Abstract
We have developed ExAtlas, an on-line software tool for meta-analysis and visualization of gene expression data. In contrast to existing software tools, ExAtlas compares multi-component data sets and generates results for all combinations (e.g., all gene expression profiles vs. all Gene Ontology annotations). ExAtlas handles both users’ own data and data extracted semi-automatically from the public repository (GEO/NCBI database). ExAtlas provides a variety of tools for meta-analyses: (1) standard meta-analysis (fixed effects, random effects, z-score, and Fisher’s methods); (2) analyses of global correlations between gene expression data sets; (3) gene set enrichment; (4) gene set overlap; (5) gene association by expression profile; (6) gene specificity; and (7) statistical analysis (ANOVA, pair-wise comparison, and PCA). ExAtlas produces graphical outputs, including heatmaps, scatter-plots, bar-charts, and 3-deminsional images. Some of the most widely used public data sets (e.g., GNF/BioGPS, Gene Ontology, KEGG, GAD phenotypes, BrainScan, ENCODE ChIP-seq, and protein-protein interaction) are pre-loaded and can be used for functional annotations.
Keywords: Meta-analysis, Correlation matrix, Gene set enrichment, ANOVA, PCA, Expected proportion of false positives
1. Introduction
Statistical analysis of global gene expression profiles is a well established discipline 1, supported by many versatile software applications (see reviews 2–4). The most widely used programs include TM4 Microarray Software Suite 5, SAM (Significance Analysis of Microarrays) 6, NIA Array Analysis 7, GenMAPP 8, 9, Onto-Express 10, 11, DAVID 12, GSEA 13, GenomatixSuite (commercial program), and Pathway Studio (commercial program). Meta-analysis programs such as GeneMeta 14, metaArray 15, and MAMA 16 are also available. However, these tools are not designed to seamlessly extract data sets from the public depositories such as GEO17 and ArrayExpress18 databases, and cannot process multi-component data sets in all combinations -- for example, estimating global correlation matrixes for different gene expression projects or evaluating gene set enrichment/overlap for multiple gene expression profiles. Although “Microarray retriever” allows downloading gene expression data from GEO and ArrayExpress databases19, that software does not include any analysis tools and downloaded data require additional pre-processing. In addition, as far as we know, there is no software for estimating correlation matrixes for different gene expression projects, and all programs for gene set enrichment or overlap handle one gene expression profile at a time (e.g., as in DAVID or GSEA). Our motivation for developing ExAtlas software (http://lgsun.grc.nia.nih.gov/exatlas), presented in this paper, is to overcome these limitations, thereby providing a versatile tool for comparative analysis of gene expression profile projects and functionally-annotated gene sets.
2. Overview of the Program Workflow
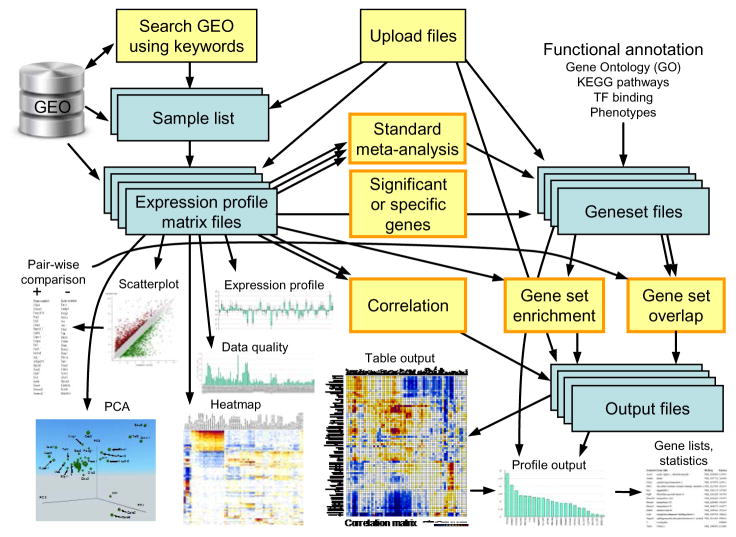
A typical workflow in the use of ExAtlas is shown in Fig. 1. Expression profile data is extracted from the GEO database or uploaded manually. After evaluation of data quality some low-quality samples can be removed. Each data file with gene expression profiles can be used for standard statistical analysis [e.g., ANOVA, pair-wise comparison between tissues or cell types, Principal Component Analysis (PCA)], or to generate lists of genes with significant expression change, plotting of heatmaps, scatter-plots, and expression profiles for individual genes), or to compare with other data files via meta-analysis, correlation analysis, or gene set enrichment analysis. ANOVA and PCA modules were adopted with minor modifications from NIA Array Analysis 7. Lists of differentially-expressed genes can be saved as a gene set file.
