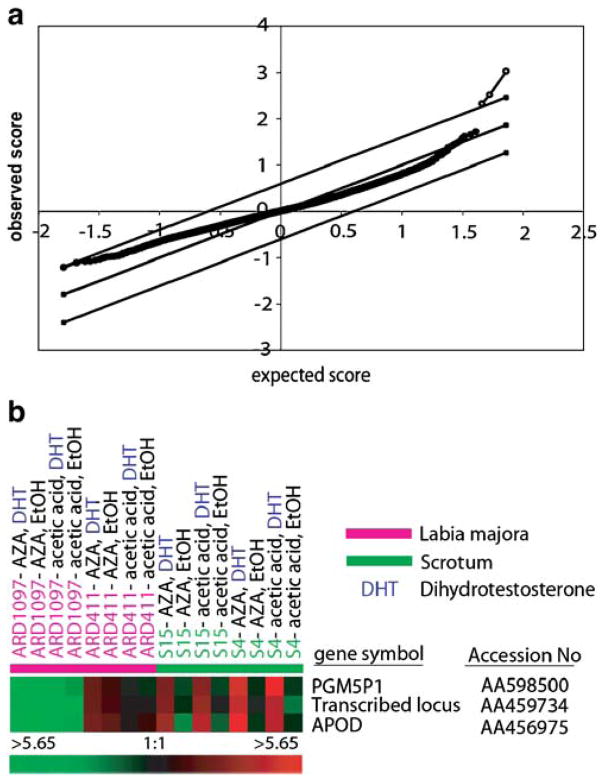
Since unsupervised analysis did not show obvious DHT-effects, we performed a supervised statistical search for significant DHT-regulated genes using the well-established SAM procedure [23]. Using this algorithm, we compared all DHT-treated normal scrotum fibroblast experiments with all non-DHT-treated normal scrotum fibroblast experiments, independent of the presence or absence of AZA. Based on analysis of all expressed genes with 80% interpretable data in the fibroblasts (23,863 genes), SAM identified three transcripts, namely, APOD (accession number AA456975), Phosphoglucomutase 5 pseudogene 1 (PGM5P1, accession number AA598500), and the transcribed locus AA459734 using a SAM filter with a false discovery rate of 0% (Fig. 2a). Figure 2b compiles the gene expression data of the above three transcripts in the 16 microarray experiments. In addition to the obvious strain-specific variability of baseline expression of all three transcripts (e.g., see ARD1097 versus ARD411), Fig. 2b illustrates the striking response in the two scrotum strains to DHT treatment, but no response in the two CAIS strains. SAM analysis did not reveal reproducible changes in gene expression in response to treatment with AZA.
Fig. 2.
a Identification of androgen induced transcripts by significance analysis of microarrays (SAM). Plot of observed scores (Y-axis) versus expected scores (X-axis) according to [23] of DHT-induced transcripts calculated by SAM analysis on the basis of 16 microarrays in experimental set 1. The open dots represent the transcripts which are significantly induced by DHT at a false positive discovery rate of 0%. b Color-coded display of expression values of DHT- induced transcripts (experimental set 1). The rows in the heatmap show the three transcripts, APOD, PGM5P1, and the transcribed locus AA459734, which were identified to be significantly induced by DHT as measured on micro-arrays using SAM. Expression values for each transcript were centered by mean. Red–green color coding has the same meaning as in Fig. 1
Quantitative RT-PCR
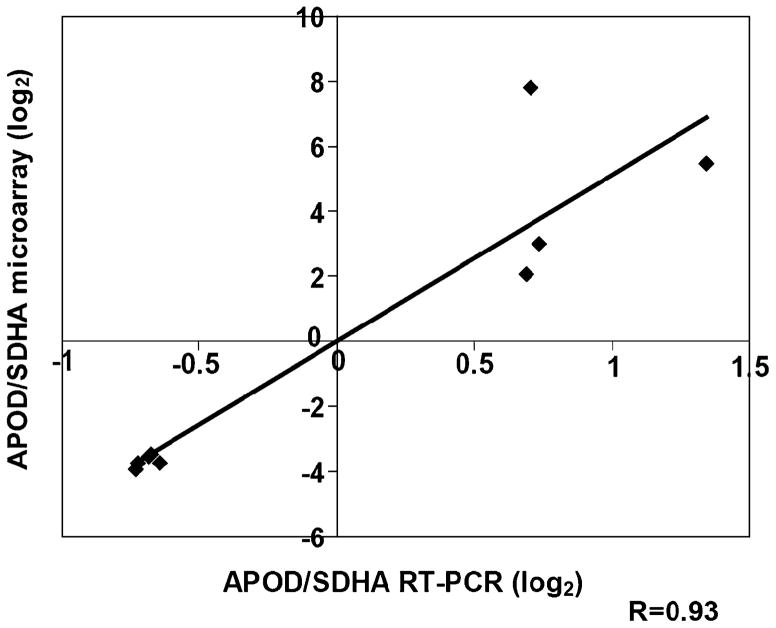
