Abstract
DNA accessibility to regulatory proteins is significantly affected by nucleosome structure and dynamics. FACT (facilitates chromatin transcription) increases the accessibility of nucleosomal DNA but the mechanism and extent of this nucleosome reorganization are unknown. We report here the effects of FACT on single nucleosomes revealed with spFRET microscopy. FACT binding results in a dramatic, ATP-independent, and reversible uncoiling of DNA that affects at least 70% of the DNA in a nucleosome. A mutated version of FACT is defective in this uncoiling, and a histone mutation that suppresses phenotypes caused by this FACT mutation in vivo restores the uncoiling activity in vitro. Thus FACT-dependent nucleosome unfolding modulates the accessibility of nucleosomal DNA, and this is an important function of FACT in vivo.
Introduction
Eukaryotic genomes are tightly condensed in chromatin, comprised of nucleosomes in which 147 bp segments of DNA are wrapped around octamers of histones about 1.6 times 1. This packaging significantly limits the accessibility of the DNA, providing a barrier that plays a major role in regulating DNA-dependent processes such as gene expression 2,3.
Histone chaperones promote nucleosome assembly by blocking non-productive interactions between DNA and histones 4–6. FACT (facilitates chromatin transcription) is an essential and highly conserved histone chaperone that can assist nucleosome assembly, but surprisingly it also promotes disassembly, so it can both stabilize and destabilize chromatin 7–10. FACT from the yeast Saccharomyces cerevisiae is a heterodimer of Spt16 and Pob3 proteins, whose functions are supported by the HMGB-like protein Nhp6 11,12. Consistent with roles in both forming and bypassing nucleosomes, FACT participates in a range of processes including DNA transcription, replication, and repair 13–15. Notably, FACT is needed for efficient removal of nucleosomes during induction of transcription 16,17, it facilitates nucleosome recovery during elongation and nucleosome repopulation during repression of transcription 18–20 and it promotes transcription through chromatin by RNA polymerase II in vitro 21,22, highlighting the importance of both its stabilization and destabilization activities.
FACT interacts with multiple targets in a nucleosome 23, including DNA, H3/H4 tetramers and H2A/H2B dimers 10,24,25. In particular, FACT can bind both of the H2A-H2B dimers in a nucleosome simultaneously 26. FACT can also reorganize nucleosomes, enhancing DNA accessibility to various probes in the absence of ATP hydrolysis and without permanently displacing the histones 8,27. Competition between FACT and DNA for binding histones has been proposed to be responsible for the increased DNA accessibility observed when FACT binds nucleosomes 9,21,25,26,28, however, the nature and the extent of FACT-dependent nucleosome reorganization is currently unknown.
We report here that single particle Förster resonance energy transfer (spFRET) analysis reveals FACT-induced global, large-scale, reversible, ATP-independent reorganization of nucleosomal structure. Validating the physiological relevance of the observations, we demonstrate that this reorganization activity is diminished by a FACT mutation that causes defects in replication and transcription in vivo, and that a mutation of H2A that suppresses some of the in vivo phenotypes also restores nucleosome reorganization activity in our in vitro system.