Fig. 1.
The workflow in ExAtlas.
ExAtlas has many preloaded public gene set files, including GNF/BioGPS 20, 21, BrainSpan Atlas 22, Gene Ontology (GO) 23, KEGG pathways 24, GAD phenotypes 25, ENCODE targets of transcription factors 26, and protein-protein interactions (BioGRID) 27, which can be used for functional annotation of genes. Gene set enrichment analysis is used to compare a gene expression matrix file with a gene set file. It evaluates if genes that are upregulated or downregulated in each tissue or cell type are enriched in specific gene sets. Another option is to test if the intersection of gene sets has more genes than expected by pure chance, as predicted by the hypergeometric probability distribution. Output files generated by correlation, gene set enrichment, or gene set overlap analyses can then be explored by making color-coded tables or histograms for individual rows or columns.
3. Extraction of Data from GEO Database
ExAtlas can be used to search for specific keywords (e.g., “kidney” or “heart”), authors, or GEO accession numbers, and the program returns information on series and samples where these terms are found. Samples can then be selected from one study (GEO series) or multiple studies and sample names can be edited for clarity and for specifying replications. In particular, ExAtlas requires precisely identical names for samples replicated within the same data series; otherwise, they will not be treated as replications by ExAtlas. Data files are then downloaded from GEO automatically via FTP, and information for selected samples is extracted and combined into a new file. If all selected samples belong to the same array platform, then gene expression data are stored for each probe of the array (except for control probes). But if samples belong to different array platforms, then the best probe (i.e., with highest expression signal) is selected for each gene, and gene expression data from different platforms are combined using official gene symbols. In ExAtlas, annotations of microarry platforms are automatically downloaded from GEO and gene symbols are verified using the RefSeq (NCBI) database. Extracted data are normalized using the quantile method 28. The quality of individual samples is evaluated by two criteria: (a) reproducibility of replications (i.e., deviation from the median) and (b) correlation of expression level with other data for a set of pre-selected housekeeping genes. Samples with low quality can be removed from the data set.
Gene-specific batch normalization can be used to combine two or more data sets. For example, if two data sets include the same tissue or organ (e.g., liver or cerebral cortex), then median expression levels for this common tissue/organ are equalized in two data sets. In our hands, this method usually works better than percentile normalization.
RNA-seq data cannot be retrieved automatically from the GEO database, because there is no standard format for storing processed gene expression data. Some data series include Cufflinks29 output files as a supplement, and these files that store FPKMb values for each gene can be uploaded to ExAtlas using option “Compile expression profile”. However, processing of Cufflinks files is not fully automatic but requires selection of columns with FPKM values and gene identifiers (gene symbols or GenBank/RefSeq accession numbers). In contrast to microarrays, where a high expression signal is always associated with higher statistical significance, in RNA-seq data, high FPKM values are not always associated with high statistical significance. For example, very short genes with only a few sequence reads, and thus with low statistical significance, could be erroneously represented by a high FPKM value, which is normalized to transcript length. Thus, to take full advantage of using error models in ANOVA (section 4), ExAtlas normalizes RNA-seq data by the total number of fragments sequenced but not by the length of transcripts. Because the length of transcripts is known, the FPKM values can be derived later from mean values generated by the ANOVA.