We subsequently selected APOD for our validation experiments. To directly confirm the gene expression levels found in microarray analyses, we performed qRT-PCR of APOD using eight of the 16 mRNAs that had been used for the microarray hybridizations. APOD expression values as measured by microarrays were well correlated with expression values of qRT-PCR (R=0.93; Fig. 3).
Fig. 3.
Correlation of APOD expression as measured by microarray and qRT-PCR (experimental set 1). Expression values of microarray data (ratio of APOD divided through SDHA; Y-axis) versus quantitative real time PCR (qRT-PCR; ratio of APOD divided through SDHA; X-axis). The correlation coefficient (R) is 0.93
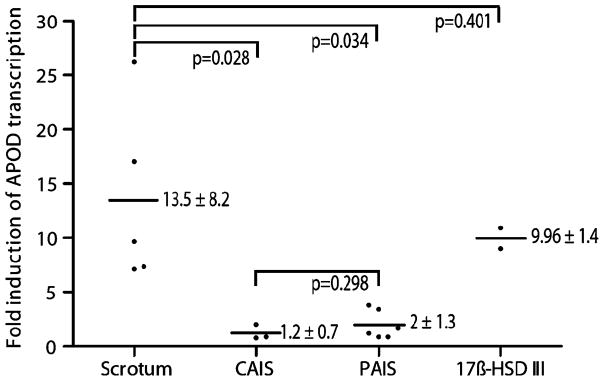
To further validate the inducibility of APOD, we carried out independent new cell culture experiments and DHT treatments of six new scrotal fibroblast strains (S3, S5, S8, S9, S12, S13), three new CAIS strains (ARD682, ARD1144, ARD402), six PAIS strains (ARD446, ARD534, ARD001, ARD084, ARD377, ARD659), and two 17β-HSDIII deficiency strains (ARD111, ARD1373; experimental set 2; see also Table 1). qRT-PCR showed upregulation of APOD to 13.5±8.2-fold (mean±SD) in five of the six control strains, while one of the control strains (S8) showed little or no relevant DHT-mediated APOD upregulation (1.28-fold; Fig. 4). Intriguingly, when we sequenced the AR gene of the S8 fibroblast strain (Table 1) without APOD response, we detected an inactivating I841S mutation, suggesting that this strain held a clinically occult mutation identical to a mutation our group had identified in a PAIS patient previously [24]. We therefore had to exclude this sample from our analysis as part of the normal controls. The three new CAIS strains showed no obvious response of APOD to DHT treatment (1.2±0.7-fold; Fig. 4). In the six PAIS fibroblast samples, we observed a comparably slight upregulation of APOD (2±1.3-fold; Fig. 4). The differences between the normal scrotum and CAIS fibroblasts and between the normal scrotum and PAIS fibroblasts were statistically significant (p=0.028 and 0.034, respectively; t test). 17β-HSDIII fibroblast strains showed APOD upregulation to be 9.96±1.4-fold in response to DHT, and there was no significant difference compared to normal scrotal cells (p=0.401; t test; Fig. 4).