Results
Measuring the effects of FACT on nucleosome structure and dynamics using spFRET
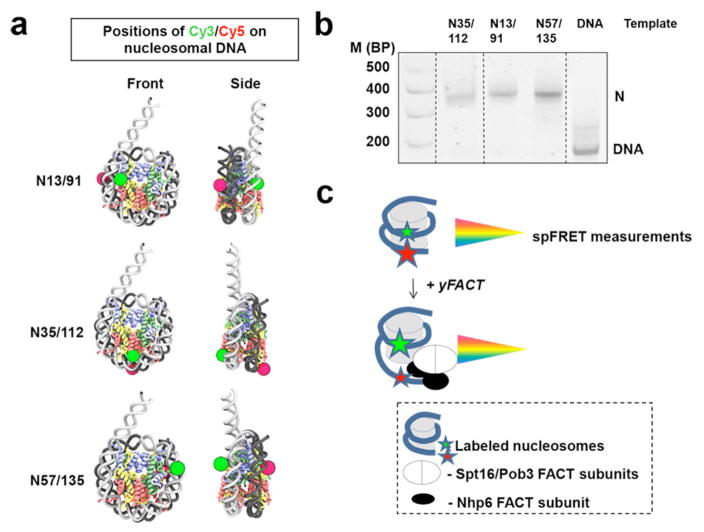
To study the reorganization of nucleosomal DNA by yeast FACT, we used three mononucleosomal templates with pairs of fluorescent labels in different regions of the DNA (Fig. 1a). Nucleosomes were assembled with chicken or recombinant Xenopus laevis histones on the previously characterized 603 nucleosome positioning sequence 29,30 with a 20 bp linker, then gel purified. Label positions were directed by the known nucleosome structure 31 to allow FRET between Cy3 and Cy5 labels without disrupting histone-DNA contacts (Fig. 1a). Label pairs were introduced at +35/+112 (template N35/112), +13/+91 (N13/91), and +57/+135 (N57/135) bp relative to the 603 sequence boundary 32. These correspond to the point of contact between the two H2A-H2B dimers, and the entry/exit points adjacent to and distal from the linker segment, respectively. The nucleosomes were about 90% pure, with minor amounts of hexasomes (lacking one H2A-H2B dimer) and free DNA (Fig. 1b).
Fig. 1. Experimental approach for analysis of the effect of FACT on nucleosome structure and dynamics.
a. Mononucleosomes were assembled on DNA having the 147 bp 603 nucleosome positioning sequence 32. The N13/91, N35/112 and N57/135 DNAs contain pairs of fluorescent labels (Cy3 and Cy5) on the adjacent gyres of nucleosomal DNA at +13/+91, +35/+112 and +57/+135 bp relative to the 603 sequence boundary, respectively, allowing FRET in the context of the expected nucleosomal structure.
b. Analysis of gel-purified nucleosomes by native PAGE. Fluorescence of Cy5 (nucleosomal templates) or FAM (DNA markers, M) was detected using a PhosphorImager.
c. Fluorescently labeled nucleosomes were used for spFRET measurements in solution to analyze the effect of yeast FACT on nucleosomal DNA structure. FRET efficiency decreases when the distance between labeled DNA sites increases, allowing analysis of proximity of the labeled gyres of nucleosomal DNA in single nucleosomes.
A laser of the 514.5 nm wavelength was used to excite the donor fluorophore (Cy3) in single nucleosomal complexes diffusing freely in solution as they traversed the focal volume of the microscope, and fluorescence intensities of both donor (Cy3) and acceptor (Cy5) dyes were measured (Fig 1c) as described previously 33. The proximity ratio (EPR) was then calculated in the absence or presence of FACT, revealing changes in the distance between the labeled DNA sites through changes in FRET efficiency.
Nucleosome unfolding by yFACT is extensive and reversible
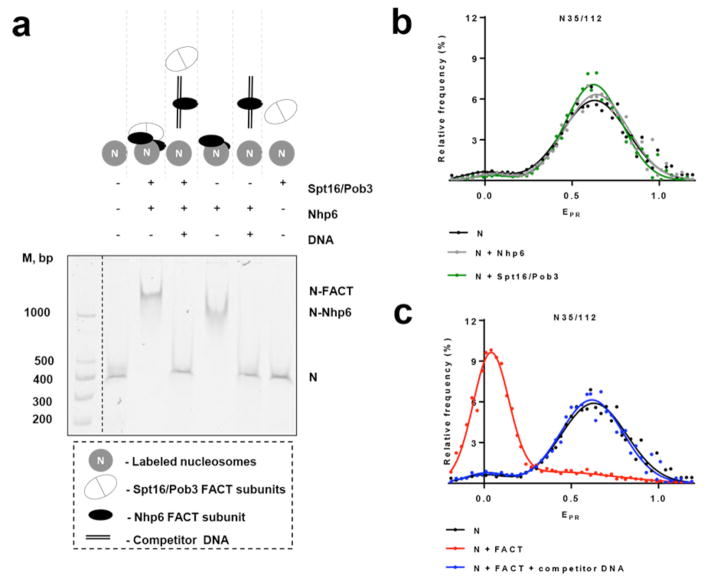
Addition of yeast FACT (Spt16/Pob3 with Nhp6) to fluorescently labeled nucleosomes resulted in formation of protein-nucleosome complexes as detected by non-denaturing PAGE (Fig 2a). In accordance with previous results 12,26, formation of complexes required binding of nucleosomal DNA by Nhp6 (Fig. 2a), so complex formation was reversed by the addition of excess of unlabeled competitor DNA (Fig. 2a and refs. 12,26). The fluorescent labels therefore did not alter FACT’s interactions with nucleosomes.
