Abstract
Background:
Statin use after colorectal cancer diagnosis may improve survival but evidence from observational studies is conflicting. The anti-cancer effect of statins may be restricted to certain molecular subgroups. In this population-based cohort study, the interaction between p53 and 3-hydroxy-3-methylglutaryl coenzyme-A reductase (HMGCR) expression, KRAS mutations, and the association between statin use and colon cancer survival was assessed.
Methods:
The cohort consisted of 740 stage II and III colon cancer patients diagnosed between 2004 and 2008. Statin use was determined through clinical note review. Tissue blocks were retrieved to determine immunohistochemical expression of p53 and HMGCR in tissue microarrays and the presence of KRAS mutations in extracted DNA. Cox proportional hazards models were used to calculate hazard ratios (HRs) and 95% confidence intervals (CIs) for colorectal cancer-specific and overall survival.
Results:
Statin use was not associated with improved cancer-specific survival in this cohort (HR=0.91, 95% CI 0.64–1.28). Statin use was also not associated with improved survival when the analyses were stratified by tumour p53 (wild-type HR=1.31, 95% CI 0.67–2.56 vs aberrant HR=0.80, 95% CI 0.52–1.24), HMGCR (HMGCR-high HR=0.69, 95% CI 0.40–1.18 vs HMGCR-low HR=1.10, 95% CI 0.66–1.84), and KRAS (wild-type HR=0.73, 95% CI 0.44–1.19 vs mutant HR=1.21, 95% CI 0.70–2.21) status.
Conclusions:
Statin use was not associated with improved survival either independently or when stratified by potential mevalonate pathway biomarkers in this population-based cohort of colon cancer patients.
Keywords: hydroxymethylglutaryl-CoA reductase inhibitors, tumour suppressor protein p53, HMGCR protein, KRAS, colonic neoplasms, survival
Statins are commonly prescribed lipid lowering medications that inhibit the enzyme 3-hydroxy-3-methylglutaryl coenzyme-A reductase (HMGCR) (Ng et al, 2011). In addition to their cholesterol-lowering action, they may have pleiotropic anti-cancer effects through inhibition of the mevalonate pathway (Bardou et al, 2010; Thurnher et al, 2012). However, observational data assessing the association between post-diagnostic statin use and colorectal cancer survival lacks consistency (Ng et al, 2011; Mace et al, 2013; Cardwell et al, 2014a; Krens et al, 2014; Hoffmeister et al, 2015; Kim et al, 2015; Lim et al, 2015; Zanders et al, 2015). As a robust association has yet to be identified, caution is required in proceeding with clinical trials assessing the role of statins as novel adjuvant agents. In addition, colorectal tumours are known to display significant molecular heterogeneity (Ogino et al, 2012). A molecular pathological epidemiology approach is therefore required to determine whether the potential anti-cancer effect of statins is confined to specific molecular subgroups (Ogino et al, 2011). Candidate mevalonate pathway biomarkers that may differentiate tumours more likely to respond to statin therapy include HMGCR, p53, and KRAS.
The seminal work by Freed-Pastor et al (2012) implicates the mevalonate pathway as a potential therapeutic target for tumours bearing mutations in TP53. They demonstrated that statins were able to reverse the malignant phenotype of p53 mutant but not p53 wild-type breast cancer cells in vitro. Similarly, an in vivo breast cancer study demonstrated that the anti-proliferative effect of statins was limited to tumours that overexpressed HMGCR (Bjarnadottir et al, 2013). Finally, RAS signalling may be inhibited by statin-induced depletion of downstream isoprenoids required for posttranslational prenylation of small GTPases such as ras and rho (Bardou et al, 2010; Ng et al, 2011; Thurnher et al, 2012). Prenylation of k-ras makes the protein lipophilic and ensures translocation to the cell membrane where it can exert its proliferative effects (Konstantinopoulos et al, 2007; Krens et al, 2014). Based on this hypothesis, the effect of statins on colorectal cancer survival may differ according to KRAS gene mutation status (Ng et al, 2011). The aim of this study was therefore to assess the interaction between statin use, the potential mevalonate pathway biomarkers p53, HMGCR and KRAS, and survival in a population-based cohort study of patients with stage II and III colon cancer.
Materials and methods
Study cohort
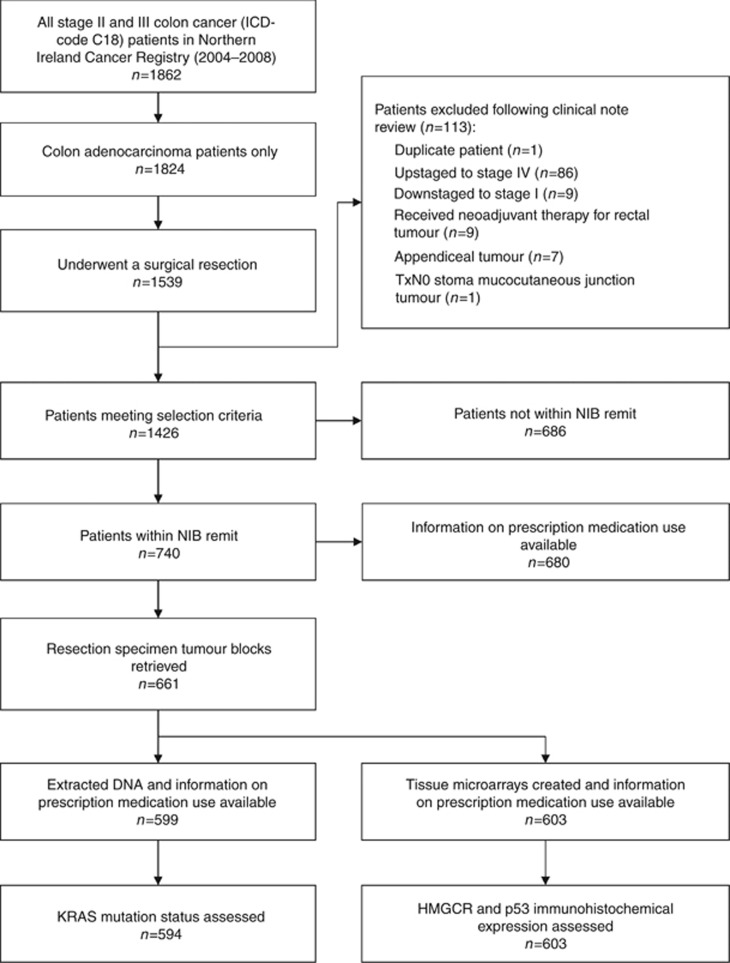
The Northern Ireland Cancer Registry was used to identify 1426 stage II and III colon cancer patients undergoing surgical resection between 2004 and 2008 (Figure 1). Rectal cancers were excluded as neoadjuvant radiotherapy could potentially alter tumour expression profiles. Ethical approval through the Northern Ireland Biobank (NIB reference number 13–0087) permitted retrieval of formalin-fixed, paraffin-embedded (FFPE) tissue blocks for patients within two of the five regional Health and Social Care trusts. For this molecular pathological epidemiology study, the final cohort was subsequently restricted to only include patients within the biobank remit (n=740, 51.9%). These patients were representative of the overall Northern Ireland cohort with respect to age, sex, stage, and adjuvant chemotherapy receipt (Supplementary Table 1).