Fig. 4.
DHT-mediated induction of APOD transcription in normal male scrotum, PAIS, CAIS, and individuals with 17β-HSDIII defect (experimental set 2). The Y-axis represents the fold induction of APOD transcription. The X-axis represents the different groups of investigated cell strains. Black dots represent the APOD induction in each of the normal scrotum (five individuals), CAIS (three individuals), PAIS (six individuals), and 17β-HSDIII deficiency (two individuals) cell strains. The black lines represent the mean fold induction of APOD transcription per group based on these values
Discussion
To date, no androgen-induced genes have been identified in primary human genital skin fibroblasts. Using a genome-wide microarray strategy, we demonstrated for the first time that cultured normal male scrotum skin fibroblasts are transcriptionally responsive to DHT treatment in vitro. We found that APOD, a documented androgen-regulated gene in the prostate cancer cell line LNCaP [25], is significantly induced by DHT in these cells. The presence of steroid hormone regulatory elements in the APOD gene promoter [26] corresponds with our findings. Severe reduction or complete absence of APOD induction in PAIS and CAIS samples, as assessed by qRT-PCR, supports specificity of DHT-mediated APOD upregulation for the AR signaling pathway in the genital skin fibroblasts. High APOD response to DHT treatment in labia majora fibroblast strains derived from two different females with 17β-HSDIII deficiency (Table 1) but intact AR confirmed that the AR status, but not labial, versus scrotal anatomy per se is the determinant of APOD response to DHT in fibroblasts. Therefore, qRT PCR of DHT-induced APOD transcription in labioscrotal fibroblasts is a first molecular strategy for the functional identification of AIS assessing the degree of AR-mediated transcription activation in a primary human tissue culture model.
Our data on scrotum fibroblasts contrast with our previous observations in foreskin fibroblasts which failed to show a reproducible androgen response [17, 18]. However, foreskin fibroblasts and scrotal skin fibroblasts originate from different embryonic origin within the bipotent external genitalia and differ significantly in their postnatal baseline expression signatures [19]. These differences in the baseline transcriptome might have contributed to the observed differences in androgen responsiveness. Since epigenetic mechanisms such as gene methylation [20] and chromatin structure modulation due to histone modification [21] play important roles during control of cell and tissue differentiation and since we initially suspected only marginal response of scrotum cells based on our previous experience with foreskin fibroblasts, we performed parallel experiments with the demethylating agent AZA. However, AZA did neither affect DHT-induced APOD upregulation nor did it lead to the identification of additional androgen-regulated genes silenced during development by methylation. We were surprised not to see significant changes in gene expression induced by AZA since we used doses that have been documented to change methylation in cell culture [27]. A possible explanation of the lack of transcriptional changes due to AZA could be that we did not treat for sufficient time so that the number of cell divisions prior to confluency was inadequate to incur promoter demethylation.