Fig. 2. Nucleosome unfolding by yFACT is extensive and reversible.
a, Binding of FACT and different combinations of its subunits to fluorescently labeled N35/112 nucleosomes analyzed by native PAGE. Fluorescence of Cy5 (nucleosomal templates) or FAM (DNA markers, M) was detected using a PhosphorImager. An excess of competitor DNA was added to remove FACT from FACT-nucleosome complexes.
b,c, Typical frequency distributions of proximity ratios (EPR) for nucleosomes N35/112 in the presence/absence of Nhp6, Spt16/Pob3 or FACT with/without competitor DNA. Analysis by spFRET microscopy. The sample sizes (n, single particle events) were the following: (N) – 4622; (N+Nhp6) – 14128; (N+Spt16/Pob3) – 15779; (N+FACT) – 5958; (N+FACT + competitor DNA) – 9297. The mean values of EPR peaks and the standard errors were the following: (N) – 0.02±0.03, 0.653±0.004; (N+Nhp6) – 0.03±0.03, 0.620±0.004; (N+Spt16/Pob3) – 0.01±0.07, 0.64±0.01; (N+FACT) – 0.033±0.003, 0.51±0.03; (N+FACT+ competitor DNA) – 0.02±0.03, 0.65±0.06. b, Addition of Nhp6 or Spt16/Pob3 to the nucleosomes induced minor changes in EPR of the majority of nucleosomes that indicates minimal changes in the folding of nucleosomal DNA near the contact of H2A-H2B dimers.
c, FACT induces unfolding of the nucleosomal DNA which can be reversed by addition of an excess of competitor DNA disrupting the FACT:nucleosome complexes.
Figure 2b,c shows a typical frequency distribution of EPR measured by spFRET for N35/112 nucleosomes in solution. The distribution is described by two Gaussian peaks: a minor peak (mean EPR of 0.02±0.03) that is likely to be due to free DNA with a large distance between labels (low EPR) and a major peak (mean EPR of 0.653±0.004) corresponding to nucleosomes (typically >85% of the individual signals) with the expected close proximity of labeled DNA sites (high EPR). Addition of Nhp6 or Spt16/Pob3 alone induced only minor changes in the distance between the two DNA gyres in this region of a nucleosome, resulting in little change to the EPR frequency distributions (Fig. 2b). In contrast, adding complete FACT dramatically changed the EPR frequency distribution (Fig. 2c), with the low EPR peak becoming dominant, suggesting a large increase in the distance between gyres of the nucleosomal DNA upon binding by FACT. This supports the model that FACT promotes a structural change in nucleosomes, and that the three subunits of FACT act cooperatively to induce this change. An alternative explanation for the decreased proximity ratio is that FACT might have quenched the fluorescence of the Cy5 acceptor. To evaluate this possibility, we measured the fluorescence of Cy5 using its inherent excitation wavelength (633 nm) instead of indirect excitation through Cy3. Measurements taken before and after FACT binding showed no quenching (data not shown), making this possibility unlikely. We note that these changes in FRET proximity ratio were observed in the absence of ATP, distinguishing this alteration from ATP-dependent remodeling. Importantly, the change in structure was reversible, as disruption of the FACT:nucleosome complexes by addition of an excess of competitor DNA restored the nucleosomes to the form with high EPR (Fig. 2c). This suggests that the core histones remain constrained or tethered together within the FACT:nucleosome complexes, as they are readily reassembled into apparently intact nucleosomes.