Figure 1.
Selection of stage II and III colon cancer (adenocarcinoma) patients and samples. ICD=International Classification of Disease; NIB=Northern Ireland Biobank.
Clinicopathological variables and follow-up
The Clinical Oncology Information System (COIS), a prospective electronic record of cancer patient management, was used to collect clinical variables including adjuvant chemotherapy use, prescription medication use, family history of colorectal cancer and Eastern Cooperative Oncology Group (ECOG) performance status. This process was supplemented by a manual chart review when insufficient information was recorded on COIS or no record was present. Pathological variables were retrieved from full pathology reports. Occurrence and cause of death were assessed via data linkage to the Northern Ireland Registrar General’s Office (follow-up censored 31 December 2013). Colorectal cancer-specific deaths were defined as those with an underlying cause of death International Classification of Disease code C18, C19, C20 (anus), and/or C26 (other and ill-defined digestive organs).
Drug exposure assessment
Statin exposure (user vs non-user) based on current prescription medications was assessed at a single perioperative time point for all patients. When medication information was available on COIS, this time point was the initial post-operative oncology review. When medication information was not available on COIS, statin exposure was determined from the post-operative hospital discharge letter. Information on medication dosage was not consistently recorded on COIS and therefore not considered. Information on aspirin exposure was also assessed using these methods. Our research group has previously demonstrated that 98.5% of aspirin prescriptions after colorectal cancer diagnosis in the United Kingdom are for low-dose (75 mg) aspirin (Cardwell et al, 2014b). Aspirin exposure in this study is therefore considered representative of low-dose aspirin.
Tumour molecular analysis
FFPE blocks were retrieved for 89.3% of the cohort (661 of 740). Three 1.0 mm diameter tissue cores were extracted from representative areas within donor blocks and inserted into recipient blocks using a manual tissue arrayer (Estigen, Tartu, and Estonia) as described previously (Zhang et al, 2003; Boyle et al, 2014). The immunohistochemistry methods for p53 (DO-7 antibody clone to p53, Dako UK Ltd, Ely, UK—catalogue number M7001) and HMGCR (Atlas Antibodies AB, Stockholm, Sweden—catalogue number HPA008338) staining are described in the Supplementary Methods. QuPath (Queen’s University Belfast, Northern Ireland) image analysis software facilitated digital immunoscoring (Supplementary Methods). An H-score was calculated based on the extent and intensity of cytoplasmic or nuclear staining where appropriate (H-score=3 × % of strongly staining cytoplasm+2 × % of moderately staining cytoplasm+1 × % of weakly staining cytoplasm, giving a range of 0–300) (McCarty et al, 1986).
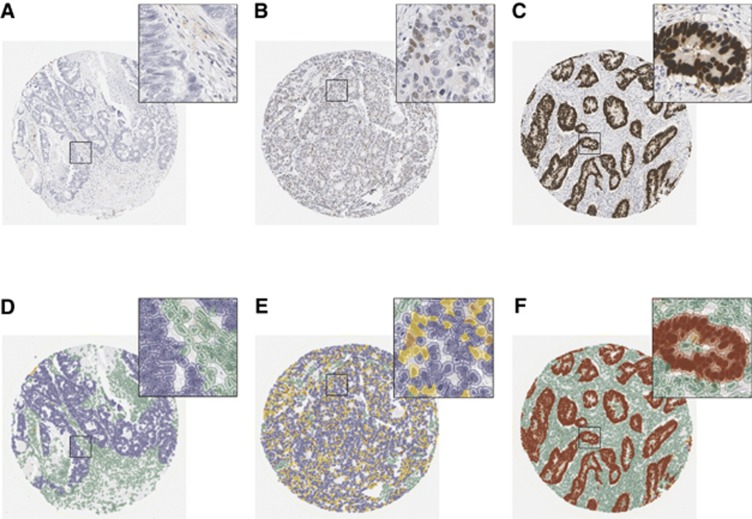
A three-tier scoring system was applied to differentiate normal (non-extreme) from aberrant (extreme positive or extreme negative) patterns of p53 expression (Figure 2). Cores were designated as extreme negative if there was confluent negative staining within the represented population of tumour nuclei. Diffuse strong positivity was considered representative of extreme positive expression and intermediate heterogeneous expression was considered a non-extreme (normal) pattern of staining (Boyle et al, 2014). Selection and validation of the p53 cut points are described in the Supplementary Methods. 3-Hydroxy-3-methylglutaryl coenzyme-A reductase H-scores were categorised into tertiles for prognostic analyses and dichotomised around the median value for survival analyses that tested the interaction between statin use and HMGCR expression. Representative images of HMGCR expression are shown in Supplementary Figure 1. Based on these methods, the final p53 and HMGCR categories were determined using the median of the three available H-scores for each case.