Our data imply that APOD could be a potential “biomarker” of androgen receptor function in genital tissues that could be used to establish the diagnosis of AIS. APOD response to DHT was extremely reduced or absent in all of our PAIS and CAIS samples, but was present in the 17β-HSDIII deficiency fibroblasts. The utility of measuring the APOD response to DHT stimulation was suggested by the surprising finding of non-responsiveness in one control, S8. The S8 cell line had been considered to be derived from a normal male control because of phenotypic normal male external genitalia. This subject had initially come to medical attention because of infertility, and scrotal fibroblasts had been harvested at the time of a testicular sperm extraction surgical procedure. Interestingly, S8 fibroblasts showed severely reduced APOD responsiveness to DHT treatment, and sequence analysis showed that they had an inactivating I841S mutation within the ligand-binding domain of the AR gene. We have previously described this mutation in a different patient having PAIS and ambiguous external genitalia [24]. Therefore, reduced APOD transcription in S8 uncovered the diagnosis of AIS in this subject (MAIS, minimal androgen insensitivity syndrome) which was later proven by DNA sequence analysis. On the one hand, this surprising finding supports the suitability of APOD as a potential molecular “biomarker” for the transcription regulatory function of the AR in the genital tissue, allowing a function-based diagnosis of AIS. This also offers valuable perspectives for future categorization of patients with clinically presumed AIS but without currently identified mutations in the AR gene. On the other hand, the degree of genital virilization as part of the AIS phenotype seemed not to be directly reflected by the level of DHT-induced APOD transcription in our study, since we did not observe a statistically significant difference between CAIS and PAIS (Fig. 4) and since the above-described I841S patient demonstrated a normal male external genital phenotype. Therefore, more data are needed to better define the relationship between DHT-induced APOD response and the phenotypic degree of AIS.
APOD plays physiological roles in growth control and growth arrest [25, 28]. High APOD expression in fibroblasts has been described in quiescent cells under conditions of confluency and serum starvation [29]. Interestingly, all four cultures of ARD411 showed higher baseline APOD expression on microarrays compared to the four ARD1097 experiments and to the untreated S4 and S15 experiments (Fig. 2b). Since all these cells were cultured at comparable passage numbers (Table 1), the baseline transcript level differences are not likely to reflect relevant differences in the state of senescence. Rather, we hypothesize that these differences reflect inter-individual differences in gene expression.
APOD belongs to the alpha2mu-microglobulin superfamily, also called lipocalins, an evolutionary conserved superfamily of proteins that function in the binding and transport of many physiologically important ligands including pheromones [30]. E-3-methyl-2-hexenoic acid is the most abundant axillary odor constituent in males and is carried to the skin surface by APOD before it is liberated from non-odorous apocrine secretions by axillary micro-organisms [31]. Pheromonal communication is an important pathway for the transmission of information on gender and social status in mammals [32]. There is evidence that male axillary pheromones can even mediate female menstrual synchrony by potential modulation of the hypothalamic pituitary–gonadal axis [33]. These considerations are intriguing in the context of androgen regulation of APOD transcription in the human genital skin, AIS, and disorders of sex development, but further studies are necessary to investigate the potential role of APOD therein.
Supplementary Material
Acknowledgments
Funding This study was funded by the German Research Foundation (DFG, KFO111-C, and Ho2073/5-1 and Ho2073/5-2 to PMH). We thank Brigitte Karvelis, Gila Hoffman, Kerstin Övermöhle, and Tanja Dahm for excellent technical assistance. We are indebted to the staff of the Stanford Microarray Database (SMD), without whom the project would not have been realized.
Abbreviations
- APOD
Apolipoprotein D
- AR
Androgen receptor
- AIS
Androgen insensitivity syndrome
- PAIS
Partial androgen insensitivity syndrome
- CAIS
Complete androgen insensitivity syndrome
- DHT
Dihydrotestosterone
- AZA
5-Aza-deoxy-cytidine
Footnotes
Supplementary information Microarray data can be accessed in the GEO database (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=lnctdkqeqgeouvc&acc=GSE11847).
Contributor Information
Mahesh Appari, Division of Pediatric Endocrinology and Diabetes, Department of Pediatrics, University-Hospital Schleswig-Holstein, Christian Albrechts University of Kiel, Campus Kiel, Schwanenweg 20, 24105 Kiel, Germany.
Ralf Werner, Department of Pediatric and Adolescent Medicine, University-Hospital Schleswig-Holstein, University of Lübeck, Campus Lübeck, Lübeck, Germany.