4. Standard Statistics: ANOVA, Significance of Expression Change, and PCA
ExAtlas uses the same algorithm for statistical analysis as NIA Array Analysis 7. Gene expression values are log-transformed and used for ANOVA, which is modified for the multiple hypotheses testing case. Because of the large number of tested probes/genes the error variance for some genes may appear very low by pure chance. The error model attempts to get a better estimate for the true error variance than the error variance estimated from data (we call it ‘empirical error variance’). By default, ExAtlas uses the maximum of empirical error variance and expected error variance averaged for 500 genes with similar expression level. Genes are first sorted according to their average log-expression, and then the error variance is averaged in a sliding window of 500 genes. Because the variation in low-expressed genes could be underestimated due to cutoffs applied during data processing, we force the average error variance in the sliding window to increase monotonically with decreasing average log-expression of genes below the median. Other error models are also available in ExAtlas, which include empirical error variance, error variance averaged in a sliding window of 500 genes, Bayesian correction of error variance 29, and the maximum between empirical error variance and its Bayesian correction. In addition, the False Discovery Rate (FDR) 30 is used to assess the significance of gene expression change instead of p-values.
ExAtlas can run modified ANOVA even for data sets with no replications (this option was not available in the NIA Array Analysis). In this case, the error variance is estimated based on the half-normal probability plot method 31. We assume that at least half of gene expression values (log-transformed) represent random deviations from the average. Then the standard deviation, σ, of random effects can be estimated as a median value of absolute deviations (i.e., di,j = |xi,j − Mi|, where xi,j is log-expression of gene i in sample j, and Mi is median log-expression of gene i) divided by 0.675, which is the median of inverse cumulative standard half-normal distribution. The error variance in ANOVA is then set to σ2. Because standard deviation usually depends on expression levels of genes, this method is applied to a sliding window of 500 genes sorted by their average log-expression value.
Lists of genes with significant change of expression relative to the median value or user-defined control sample (based on FDR and fold-change thresholds) can be saved as a new gene set file for future comparison with other data sets. There is also an option available to use gene specificity as an additional quantitative criterion of significance. Gene specificity is measured by z-value that compares log-transformed expression in a given tissue with average expression in other tissues that are not correlated with this tissue. Tissues are considered correlated if the multi-dimensional distance of their log-transformed expression profiles is <1/3 of the maximum distance between tissues. Low specificity corresponds to z-values >3, and high specificity corresponds to z-values >5.
5. Meta-Analysis
The goal of standard meta-analysis is to integrate information from multiple independent studies. It can increase statistical power and reduce false-positive effects. ExAtlas implements four most popular methods: Fisher’s, z-score, fixed effects, and random effects. The first three methods are relevant only if combined studies implement exactly the same methodology (e.g., same cell lines, same reagents, and same equipment). In practice, the methodologies often differ between studies, and thus the random effect method appears most relevant. Fisher’s method combines log-transformed p-values from m studies and generates a chi-square statistics with 2m degrees of freedom32. The z-score method combines z-scores (i.e., the ratio of mean effect to the S.D. of effect) of different studies with weights equal to square root of sample size33. Here the term “effect” means logratio of gene expression change/difference compared to control or study-wide mean or median. The fixed effects method estimates a weighted sum of effects (i.e., logratio of gene expression change), where weights are inverse to the variance. The random effects method takes into account the variance of heterogeneity between studies, T2, which is added to the variance of individual effects34.
Meta-analysis in ExAtlas starts by selecting gene expression data to be combined. The button to start meta-analysis can be found after opening a file with gene expression profiles, in section 2 (Pairwise comparison). Then a new screen appears where the user can add more data for meta-analysis. If all data sets use the same array platform, then the meta-analysis is done for each probe ID; alternatively, the meta-analysis is done for each gene symbol. It is possible to integrate data sets that belong to a different species; in this case, gene symbols are converted using HomoloGene35.