Figure 2.
p53 immunohistochemistry in colon cancer tissue microarrays and associated markup for digital immunoscoring using QuPath image analysis software. Detected cells are colour-coded according to their classification: green=non-tumour; blue=negatively staining tumour; yellow=weakly staining tumour; orange=moderately staining tumour; red=strongly staining tumour. (A) Original core from a tumour demonstrating aberrant negative p53 immunostaining. (B) Original core from a tumour deomonstrating non-extreme (normal) p53 immunostaining. (C) Original core from a tumour demonstrating aberrant positive p53 immunostaining. (D) QuPath cellular markup in the aberrant negative p53 core. (E) QuPath cellular markup in the non-extreme (normal) p53 core. (F) QuPath cellular markup in the aberrant positive p53 core.
Detailed methods for DNA extraction, KRAS mutation analysis, and microsatellite instability (MSI) status using commercially available kits are provided as Supplementary Methods. Briefly KRAS mutation status was assessed using previously described methods for the ColoCarta Panel (Agena Bioscience, Hamburg, Germany; Fumagalli et al, 2010), whereas MSI status was determined using five mononucleotide repeat markers (BAT-25, BAT-26, NR-21, NR-24, and MONO-27).
Statistical analysis
All statistical analysis was performed using Stata 13 (StataCorp, College Station, TX, USA). The χ2-test was used to compare characteristics between statin users and non-users. The primary outcome of this study was colorectal cancer-specific survival and the secondary outcome was overall survival. The association between statin use and survival was assessed in the whole cohort and then in analyses stratified by biomarker status. Only cases with information on statin exposure (known user vs known non-user) were included in the former analysis. Only cases with available exposure information and tissue for biomarker assessment were included in the subsequent stratified analyses. Other missing categorical data were coded as unknown.
Survival analysis was performed using the Cox proportional hazards model to calculate hazard ratios (HRs) and associated 95% confidence intervals (CIs). The multivariable models for colorectal cancer-specific survival adjusted for age, gender, year of diagnosis, grade, MSI status, ECOG performance status, family history of colorectal cancer, adjuvant chemotherapy use, stage, and aspirin use. In addition to the these variables, the overall survival multivariable model also adjusted for Charlson Comorbidity Index score as a continuous variable (Khan et al, 2010). Analyses were stratified by biomarker status. Interaction terms for statin use and p53, HMGCR, or KRAS were then included in the Cox model and Wald’s test was used to assess for statistical interaction. Sensitivity analysis was performed using complete-case data (cases with missing data were excluded). All P-values were two-sided and a value <0.05 was considered statistically significant.
Results
Patients
Information on prescription medication use was available for 91.9% (n=680) of patients in this population-based cohort study. Overall, 25.3% (n=172) used statins. Compared with those with available information on medication use, patients with no information on medication use were older and more likely to be diagnosed in the earlier years of the study. However, there was no difference in stage, grade of tumour differentiation, or MSI status (Supplementary Table 2). After a mean follow-up of 5.7 years (range 0–10), there were 299 all-cause and 212 colorectal cancer-specific deaths among these patients.
Statin use and survival
Table 1 summarises the baseline characteristics between statin users and non-users. Statin users were more likely to be older, male, and diagnosed later in the cohort compared with statin non-users. Statin users were also more likely to concomitantly use aspirin. There were a smaller proportion of MSI-high tumours among statin users but the proportion of patients with unknown MSI status was higher in statin non-users. However, there was no difference in tumour differentiation grade, stage, or the proportion of patients receiving adjuvant chemotherapy between users and non-users. There was also no difference in the proportion of right-sided tumours in statin users compared with non-users (58.1 vs 56.1%).
Table 1. Clinical and demographic characteristics according to statin use.
| Characteristic | Statin non-user (n=508) number (%) | Statin user (n=172) number (%) | P-value |
|---|---|---|---|
|
Age category | |||
| <50 | 44 (8.7) | 3 (1.7) | 0.001 |
| 50–<60 | 61 (12.0) | 10 (5.8) | |
| 60–<70 | 134 (26.4) | 63 (36.6) | |
| 70–<80 | 173 (34.1) | 66 (38.4) | |
| >80 | 96 (18.9) | 30 (17.4) | |
|
Gender | |||
| Male | 265 (52.2) | 105 (61.1) | 0.04 |
| Female | 243 (47.8) | 67 (39.0) | |
|
Year of diagnosis | |||
| 2004 | 77 (15.2) | 24 (14.0) | 0.003 |
| 2005 | 116 (22.8) | 18 (10.5) | |
| 2006 | 98 (19.3) | 33 (19.2) | |
| 2007 | 104 (20.5) | 42 (24.4) | |
| 2008 | 113 (22.2) | 55 (32.0) | |
|
Stage | |||
| II | 290 (57.1) | 99 (57.6) | 0.91 |
| III | 218 (42.9) | 73 (42.4) | |
|
Grade | |||
| Well-moderate | 429 (84.5) | 148 (86.1) | 0.88 |
| Poor | 76 (15.0) | 23 (13.4) | |
| Unknown | 3 (0.6) | 1 (0.6) | |
|
Location | |||
| Right colon | 285 (56.1) | 100 (58.14) | 0.62 |
| Left colon | 218 (42.9) | 69 (40.1) | |
| Not specified | 5 (1.0) | 3 (1.7) | |
|
MSI status | |||
| Non MSI-high | 312 (61.4) | 125 (72.7) | 0.02 |
| MSI-high | 96 (18.9) | 27 (15.7) | |
| Unknown | 100 (19.7) | 20 (11.6) | |
|
Adjuvant chemotherapy | |||
| No | 332 (65.4) | 123 (71.5) | 0.14 |
| Yes | 176 (34.7) | 49 (28.5) | |
|
ECOG performance status | |||
| 0–1 | 285 (56.1) | 87 (50.6) | 0.10 |
| 2 | 30 (5.9) | 17 (9.9) | |
| 3–4 | 20 (3.9) | 12 (7.0) | |
| Unknown | 173 (34.1) | 56 (32.6) | |
|
Family history of colorectal cancer | |||
| Yes | 276 (54.3) | 91 (52.9) | 0.85 |
| No | 78 (15.4) | 25 (14.5) | |
| Unknown | 154 (30.3) | 56 (32.6) | |
|
Aspirin use | |||
| Non-user | 435 (85.6) | 99 (57.6) | <0.001 |
| User | 73 (14.4) | 73 (42.4) | |
Abbreviations: ECOG=Eastern Cooperative Oncology Group; MSI=microsatellite instability.