Lutz Wünsch, Department of Pediatric Surgery, University Hospital Schleswig-Holstein, Campus Lübeck, Lübeck, Germany.
Gunnar Cario, Department of Pediatrics, University-Hospital Schleswig-Holstein, Christian Albrechts University of Kiel, Campus Kiel, Schwanenweg 20, 24105 Kiel, Germany.
Janos Demeter, Department of Biochemistry, Stanford University School of Medicine, Stanford, CA, USA.
Olaf Hiort, Department of Pediatric and Adolescent Medicine, University-Hospital Schleswig-Holstein, University of Lübeck, Campus Lübeck, Lübeck, Germany.
Felix Riepe, Division of Pediatric Endocrinology and Diabetes, Department of Pediatrics, University-Hospital Schleswig-Holstein, Christian Albrechts University of Kiel, Campus Kiel, Schwanenweg 20, 24105 Kiel, Germany.
James D. Brooks, Department of Urology, Stanford University School of Medicine, Stanford, CA, USA
Paul-Martin Holterhus, Division of Pediatric Endocrinology and Diabetes, Department of Pediatrics, University-Hospital Schleswig-Holstein, Christian Albrechts University of Kiel, Campus Kiel, Schwanenweg 20, 24105 Kiel, Germany.
References
- 1.Quigley CA, De Bellis A, Marschke KB, el-Awady MK, Wilson EM, French FS. Androgen receptor defects: historical, clinical, and molecular perspectives. Endocr Rev. 1995;16:271–321. doi: 10.1210/edrv-16-3-271. [DOI] [PubMed] [Google Scholar]
- 2.Deeb A, Mason C, Lee YS, Hughes IA. Correlation between genotype, phenotype and sex of rearing in 111 patients with partial androgen insensitivity syndrome. Clin Endocrinol (Oxf) 2005;63:56–62. doi: 10.1111/j.1365-2265.2005.02298.x. [DOI] [PubMed] [Google Scholar]
- 3.Ghali SA, Gottlieb B, Lumbroso R, Beitel LK, Elhaji Y, Wu J, Pinsky L, Trifiro MA. The use of androgen receptor amino/carboxyl-terminal interaction assays to investigate androgen receptor gene mutations in subjects with varying degrees of androgen insensitivity. J Clin Endocrinol Metab. 2003;88:2185–2193. doi: 10.1210/jc.2002-021324. [DOI] [PubMed] [Google Scholar]
- 4.Quigley CA, Tan JA, He B, Zhou ZX, Mebarki F, Morel Y, Forest MG, Chatelain P, Ritzén EM, French FS, Wilson EM. Partial androgen insensitivity with phenotypic variation caused by androgen receptor mutations that disrupt activation function 2 and the NH(2)- and carboxyl-terminal interaction. Mech Ageing Dev. 2004;125:683–695. doi: 10.1016/j.mad.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Umar A, Berrevoets CA, Van NM, van Leeuwen M, Verbiest M, Kleijer WJ, Dooijes D, Grootegoed JA, Drop SL, Brinkmann AO. Functional analysis of a novel androgen receptor mutation, Q902K, in an individual with partial androgen insensitivity. J Clin Endocrinol Metab. 2005;90:507–515. doi: 10.1210/jc.2004-0057. [DOI] [PubMed] [Google Scholar]
- 6.Adachi M, Takayanagi R, Tomura A, Imasaki K, Kato S, Goto K, Yanase T, Ikuyama S, Nawata H. Androgen-insensitivity syndrome as a possible coactivator disease. N Engl J Med. 2000;21:856–862. doi: 10.1056/NEJM200009213431205. [DOI] [PubMed] [Google Scholar]
- 7.Holterhus PM, Werner R, Hoppe U, Bassler J, Korsch E, Ranke MB, Dörr HG, Hiort O. Molecular features and clinical phenotypes in androgen insensitivity syndrome in the absence and presence of androgen receptor gene mutations. J Mol Med. 2005;83:1005–1013. doi: 10.1007/s00109-005-0704-y. [DOI] [PubMed] [Google Scholar]