6. Gene Set Enrichment
Gene set enrichment analysis is used to evaluate if specific gene sets (such as GO or KEGG) are over-represented among upregulated and/or downregulated genes. The advantage of gene set enrichment analysis compared to a simple overlap of gene sets (which is also supported by ExAtlas) is that no predefined threshold is used for selecting differentially expressed genes. In particular, gene set enrichment analysis can find significant associations with functional gene sets even if there are no significantly upregulated genes based on standard criteria (e.g., FDR ≤ 0.05 and fold change ≥ 2). Among various existing methods for gene set enrichment analysis we use Parametric Analysis of Gene Enrichment (PAGE) 36 because of its simplicity and reliability 25. PAGE is based on the comparison of the average log-transformed expression change in a specific subset of genes, xset, with the average expression change in all genes, xall:
| (1) |
where nset is the size of the gene set and SDall is standard deviation of expression change among all genes. In ExAtlas, this method is modified by applying eq. (1) to the subset of N top upregulated and another subset of N top downregulated genes rather than to all genes (here we use N = 25% of all genes). This modification is designed to detect enrichment of each gene set separately among upregulated and downregulated genes. Upregulation or downregulation is estimated relative to the median expression of each gene or to a user-specified baseline (e.g., control samples).
7. Correlation between Gene Expression Profiles
Correlation between gene expression profiles may be indicative of functional changes in cells or tissues, and thus it can be used as a tool for functional annotation in global transcriptome analyses. For example, it was used to identify directions of mouse ESC differentiation two days after the induction of each of 137 tested transcription factors37. Positive correlation of gene expression profiles of manipulated cells with those in specific organs (e.g., brain, muscles, intestine, liver), indicates that the induction of a speccific transcription factor facilitates cell differentiation into specific cell types. For example, the transcriptome of ES cells shifted toward neural lineages after the induction of Ascl1, Sox9, and Foxg1; toward endoderm after the induction of Hnf4a, Gata2, and Gata3, or Esx1; toward skeletal muscle and heart after the induction of Myod1 or Mef2c, and toward hematopoietic cell lineages after the induction of Sfpi1, Elf1, or Irf2 37. Association of gene expression changes with tissue-specific expression profiles can be quantified by other methods, such as gene set enrichment analysis; however, we expect that correlation analysis provides a more balanced result, because it accounts for both upregulated and downregulated genes and does not use any arbitrarily-chosen criteria for selection of tissue-specific gene sets.
Because gene expression profiles are often identified using different array platforms or sequencing technologies, it makes sense to limit comparison to those genes that show significant changes of expression in both data sets assessed in correlation analysis. For example, if a certain gene is properly measured in one data set, but shows no signal or only noise in another data set, then its use in correlation analysis would be misleading. ExAtlas estimates correlation as follows: (a) the best probe is selected for each gene (with the highest F-statistics, ANOVA); (b) genes with significant change of expression (based on FDR and fold-change thresholds) are identified in each data set; (c) gene expression change is estimated as the difference in log-transformed expression values relative to median expression or specific user-defined baseline sample; (d) Pearson or Spearman correlation between gene expression changes in two data sets is then estimated for the subset of common significant genes. ExAtlas supports correlation analysis between data sets from different species; in this case gene symbols are converted using HomoloGene.
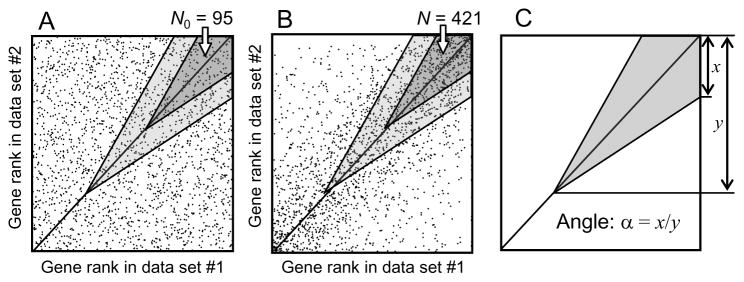
In addition to estimating correlation and its statistical significance, ExAtlas provides an option to identify coregulated genes. If a corresponding box is checked in the page for starting correlation analysis, then ExAtlas will identify lists of genes that are both upregulated or both downregulated in two data files. Coregulated genes are detected only if correlation is positive and significant (z ≥ 2), and the Expected Proportion of False Positives (EPFP) is above a specified threshold. EPFP is similar to FDR, but estimated differently; it shows the proportion of false positives (i.e., genes associated by chance) in the list of coregulated genes38. The algorithm for finding positively coregulated genes is based on the analysis of data points in the positive quadrant (i.e. x>0 and y>0). Negatively coregulated genes are identified in the same way in the negative quadrant. First, gene expression changes are all replaced by their rank. If the null hypothesis holds (no correlation), then genes are expected to have a uniform random distribution in the positive quadrant (Fig. 2A). Coregulated genes can be found close to the diagonal in the wedge-shaped shaded area (Fig. 2B) and their numbers are compared with the expected number of genes in the same area under the null hypothesis (Fig. 2A). The inverse value of gene enrichment in the wedge area is equal to EPFP. As the wedge area slides along the diagonal from the upper right corner towards the zero corner (and possibly below it), the EPFP increases and at some point reaches the threshold value. All genes in the wedge area are then considered coregulated. Because EPFP may appear to vary from monotonic increase, it is constrained to be monotonic by using the following correction: if a bigger wedge area #2 includes wedge area #1 and EPFP(2) < EPFP(1), then EPFP(1) is set to EPFP(2). The shape of the wedge is controlled by one parameter, called “angle”, which is estimated as a ratio of the short side of the wedge to its height/width (Fig. 2C).