Statin use at the time of diagnosis was not associated with a significant reduction in colorectal cancer-specific (adjusted HR=0.91, 95% CI 0.64–1.28) or overall mortality (adjusted HR=0.83, 95% CI 0.61–1.12) compared with non-use (Table 2).
Table 2. Colorectal cancer-specific and overall survival according to statin use, p53 and HMGCR immunohistochemistry, and KRAS mutation status.
|
Colorectal cancer-specific survival |
Overall survival |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Characteristic | No. of CRC deaths/at risk | Unadjusted HR (95% CI) | P-value | Adjusted HR (95% CI)a | P-value | No. of deaths/at risk | Unadjusted HR (95% CI) | P-value | Adjusted HR (95% CI)b | P-value |
|
All participants | ||||||||||
| Statin non-user | 163/508 | 1 (Reference) | 1 (Reference) | 227/508 | 1 (Reference) | 1 (Reference) | ||||
| Statin user | 49/172 | 0.90 (0.65–1.24) | 0.51 | 0.91 (0.64–1.28) | 0.58 | 72/172 | 0.96 (0.74–1.25) | 0.76 | 0.83 (0.61–1.12) | 0.22 |
|
Patterns of p53 immunostaining | ||||||||||
| Normal | 59/242 | 1 (Reference) | 1 (Reference) | 97/242 | 1 (Reference) | 1 (Reference) | ||||
| Aberrant | 128/361 | 1.53 (1.13–2.09) | 0.007 | 1.38 (0.97–1.95) | 0.07 | 173/361 | 1.27 (0.99–1.62) | 0.06 | 1.13 (0.85–1.49) | 0.41 |
|
HMGCR tertiles | ||||||||||
| 1 (lowest) | 72/197 | 1 (Reference) | 1 (Reference) | 102/197 | 1 (Reference) | 1 (Reference) | ||||
| 2 | 61/200 | 0.84 (0.60–1.19) | 0.33 | 0.84 (0.58–1.21) | 0.35 | 91/200 | 0.91 (0.69–1.21) | 0.53 | 0.89 (0.66–1.21) | 0.47 |
| 3 (highest | 55/206 | 0.73 (0.51–1.04) | 0.08 | 0.76 (0.51–1.14) | 0.18 | 78/206 | 0.76 (0.56–1.02) | 0.07 | 0.77 (0.55–1.07) | 0.12 |
|
KRAS | ||||||||||
| Wild-type | 109/365 | 1 (Reference) | 1 (Reference) | 156/365 | 1 (Reference) | 1 (Reference) | ||||
| Mutated | 76/229 | 1.16 (0.86–1.55) | 0.33 | 1.12 (0.82–1.53) | 0.49 | 111/229 | 1.18 (0.92–1.51) | 0.18 | 1.14 (0.88–1.48) | 0.32 |
Abbreviations: CI=confidence interval; CRC=colorectal cancer; ECOG=Eastern Cooperative Oncology Group; HR=hazard ratio; MSI=microsatellite instability.
Multivariable model adjusted for age, gender, year of diagnosis, grade, MSI status, ECOG performance status, family history of colorectal cancer, adjuvant chemotherapy use, stage, and aspirin use.
Multivariable model adjusted for all variables in footnote a and also adjusted for Charlson Comorbidity Index score.
Immunohistochemical expression of p53
In total, n=361 (59.9%) tumours demonstrated an aberrant pattern of p53 immunostaining (extreme positive or extreme negative). The proportion of tumours with aberrant patterns of p53 immunostaining was similar between statin users and statin non-users (59.1 vs 62.0%, P=0.52). Compared with wild-type p53 expression, aberrant p53 immunostaining was associated with a 53% increase in hazard for unadjusted colorectal cancer-specific mortality (HR=1.53, 95% CI 1.13–2.09). However, this association was attenuated when potential confounding variables were included in the multivariable model (adjusted HR=1.38, 95% CI 0.97–1.95, Table 2).
In stratified analysis, there was no evidence that the association between statin use and colorectal cancer-specific survival differed by p53 immunostaining patterns (wild-type adjusted HR=1.31, 95% CI 0.67–2.56 vs aberrant adjusted HR=0.80, 95% CI 0.52–1.24). Similar results were observed for overall survival (Table 3).
Table 3. Statin use and colorectal cancer-specific / overall survival stratified by biomarker subgroups.
|
Colorectal cancer-specific survival |
Overall survival |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Statin non-user | Statin user | HR (95%CI)a user vs non-user | P-value | P for interaction | Statin non-user | Statin user | HR (95%CI)b user vs non-user | P-value | P for interaction | |
| No. of deaths/patients | No. of deaths/patients | |||||||||
|
Stratified by patterns of p53 immunostaining | ||||||||||
| Normal | 43/182 | 16/60 | 1.31 (0.67–2.56) | 0.43 | 0.51 | 75/182 | 22/60 | 0.67 (0.34–1.30) | 0.24 | 0.60 |
| Aberrant | 98/263 | 30/98 | 0.80 (0.52–1.24) | 0.33 | 127/263 | 46/98 | 0.91 (0.63–1.32) | 0.61 | ||
|
Stratified by HMGCR expression | ||||||||||
| H-score <median | 79/231 | 26/69 | 1.10 (0.66–1.84) | 0.71 | 0.08 | 118/231 | 34/69 | 0.78 (0.49–1.25) | 0.30 | 0.28 |
| H-score ⩾median | 63/214 | 20/89 | 0.69 (0.40–1.18) | 0.17 | 85/214 | 34/89 | 0.77 (0.49–1.22) | 0.26 | ||
|
Stratified by KRAS mutation status | ||||||||||
| Wild-type | 83/262 | 26/103 | 0.73 (0.44–1.19) | 0.20 | 0.05 | 116/262 | 40/103 | 0.68 (0.44–1.04) | 0.08 | 0.07 |
| Mutated | 57/176 | 19/53 | 1.21 (0.70–2.21) | 0.50 | 84/176 | 27/53 | 1.00 (0.61–1.66) | 0.98 | ||
Abbreviations: CI=confidence intervals; ECOG=Eastern Cooperative Oncology Group; HR=hazard ratio; MSI=microsatellite instability.