- 8.Rodien P, Mebarki F, Mowszowicz I, Chaussain JL, Young J, Morel Y, Schaison G. Different phenotypes in a family with androgen insensitivity caused by the same M780I point mutation in the androgen receptor gene. J Clin Endocrinol Metab. 1996;81:2994–2998. doi: 10.1210/jcem.81.8.8768864. [DOI] [PubMed] [Google Scholar]
- 9.Evans BA, Hughes IA, Bevan CL, Patterson MN, Gregory JW. Phenotypic diversity in siblings with partial androgen insensitivity syndrome. Arch Dis Child. 1997;76:529–531. doi: 10.1136/adc.76.6.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holterhus PM, Sinnecker GH, Hiort O. Phenotypic diversity and testosterone-induced normalization of mutant L712F androgen receptor function in a kindred with androgen insensitivity. J Clin Endocrinol Metab. 2000;85:3245–3250. doi: 10.1210/jcem.85.9.6812. [DOI] [PubMed] [Google Scholar]
- 11.McPhaul MJ, Marcelli M, Zoppi S, Griffin JE, Wilson JD. Genetic basis of endocrine disease. 4. The spectrum of mutations in the androgen receptor gene that causes androgen resistance. J Clin Endocrinol Metab. 1993;76:17–23. doi: 10.1210/jcem.76.1.8421085. [DOI] [PubMed] [Google Scholar]
- 12.Sinnecker G, Köhler S. Sex hormone-binding globulin response to the anabolic steroid stanozolol: evidence for its suitability as a biological androgen sensitivity test. J Clin Endocrinol Metab. 1989;68:1195–1200. doi: 10.1210/jcem-68-6-1195. [DOI] [PubMed] [Google Scholar]
- 13.Sinnecker GH, Hiort O, Nitsche EM, Holterhus PM, Kruse K. Functional assessment and clinical classification of androgen sensitivity in patients with mutations of the androgen receptor gene. German Collaborative Intersex Study Group. Eur J Pediatr. 1997;156:7–14. doi: 10.1007/s004310050542. [DOI] [PubMed] [Google Scholar]
- 14.Berkovitz GD, Carter KM, Brown TR, Migeon CJ. Testosterone lowers aromatase activity in cultured human genital skin fibroblasts. Mol Cell Endocrinol. 1990;69:187–197. doi: 10.1016/0303-7207(90)90012-w. [DOI] [PubMed] [Google Scholar]
- 15.Stillman SC, Evans BA, Hughes IA. Androgen dependent stimulation of aromatase activity in genital skin fibroblasts from normal and patients with androgen insensitivity. Clin Endocrinol (Oxf) 1991;35:533–538. doi: 10.1111/j.1365-2265.1991.tb00940.x. [DOI] [PubMed] [Google Scholar]
- 16.Nitsche EM, Moquin A, Adams PS, Guenette RS, Lakins JN, Sinnecker GH, Kruse K, Tenniswood MP. Differential display RT PCR of total RNA from human foreskin fibroblasts for investigation of androgen-dependent gene expression. Am J Med Genet. 1996;63:231–238. doi: 10.1002/(SICI)1096-8628(19960503)63:1<231::AID-AJMG40>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 17.Holterhus PM, Hiort O, Demeter J, Brown PO, Brooks JD. Differential gene-expression patterns in genital fibroblasts of normal males and 46,XY females with androgen insensitivity syndrome: evidence for early programming involving the androgen receptor. Genome Biol. 2003;4:R37. doi: 10.1186/gb-2003-4-6-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bebermeier JH, Brooks JD, DePrimo SE, Werner R, Deppe U, Demeter J, Hiort O, Holterhus PM. Cell-line and tissue-specific signatures of androgen receptor-coregulator transcription. J Mol Med. 2006;84:919–931. doi: 10.1007/s00109-006-0081-1. [DOI] [PubMed] [Google Scholar]