Fig. 2.
Estimating Expected Proportion of False Positives (EPFP) for positively coregulated genes: (A) no coregulation (null hypothesis); (B) coregulated genes are counted in a shaded wedge area; (C) using angle to define the shape of the wedge area. For the dark-shaded wedge area, EPFP = N0/N = 0.225, where N0 and N are the numbers of data points in the area on conditions (A) and (B), respectively. The wedge shifts along the diagonal (i.e., down and left, to light-shaded area) till EPFP value reaches the threshold (e.g., EPFP = 1/3). Then all genes in the shaded area are considered coregulated.
8. Conclusions
ExAtlas software offers a set of computational and file-management tools for meta-analysis of gene expression data stored in the GEO database as well as a user’s own data. It does not require any programming or database management skills, and therefore it can be used by biologists with little or no expertise in bioinformatics. All programs are open source and their codes are available at the website (http://lgsun.grc.nia.nih.gov/exatlas), which allows establishment of mirror sites as well as modifications and improvements.
Acknowledgments
This research was supported entirely by the Intramural Research Program of the National Institutes of Health, National Institute on Aging.
Footnotes
FPKM equals the number of DNA fragments that match to mRNA per 1 million total fragments sequenced and per 1000 bp of transcript length.
Contributor Information
ALEXEI A. SHAROV, Laboratory of Genetics, National Institute on Aging, National Institutes of Health, Baltimore, MD 21224, USA
DAVID SCHLESSINGER, Laboratory of Genetics, National Institute on Aging, National Institutes of Health, Baltimore, MD 21224, USA.
MINORU S.H. KO, Department of Systems Medicine, The Sakaguchi Laboratory, Keio University School of Medicine, Tokyo 160-8582, Japan.
References
- 1.Tseng GC, Ghosh D, Feingold E. Comprehensive literature review and statistical considerations for microarray meta-analysis. Nucleic Acids Res. 2012;40(9):3785–99. doi: 10.1093/nar/gkr1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grant GR, Manduchi E, Stoeckert CJ, et al. Analysis and management of microarray gene expression data. Curr Protoc Mol Biol. 2007;Chapter 19(Unit 19.6) doi: 10.1002/0471142727.mb1906s77. [DOI] [PubMed] [Google Scholar]
- 3.Grewal A, Lambert P, Stockton J. Analysis of expression data: an overview. Curr Protoc Bioinformatics. 2007;Chapter 7(Unit 7.1) doi: 10.1002/0471250953.bi0701s17. [DOI] [PubMed] [Google Scholar]
- 4.Mehta JP, Rani S. Software and tools for microarray data analysis. Methods Mol Biol. 2011;784:41–53. doi: 10.1007/978-1-61779-289-2_4. [DOI] [PubMed] [Google Scholar]
- 5.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34(2):374–8. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 6.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98(9):5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharov AA, Dudekula DB, Ko MS. A web-based tool for principal component and significance analysis of microarray data. Bioinformatics. 2005;21(10):2548–9. doi: 10.1093/bioinformatics/bti343. [DOI] [PubMed] [Google Scholar]
- 8.Dahlquist KD, Salomonis N, Vranizan K, Lawlor SC, Conklin BR. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat Genet. 2002;31(1):19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- 9.Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4(1):R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khatri P, Draghici S. Ontological analysis of gene expression data: current tools, limitations, and open problems. Bioinformatics. 2005;21(18):3587–95. doi: 10.1093/bioinformatics/bti565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khatri P, Voichita C, Kattan K, Ansari N, Khatri A, Georgescu C, Tarca AL, Draghici S. Onto-Tools: new additions and improvements in 2006. Nucleic Acids Res. 2007;35(Web Server issue):W206–11. doi: 10.1093/nar/gkm327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gentleman R, Ruschhaupt M, Huber W, Lusa L. Meta-analysis for microarray experiments. http://www.bioconductor.org/packages/release/bioc/vignettes/GeneMeta/inst/doc/GeneMeta.pdf.