Multivariable model adjusted for age, gender, year of diagnosis, grade, MSI status, ECOG performance status, family history of colorectal cancer, adjuvant chemotherapy use, stage, and aspirin use.
Multivariable model adjusted for all variables in footnote a and also adjusted for Charlson Comorbidity Index score.
3-Hydroxy-3-methylglutaryl coenzyme-A reductase immunohistochemical expression
Statin users were more likely to have tumours in the highest HMGCR tertile compared with non-users (43.7 vs 30.8%, P=0.01). As shown in Table 2, however, there was no evidence that higher levels of HMGCR expression were associated with significantly better colorectal cancer-specific or overall survival (adjusted P for trend=0.18 and 0.12, respectively).
There was no evidence of significant associations with colorectal cancer-specific survival among statin users compared with non-users when the cohort was stratified by tumour HMGCR expression level, although the direction of HRs did differ (HMGCR-high adjusted HR=0.69, 95% CI 0.40–1.18 vs HMGCR-low adjusted HR=1.10, 95% CI 0.66–1.84). Similarly, there was no evidence of a differential benefit for overall survival in statin users compared with non-users when the cohort was stratified by HMGCR status (Table 3).
KRAS mutations
KRAS mutation status (mutant vs wild-type) was available for 99.2% of these samples with extracted DNA (594 of 599). Statin users had slightly less KRAS mutant tumours compared with statin non-users, although the difference was not statistically significant (34.0 vs 40.2%, P=0.17). Compared with wild-type KRAS, the presence of a KRAS mutation was not associated with significantly worse colorectal cancer-specific survival (adjusted HR=1.12, 95% CI 0.82–1.53, Table 2).
There was no evidence of an improvement in colorectal cancer-specific survival among statin users compared with non-users when the cohort was stratified by KRAS mutation status, although again the direction of effect differed between wild-type and mutant KRAS tumours (KRAS wild-type adjusted HR=0.73, 95% CI 0.44–1.19 vs KRAS mutant tumour adjusted HR=1.21, 95% CI 0.70–2.21). Similar nonsignificant results were observed for overall survival (Table 3).
Sensitivity analysis
In general, the associations described above were not markedly altered when a complete case data set (n=372) was used (Table 4). There was some evidence though that statin use was associated with improved colorectal cancer-specific survival in tumours that had higher levels of HMGCR expression (HMGCR-high adjusted HR=0.51, 95% CI 0.26–0.97 vs HMGCR-low adjusted HR=0.92, 95% CI 0.45–1.87, P for interaction=0.05). Previously observed null associations by KRAS status and p53 expression remained. All associations became attenuated in analyses evaluating overall survival.
Table 4. Sensitivity analysis using complete-case data.
|
Colorectal cancer-specific survival |
Overall survival |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Statin non-user | Statin user | HR (95%CI)a user vs non-user | P-value | P for interaction | Statin non-user | Statin user | HR (95%CI)b user vs non-user | P-value | P for interaction | |
| No. of deaths/patients | No. of deaths/patients | |||||||||
|
Overall | ||||||||||
| n=372 | 95/238 | 27/88 | 0.75 (0.48–1.20) | 0.24 | − | 127/270 | 41/102 | 0.71 (0.47–1.06) | 0.10 | − |
|
Stratified by patterns of p53 immunostaining | ||||||||||
| Normal | 25/110 | 9/39 | 1.14 (0.48–2.72) | 0.77 | 0.42 | 40/110 | 13/39 | 0.54 (0.22–1.30) | 0.17 | 0.68 |
| Aberrant | 69/159 | 18/63 | 0.63 (0.36–1.11) | 0.11 | 86/159 | 28/63 | 0.73 (0.45–1.18) | 0.20 | ||
|
Stratified by HMGCR expression | ||||||||||
| H-score <median | 48/138 | 14/40 | 0.92 (0.45–1.87) | 0.82 | 0.05 | 71/138 | 18/40 | 0.56 (0.28–1.09) | 0.09 | 0.32 |
| H-score ⩾median | 47/131 | 13/62 | 0.51 (0.26–0.97) | 0.04 | 56/131 | 23/62 | 0.66 (0.39–1.14) | 0.14 | ||
|
Stratified by KRAS mutation status | ||||||||||
| Wild-type | 57/171 | 15/66 | 0.66 (0.34–1.28) | 0.22 | 0.21 | 76/171 | 24/66 | 0.60 (0.34–1.05) | 0.07 | 0.23 |
| Mutated | 38/99 | 12/36 | 1.14 (0.56–2.33) | 0.72 | 51/99 | 17/36 | 0.91 (0.48–1.74) | 0.78 | ||
Abbreviations: CI=confidence intervals; ECOG=Eastern Cooperative Oncology Group; HR=hazard ratio; MSI=microsatellite instability.
Multivariable model adjusted for age, gender, year of diagnosis, grade, MSI status, ECOG performance status, family history of colorectal cancer, adjuvant chemotherapy use, stage, and aspirin use.
Multivariable model adjusted for all variables in footnote a and also adjusted for Charlson Comorbidity Index score.
Discussion
In this population-based cohort study of stage II and III colon cancer, perioperative statin prescription was not associated with significantly improved colorectal cancer-specific survival. Similarly, statin use was not associated with better colorectal cancer-specific or overall survival when the cohort was stratified by tumour biomarkers related to the mevalonate pathway.