- 19.Holterhus PM, Deppe U, Werner R, Richter-Unruh A, Bebermeier JH, Wünsch L, Krege S, Schweikert HU, Demeter J, Riepe F, Hiort O, Brooks JD. Intrinsic androgen-dependent gene expression patterns revealed by comparison of genital fibroblasts from normal males and individuals with complete and partial androgen insensitivity syndrome. BMC Genomics. 2007;8:376. doi: 10.1186/1471-2164-8-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Izbicka E, MacDonald JR, Davidson K, Lawrence RA, Gomez L, Von Hoff DD. 5, 6 Dihydro-5′-azacytidine (DHAC) restores androgen responsiveness in androgen-insensitive prostate cancer cells. Anticancer Res. 1999;19:1285–1291. [PubMed] [Google Scholar]
- 21.Metzger E, Wissmann M, Yin N, Müller JM, Schneider R, Peters AH, Günther T, Buettner R, Schüle R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;15:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 22.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hiort O, Sinnecker GH, Holterhus PM, Nitsche EM, Kruse K. The clinical and molecular spectrum of androgen insensitivity syndromes. Am J Med Genet. 1996;63:218–222. doi: 10.1002/(SICI)1096-8628(19960503)63:1<218::AID-AJMG38>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 25.Simard J, Veilleux R, de Launoit Y, Haagensen DE, Labrie F. Stimulation of apolipoprotein D secretion by steroids coincides with inhibition of cell proliferation in human LNCaP prostate cancer cells. Cancer Res. 1991;51:4336–4341. [PubMed] [Google Scholar]
- 26.Lambert J, Provost PR, Marcel YL, Rassart E. Structure of the human apolipoprotein D gene promoter region. Biochim Biophys Acta. 1993;1172:190–192. doi: 10.1016/0167-4781(93)90292-l. [DOI] [PubMed] [Google Scholar]
- 27.Jüttermann R, Li E, Jaenisch R. Toxicity of 5-aza-2′-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc Natl Acad Sci USA. 1994;91:11797–11801. doi: 10.1073/pnas.91.25.11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Do Carmo S, Séguin D, Milne R, Rassart E. Modulation of apolipoprotein D and apolipoprotein E mRNA expression by growth arrest and identification of key elements in the promoter. J Biol Chem. 2002;277:5514–5523. doi: 10.1074/jbc.M105057200. [DOI] [PubMed] [Google Scholar]
- 29.Provost PR, Marcel YL, Milne RW, Weech PK, Rassart E. Apolipoprotein D transcription occurs specifically in nonproliferating quiescent and senescent fibroblast cultures. FEBS Lett. 1991;290:139–141. doi: 10.1016/0014-5793(91)81244-3. [DOI] [PubMed] [Google Scholar]
- 30.Flower DR. The lipocalin protein family: structure and function. Biochem J. 1996;318:1–14. doi: 10.1042/bj3180001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeng C, Spielman AI, Vowels BR, Leyden JJ, Biemann K, Preti G. A human axillary odorant is carried by apolipoprotein D. Proc Natl Acad Sci USA. 1996;93:6626–6630. doi: 10.1073/pnas.93.13.6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bigiani A, Mucignat-Caretta C, Montani G, Tirindelli R. Pheromone reception in mammals. Rev Physiol Biochem Pharmacol. 2005;154:1–35. doi: 10.1007/s10254-004-0038-0. [DOI] [PubMed] [Google Scholar]
- 33.Preti G, Cutler WB, Garcia CR, Huggins GR, Lawley HJ. Human axillary secretions influence women’s menstrual cycles: the role of donor extract from men. Horm Behav. 1986;20:463–473. doi: 10.1016/0018-506x(86)90008-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.