- 15.Choi H, Shen R, Chinnaiyan AM, Ghosh D. A latent variable approach for meta-analysis of gene expression data from multiple microarray experiments. BMC Bioinformatics. 2007;8:364. doi: 10.1186/1471-2105-8-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Z, Fenstermacher D. An Introduction to MAMA (Meta-Analysis of MicroArray data) System. Conf Proc IEEE Eng Med Biol Soc. 2005;7:7730–3. doi: 10.1109/IEMBS.2005.1616304. [DOI] [PubMed] [Google Scholar]
- 17.Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Muertter RN, Holko M, Ayanbule O, Yefanov A, Soboleva A. NCBI GEO: archive for functional genomics data sets--10 years on. Nucleic Acids Res. 2011;39(Database issue):D1005–10. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolesnikov N, Hastings E, Keays M, Melnichuk O, Tang YA, Williams E, Dylag M, Kurbatova N, Brandizi M, Burdett T, Megy K, Pilicheva E, Rustici G, Tikhonov A, Parkinson H, Petryszak R, Sarkans U, Brazma A. ArrayExpress update-simplifying data submissions. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivliev AE, t Hoen PA, Villerius MP, den Dunnen JT, Brandt BW. Microarray retriever: a web-based tool for searching and large scale retrieval of public microarray data. Nucleic Acids Res. 2008;36(Web Server issue):W327–31. doi: 10.1093/nar/gkn213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, Patapoutian A, Hampton GM, Schultz PG, Hogenesch JB. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci U S A. 2002;99(7):4465–70. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, 3rd, Su AI. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10(11):R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, Riley ZL, Royall JJ, Aiona K, Arnold JM, Bennet C, Bertagnolli D, Brouner K, Butler S, Caldejon S, Carey A, Cuhaciyan C, Dalley RA, Dee N, Dolbeare TA, Facer BA, Feng D, Fliss TP, Gee G, Goldy J, Gourley L, Gregor BW, Gu G, Howard RE, Jochim JM, Kuan CL, Lau C, Lee CK, Lee F, Lemon TA, Lesnar P, McMurray B, Mastan N, Mosqueda N, Naluai-Cecchini T, Ngo NK, Nyhus J, Oldre A, Olson E, Parente J, Parker PD, Parry SE, Stevens A, Pletikos M, Reding M, Roll K, Sandman D, Sarreal M, Shapouri S, Shapovalova NV, Shen EH, Sjoquist N, Slaughterbeck CR, Smith M, Sodt AJ, Williams D, Zollei L, Fischl B, Gerstein MB, Geschwind DH, Glass IA, Hawrylycz MJ, Hevner RF, Huang H, Jones AR, Knowles JA, Levitt P, Phillips JW, Sestan N, Wohnoutka P, Dang C, Bernard A, Hohmann JG, Lein ES. Transcriptional landscape of the prenatal human brain. Nature. 2014;508(7495):199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dimmer EC, Huntley RP, Alam-Faruque Y, Sawford T, O’Donovan C, Martin MJ, Bely B, Browne P, Mun Chan W, Eberhardt R, Gardner M, Laiho K, Legge D, Magrane M, Pichler K, Poggioli D, Sehra H, Auchincloss A, Axelsen K, Blatter MC, Boutet E, Braconi-Quintaje S, Breuza L, Bridge A, Coudert E, Estreicher A, Famiglietti L, Ferro-Rojas S, Feuermann M, Gos A, Gruaz-Gumowski N, Hinz U, Hulo C, James J, Jimenez S, Jungo F, Keller G, Lemercier P, Lieberherr D, Masson P, Moinat M, Pedruzzi I, Poux S, Rivoire C, Roechert B, Schneider M, Stutz A, Sundaram S, Tognolli M, Bougueleret L, Argoud-Puy G, Cusin I, Duek-Roggli P, Xenarios I, Apweiler R. The UniProt-GO Annotation database in 2011. Nucleic Acids Res. 2011;40(Database issue):D565–70. doi: 10.1093/nar/gkr1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2013;42(Database issue):D199–205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, De S, Garner JR, Smith K, Wang SA, Becker KG. Systematic analysis, comparison, and integration of disease based human genetic association data and mouse genetic phenotypic information. BMC Med Genomics. 