The HR for the association between statin use and cancer-specific survival in this cohort was similar to that reported in a recent meta-analysis of colorectal cancer observational studies. (Gray et al, 2016) The pooled estimate from four studies assessing post-diagnostic statin use was nonsignificant, despite including over 19 000 patients. It also suggests that the effect of any association is only likely to be moderate at best (pooled HR=0.84, 95% CI 0.68–1.04). Despite optimistic pre-clinical data, these findings confirm the need to evaluate biomarkers that may identify tumours more likely to respond to the potential anti-cancer effects of statins.
This is the first study to assess the interaction between statin use and HMGCR expression in colon cancer. Statin users had a higher proportion of tumours in the highest HMGCR tertile, although statin-induced inhibition of the mevalonate pathway is known to trigger a marked increase in the production of inactive HMGCR in vitro (Goldstein and Brown, 1990; Bengtsson et al, 2014). In the main analysis, there was no evidence that statin users had better survival compared with non-users in tumours with higher levels of HMGCR expression. However, in the complete-case subgroup analysis, which excluded cases with any missing data, statin use was associated with better cancer-specific survival in tumours with higher levels of HMGCR expression. This result should be interpreted with caution though, as multiple hypotheses were tested. Further exploration in additional molecular pathological epidemiology cohorts should be considered, as the complete-case subgroup analysis results are in line with an in vivo breast cancer study, which suggests that statins may have an anti-proliferative effect in tumours that overexpress HMGCR. (Bjarnadottir et al, 2013).
Overexpression of HMGCR has been proposed to be prognostic in a number of malignancies including breast (Borgquist et al, 2008; Brennan et al, 2011) and epithelial ovarian cancer (Brennan et al, 2010). However, a recent population-based breast cancer cohort study failed to demonstrate that overexpression of HMGCR was associated with better survival (Gustbée et al, 2015). Similarly, overexpression of HMGCR was not associated with improved survival after adjusting for confounding variables in colorectal cancer cases within the Malmö Diet and Cancer Study (Bengtsson et al, 2014). The present study largely corroborates this finding.
To the best of the authors’ knowledge, this is the first study to assess the interaction between statin use, p53 expression, and survival in patients with colon cancer. A significant interaction was not identified in this instance, but further work is required as TP53 mutation status was not directly assessed. Missense TP53 mutations result in stabilization of an inactive form of p53 resulting in nuclear accumulation and a correlation with the aberrant positive pattern of expression (Kaye et al, 2010; McCluggage et al, 2011). Only more recently has it been widely appreciated that the aberrant negative pattern of p53 staining is a distinct entity and not part of the spectrum of wild-type staining (Boyle et al, 2014). This pattern of staining may be attributed to a null TP53 mutation resulting in complete absence of the detectable protein (Köbel et al, 2010). Importantly though, these patterns should be viewed as a spectrum of functional protein status rather than as a surrogate for TP53 mutation status as epigenetic silencing may also contribute to aberrant negative expression (Kaye et al, 2010; Boyle et al, 2014). On this basis, mevalonate pathway gene-expression upregulation associated with mutant p53 (Freed-Pastor et al, 2012) may be specific to mutations of TP53 rather than to alternate circumstances resulting in aberrant expression of the protein. Future studies should therefore assess the interaction between the presence of TP53 mutations, statin use, and colon cancer survival before excluding the potential relevance of this biomarker.
Finally, statin use was not associated with improved colorectal cancer-specific or overall survival when the cohort was stratified by KRAS mutation status. This finding is consistent with results from a cohort of 394 patients enrolled in a chemotherapy clinical trial (CALGB 89803; Ng et al, 2011) and 1209 patients within a German population-based colorectal cancer cohort (Hoffmeister et al, 2015). Similarly, statin use was not associated with improved progression free survival in cetuximab treated metastatic colorectal cancer patients within the CAIRO2 trial, irrespective of KRAS mutation status (Krens et al, 2014). The median progression-free survival was also similar between the statin and placebo arms of a subgroup of 83 patients with KRAS mutant tumours in a randomised controlled trial of XELIRI/FOLFIRI±simvastatin in patients with metastatic colorectal cancer (Lim et al, 2015).
A major strength of this study is the inclusion of population-representative colon cancer patients. Application of a precise, automated, and validated digital immunoscoring system also ensures robust immunoexpression data that are highly reproducible. As with all observational studies, however, there may be residual confounding that we were not able to control. A more specific limitation is that data on statin prescription was also only available at a single perioperative time point and this may not reflect changes in post-diagnostic use (Paleari et al, 2016). However, a similar European colorectal cancer cohort demonstrated 88% concordance between baseline and long-term statin use (Hoffmeister et al, 2015). Statin use at this time could also alter tumour behaviour, as it has previously been reported that pre-diagnostic statin users were less likely to develop KRAS wild-type tumours (Lee et al, 2011). In the current study the opposite association (non-significant) was observed with a lower proportion of KRAS mutant tumours among statin users. Importantly though, assessing medication use at a fixed time point excludes immortal time bias (Lévesque et al, 2010). The assessment of Ras status was limited to exons 2 and 3 of KRAS in this study. Misclassification could occur for other mutations of KRAS or NRAS, although overall these mutations are uncommon (<3%), and it is unlikely that this would greatly alter the stratified analysis.
A further limitation is that information on the type and dose of statin prescribed was not available. It has previously been hypothesised that the potential anti-cancer effect of statins is restricted to lipophilic statins (Ahern et al, 2014). In addition, the serum statin concentrations achieved with cardiovascular protective doses of the medication (e.g., simvastatin 40 mg) may not be sufficient to induce the anti-cancer effects observed in preclinical studies (Lim et al, 2015). Finally, despite being population-based, this study lacks power to definitively investigate the interaction between the proposed mevalonate pathway biomarkers, statin use, and colon cancer survival.
In summary, statin use was not associated with better survival in this population-based colon cancer cohort study. In keeping with previous studies, a survival benefit for statin use was not apparent after stratification by tumour KRAS mutation status. Similar results were also observed for p53 immunohistochemical status but additional studies should assess TP53 mutation status as a potential biomarker. There was some evidence of a difference in association between statin use and colon cancer survival by tumour HMGCR expression. In general though, this finding was inconsistent and requires further investigation in additional large studies.