Nucleosome reorganization involves the majority of the nucleosomal DNA
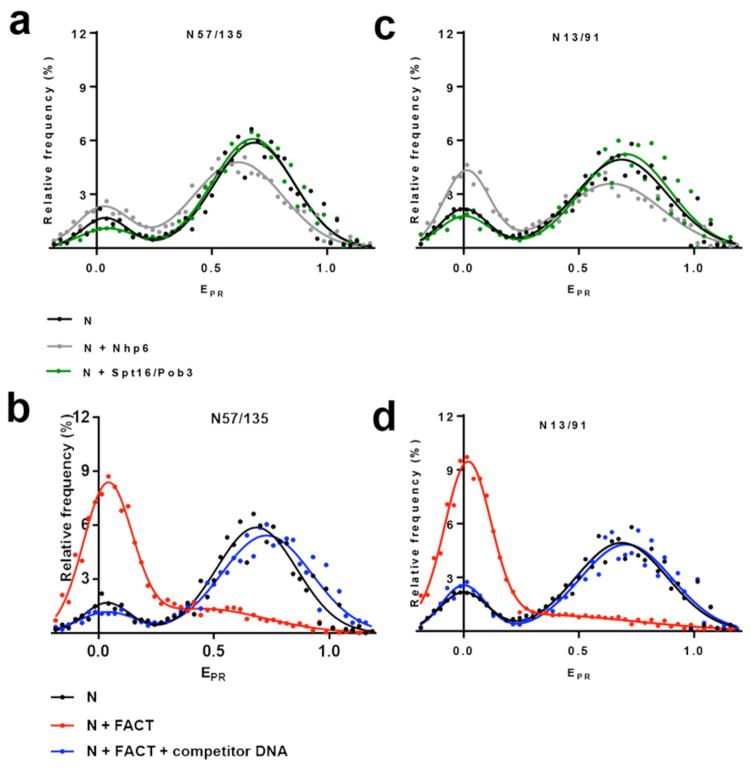
To evaluate the extent of uncoiling of nucleosomal DNA upon FACT binding, nucleosomes N13/91 and N57/135 with labels in DNA entry/exit regions were studied (Fig 3). These nucleosomes revealed major populations with high EPR, as well as an increase in the fractions of nucleosomes with low proximity ratios relative to N35/112 (Fig 2c). This is consistent with previous results showing some breathing (uncoiling) of DNA at the entry/exit points 34,35. As observed for N35/112, Spt16-Pob3 alone did not cause significant changes in the EPR distributions for N13/91 and N57/135 nucleosomes (Fig. 3a, c). Notably, Nhp6 binding alone did cause a small shift of high-EPR fractions of N57/135 (mean EPR of 0.669±0.004) and N13/91 (mean EPR of 0.71±0.01) towards the lower EPR values (mean EPR of 0.62±0.01 and 0.62±0.01, respectively) and an increase in the content of the low-EPR fractions, which was especially pronounced for N13/91. The latter effect could be explained by destabilization of DNA-histone interactions at the entry/exit points by Nhp6 binding, which is facilitated by the adjacent dsDNA linker in N13/91.
Fig. 3. Nucleosome unfolding by yFACT involves the majority of nucleosomal DNA.
a, b, c and d, Typical frequency distributions of EPR for nucleosomes N13/91 and N57/135 in the presence/absence of Nhp6, Spt16/Pob3 (a, c) or FACT with/without competitor DNA (b, d). Analysis by spFRET microscopy. a, c, Nhp6 binding increases low-EPR fractions of the nucleosomes, likely by destabilizing DNA-histone interactions at the entry/exit points. Addition of Spt16/Pob3 to the nucleosomes does not significantly affect the EPR of nucleosomes. b, d, FACT-induced unfolding of the nucleosomal DNA occurs also in the entry/exit points adjacent to and distal from the linker segment. Changes induced by FACT can be reversed by addition of an excess of competitor DNA disrupting the FACT:nucleosome complexes.