2010;3:1. doi: 10.1186/1755-8794-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, Min R, Alves P, Abyzov A, Addleman N, Bhardwaj N, Boyle AP, Cayting P, Charos A, Chen DZ, Cheng Y, Clarke D, Eastman C, Euskirchen G, Frietze S, Fu Y, Gertz J, Grubert F, Harmanci A, Jain P, Kasowski M, Lacroute P, Leng J, Lian J, Monahan H, O’Geen H, Ouyang Z, Partridge EC, Patacsil D, Pauli F, Raha D, Ramirez L, Reddy TE, Reed B, Shi M, Slifer T, Wang J, Wu L, Yang X, Yip KY, Zilberman-Schapira G, Batzoglou S, Sidow A, Farnham PJ, Myers RM, Weissman SM, Snyder M. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489(7414):91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chatr-Aryamontri A, Breitkreutz BJ, Oughtred R, Boucher L, Heinicke S, Chen D, Stark C, Breitkreutz A, Kolas N, O’Donnell L, Reguly T, Nixon J, Ramage L, Winter A, Sellam A, Chang C, Hirschman J, Theesfeld C, Rust J, Livstone MS, Dolinski K, Tyers M. The BioGRID interaction database: 2015 update. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 29.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–78. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. Journal of Royal Statistical Society, B. 1995;57:289–300. [Google Scholar]
- 31.Dyson G, Wu J. Use of the half-normal probability plot to identify significant effects for microarray data. Joint Statistical Meeting (JSM); 2002; New York, NY. 2002. [Google Scholar]
- 32.Fisher RA. Statistical Methods for Research Workers. Edinburgh, UK: Oliver and Boyd; 1925. [Google Scholar]
- 33.Mosteller F, Bush RR. Selected quantitative techniques. In: Lindzey G, editor. Handbook of Social Psychology. Vol. 1. Cambridge, MA: Addison Wesley; 1954. pp. 289–334. [Google Scholar]
- 34.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–88. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 35.Wheeler DL, Church DM, Lash AE, Leipe DD, Madden TL, Pontius JU, Schuler GD, Schriml LM, Tatusova TA, Wagner L, Rapp BA. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2001;29(1):11–6. doi: 10.1093/nar/29.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim SY, Volsky DJ. PAGE: parametric analysis of gene set enrichment. BMC Bioinformatics. 2005;6:144. doi: 10.1186/1471-2105-6-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Correa-Cerro LS, Piao Y, Sharov AA, Nishiyama A, Cadet JS, Yu H, Sharova LV, Xin L, Hoang HG, Thomas M, Qian Y, Dudekula DB, Meyers E, Binder BY, Mowrer G, Bassey U, Longo DL, Schlessinger D, Ko MSH. Generation of mouse ES cell lines engineered for the forced induction of transcription factors. Scientific Reports. 2011;1:176. doi: 10.1038/srep00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharov AA, Nishiyama A, Qian Y, Dudekula DB, Longo DL, Schlessinger D, Ko MS. Chromatin properties of regulatory DNA probed by manipulation of transcription factors. J Comput Biol. 2014;21(8):569–77. doi: 10.1089/cmb.2013.0126. [DOI] [PMC free article] [PubMed] [Google Scholar]