Acknowledgments
We acknowledge the staff of the Northern Ireland Cancer Registry, who facilitated data access, the registry for providing the secure environment for data handling, and Professor Brendan Pang (visiting professor Northern Ireland Molecular Pathology Laboratory) for a significant contribution to microsatellite instability analysis. This project was supported by a Health and Social Care (HSC) Research and Development Division of the Public Health Agency Doctoral Fellowship (EAT/4905/13 – RTG), a Cancer Research UK (CRUK) Research Bursary (C50104/A17592 – RTG), and a CRUK Population Health Postdoctoral Fellowship (C37703/A15333 – HGC). HGC, MMC, LJM, RTG and RFO’N are all co-investigators or affiliated members of the UKCRC Centre of Excellence for Public Health Northern Ireland. The Northern Ireland Cancer Registry is funded by the Public Health Agency, Northern Ireland. The Northern Ireland Biobank is funded by the HSC Research and Development Division of the Public Health Agency in Northern Ireland and CRUK through the Belfast CRUK Centre, the Northern Ireland Experimental Cancer Medicine Centre, and Friends of the Cancer Centre. The Northern Ireland Molecular Pathology Laboratory has received funding from CRUK, the Friends of the Cancer Centre, and the Sean Crummey Foundation. This work used data provided by patients and collected by the NHS as part of their care and support.
Author contributions
Study conception and design: HGC, MBL, SMcQ, CRC, JAJ, MS-T, and LJM. Data acquisition: RTG, HGC, MBL, PB, SMcQ, RFO’N, KA, VB, CMcG, and JAJ. Data analysis and interpretation: RTG, HGC, CRC, and LJM. Drafting manuscript: RTG, HGC, SMcQ, CRC, and LJM. Manuscript revision and final approval: All.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
PWH is the Founder and Director in PathXL Ltd. MS-T is a senior advisor to PathXL. All other authors declare no conflict of interest.
Supplementary Material
References
- Ahern TP, Lash TL, Damkier P, Christiansen PM, Cronin-Fenton DP (2014) Statins and breast cancer prognosis: evidence and opportunities. Lancet Oncol 15: e461–e468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardou M, Barkun A, Martel M (2010) Effect of statin therapy on colorectal cancer. Gut 59: 1572–1585. [DOI] [PubMed] [Google Scholar]
- Bengtsson E, Nerjovaj P, Wangefjord S, Nodin B, Eberhard J, Uhlén M, Borgquist S, Jirström K, Uhlen M, Borgquist S, Jirstrom K (2014) HMG-CoA reductase expression in primary colorectal cancer correlates with favourable clinicopathological characteristics and an improved clinical outcome. Diagn Pathol 9: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjarnadottir O, Romero Q, Bendahl P-O, Jirström K, Rydén L, Loman N, Uhlén M, Johannesson H, Rose C, Grabau D, Borgquist S (2013) Targeting HMG-CoA reductase with statins in a window-of-opportunity breast cancer trial. Breast Cancer Res Treat 138: 499–508. [DOI] [PubMed] [Google Scholar]
- Borgquist S, Jögi A, Pontén F, Rydén L, Brennan DJ, Jirström K (2008) Prognostic impact of tumour-specific HMG-CoA reductase expression in primary breast cancer. Breast Cancer Res 10: R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle DP, McArt DG, Irwin G, Wilhelm-Benartzi CS, Lioe TF, Sebastian E, McQuaid S, Hamilton PW, James JA, Mullan PB, Catherwood MA, Harkin DP, Salto-Tellez M (2014) The prognostic significance of the aberrant extremes of p53 immunophenotypes in breast cancer. Histopathology 65: 340–352. [DOI] [PubMed] [Google Scholar]
- Brennan DJ, Brändstedt J, Rexhepaj E, Foley M, Pontén F, Uhlén M, Gallagher WM, O’Connor DP, O’Herlihy C, Jirstrom K (2010) Tumour-specific HMG-CoAR is an independent predictor of recurrence free survival in epithelial ovarian cancer. BMC Cancer 10: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan DJ, Laursen H, O’Connor DP, Borgquist S, Uhlen M, Gallagher WM, Pontén F, Millikan RC, Rydén L, Jirström K (2011) Tumor-specific HMG-CoA reductase expression in primary premenopausal breast cancer predicts response to tamoxifen. Breast Cancer Res 13: R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardwell CR, Hicks BM, Hughes C, Murray LJ (2014. a) Statin use after colorectal cancer diagnosis and survival: a population-based cohort study. J Clin Oncol 32: 3177–3183. [DOI] [PubMed] [Google Scholar]
- Cardwell CR, Kunzmann AT, Cantwell MM, Hughes C, Baron JA, Powe DG, Murray LJ (2014. b) Low-dose aspirin use after diagnosis of colorectal cancer does not increase survival: a case-control analysis of a population-based cohort. Gastroenterology 146: 700–708. [DOI] [PubMed] [Google Scholar]
- Freed-Pastor WA, Mizuno H, Zhao X, Langerød A, Moon S-H, Rodriguez-Barrueco R, Barsotti A, Chicas A, Li W, Polotskaia A, Bissell MJ, Osborne TF, Tian B, Lowe SW, Silva JM, Børresen-Dale A-L, Levine AJ, Bargonetti J, Prives C (2012) Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell 148: 244–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli D, Gavin PG, Taniyama Y, Kim S-I, Choi H-J, Paik S, Pogue-Geile KL (2010) A rapid, sensitive, reproducible and cost-effective method for mutation profiling of colon cancer and metastatic lymph nodes. BMC Cancer 10: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JL, Brown MS (1990) Regulation of the mevalonate pathway. Nature 343: 425–430. [DOI] [PubMed] [Google Scholar]
- Gray RT, Coleman HG, Hughes C, Murray LJ, Cardwell CR (2016) Statin use and survival in colorectal cancer: results from a population-based cohort study and an updated systematic review and meta-analysis. Cancer Epidemiol 45: 71–81. [DOI] [PubMed] [Google Scholar]
- Gustbée E, Tryggvadottir H, Markkula A, Simonsson M, Nodin B, Jirström K, Rose C, Ingvar C, Borgquist S, Jernström H (2015) Tumor-specific expression of HMG-CoA reductase in a population-based cohort of breast cancer patients. BMC Clin Pathol 15: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmeister M, Jansen L, Rudolph A, Toth C, Kloor M, Roth W, Blaker H, Chang-Claude J, Brenner H (2015) Statin use and survival after colorectal cancer: the importance of comprehensive confounder adjustment. J Natl Cancer Inst 107: djv045. [DOI] [PubMed] [Google Scholar]
- Kaye PV, Haider SA, James PD, Soomro I, Catton J, Parsons SL, Ragunath K, Ilyas M (2010) Novel staining pattern of p53 in Barrett’s dysplasia-the absent pattern. Histopathology 57: 933–935. [DOI] [PubMed] [Google Scholar]
- Khan NF, Perera R, Harper S, Rose PW (2010) Adaptation and validation of the Charlson Index for Read/OXMIS coded databases. BMC Fam Pract 11: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Park SJ, Hong SP, Cheon JH, Kim WH, Kim TI (2015) The effect of prediagnostic aspirin use on the prognosis of stage III colorectal cancer. Int J Clin Exp Med 8: 13435–13445. [PMC free article] [PubMed] [Google Scholar]
- Konstantinopoulos PA, Karamouzis MV, Papavassiliou AG (2007) Post-translational modifications and regulation of the RAS superfamily of GTPases as anticancer targets. Nat Rev Drug Discov 6: 541–555. [DOI] [PubMed] [Google Scholar]
- Krens LL, Simkens LHJ, Baas JM, Koomen ER, Gelderblom H, Punt CJA, Guchelaar H-JJ (2014) Statin use is not associated with improved progression free survival in cetuximab treated KRAS mutant metastatic colorectal cancer patients: results from the CAIRO2 study. PLoS One 9: e112201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köbel M, Reuss A, du Bois A, Kommoss S, Kommoss F, Gao D, Kalloger SE, Huntsman DG, Gilks CB (2010) The biological and clinical value of p53 expression in pelvic high-grade serous carcinomas. J Pathol 222: 191–198. [DOI] [PubMed] [Google Scholar]
- Lee JE, Baba Y, Ng K, Giovannucci E, Fuchs CS, Ogino S, Chan AT (2011) Statin use and colorectal cancer risk according to molecular subtypes in two large prospective cohort studies. Cancer Prev Res 4: 1808–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SH, Kim TW, Hong YS, Han S-W, Lee K-H, Kang HJ, Hwang IG, Lee JY, Kim HS, Kim ST, Lee J, Park JO, Park SH, Park YS, Lim HY, Jung S-H, Kang WK (2015) A randomised, double-blind, placebo-controlled multi-centre phase III trial of XELIRI/FOLFIRI plus simvastatin for patients with metastatic colorectal cancer. Br J Cancer 113: 1421–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévesque LE, Hanley JA, Kezouh A, Suissa S (2010) Problem of immortal time bias in cohort studies: example using statins for preventing progression of diabetes. BMJ 340: b5087. [DOI] [PubMed] [Google Scholar]
- Mace AG, Gantt GA, Skacel M, Pai R, Hammel JP, Kalady MF (2013) Statin therapy is associated with improved pathologic response to neoadjuvant chemoradiation in rectal cancer. Dis Colon Rectum 56: 1217–1227. [DOI] [PubMed] [Google Scholar]
- McCarty KS, Szabo E, Flowers JL, Cox EB, Leight GS, Miller L, Konrath J, Soper JT, Budwit DA, Creasman WT, Seigler HF, McCarty KS (1986) Use of a monoclonal anti-estrogen receptor antibody in the immunohistochemical evaluation of human tumors. Cancer Res 46: 4244s–4248s. [PubMed] [Google Scholar]
- McCluggage WG, Soslow RA, Gilks CB (2011) Patterns of p53 immunoreactivity in endometrial carcinomas: ‘all or nothing’ staining is of importance. Histopathology 59: 786–788. [DOI] [PubMed] [Google Scholar]
- Ng K, Ogino S, Meyerhardt JA, Chan JA, Chan AT, Niedzwiecki D, Hollis D, Saltz LB, Mayer RJ, Benson AB III, Schaefer PL, Whittom R, Hantel A, Goldberg RM, Bertagnolli MM, Venook AP, Fuchs CS (2011) Relationship between statin use and colon cancer recurrence and survival: results from CALGB 89803. J Natl Cancer Inst 103: 1540–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S, Chan AT, Fuchs CS, Giovannucci E (2011) Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut 60: 397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S, Fuchs CS, Giovannucci E (2012) How many molecular subtypes? Implications of the unique tumor principle in personalized medicine. Expert Rev Mol Diagn 12: 621–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paleari L, Puntoni M, Clavarezza M, DeCensi M, Cuzick J, DeCensi A (2016) PIK3CA mutation, aspirin use after diagnosis and survival of colorectal cancer. A systematic review and meta-analysis of epidemiological studies. Clin Oncol 28: 317–326. [DOI] [PubMed] [Google Scholar]
- Thurnher M, Nussbaumer O, Gruenbacher G (2012) Novel aspects of mevalonate pathway inhibitors as antitumor agents. Clin Cancer Res 18: 3524–3531. [DOI] [PubMed] [Google Scholar]
- Zanders MMJ, van Herk-Sukel MPP, Vissers PAJ, Herings RMC, Haak HR, van de Poll-Franse LV (2015) Are metformin, statin and aspirin use still associated with overall mortality among colorectal cancer patients with diabetes if adjusted for one another? Br J Cancer 113: 403–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Salto-Tellez M, Putti TC, Do E, Koay ES-C (2003) Reliability of tissue microarrays in detecting protein expression and gene amplification in breast cancer. Mod Pathol 16: 79–84. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.