a, b, The sample sizes (n, single particle events) were the following: (N) – 3841; (N+Nhp6) – 9351; (N+Spt16/Pob3) – 7519; (N+FACT) – 13133; (N+FACT + competitor DNA) – 8080. The mean values of EPR peaks and the standard errors were the following: (N) – 0.043±0.013, 0.669±0.004; (N+Nhp6) – 0.035±0.007, 0.62±0.01; (N+Spt16/Pob3) – 0.05±0.02, 0.67±0.01; (N+FACT) – 0.036±0.004, 0.53±0.05; (N+FACT+ competitor DNA) – 0.02±0.02, 0.715±0.008.
c, d, The sample sizes (n, single particle events) were the following: (N) – 6586; (N+Nhp6) – 3587; (N+Spt16/Pob3) – 6789; (N+FACT) – 9160; (N+FACT + competitor DNA) – 4368. The mean values of EPR peaks and the standard errors were the following: (N) – 0.01±0.01, 0.71±0.01; (N+Nhp6) – 0.01±0.01, 0.62±0.01; (N+Spt16/Pob3) – 0.01±0.01, 0.68±0.01; (N+FACT) – 0.023±0.003, 0.54±0.05; (N+FACT+ competitor DNA) – 0.01±0.01, 0.71±0.01.
Once again, addition of complete FACT caused a dramatic increase in the low-EPR fractions of both N57/135 and N13/91 (Fig. 3b, d). Direct excitation of the acceptor dye was also used to confirm the absence of Cy5 quenching in these cases (not shown). Uncoiling of DNA in the complexes of FACT with N57/135 or N13/91 was also reversible as demonstrated by restoration of high EPR in the majority of nucleosomes upon addition of competitor DNA to disrupt the FACT:nucleosome complexes (Fig. 3b, d). The EPR frequency distributions for nucleosomes uncoiled by FACT and for complexes between FACT and fluorescently labeled free DNA were very similar (data not shown), suggesting that FACT promoted a significant increase in the distance between labeled DNA sites. The dramatic effect of FACT on nucleosome structure therefore extends to both entry/exit sites, consistent with a global, large-scale uncoiling of nucleosomal DNA.
Analysis of the reorganization of nucleosomal DNA by FACT: molecular modeling
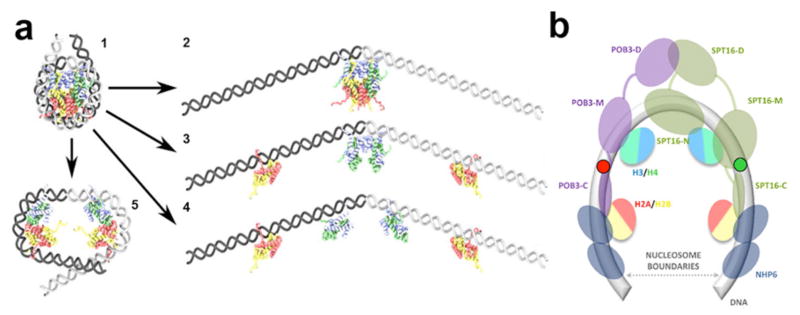
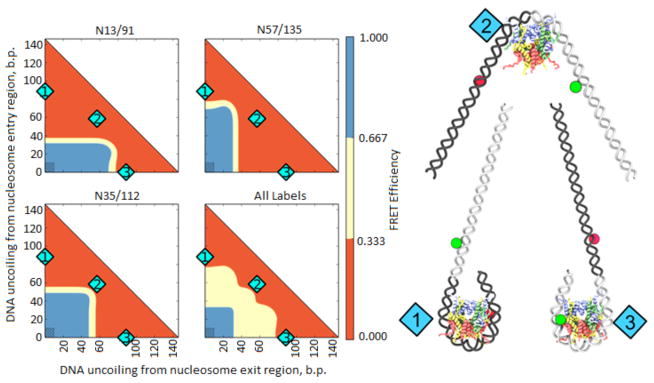
The FRET efficiency in the labeled nucleosomes depends primarily on the extent of DNA coiling in the nucleosome, as this dictates the proximity of the dyes to one another. Many different models of nucleosome unfolding have been proposed, some of them involving major changes in the histone octamer structure (36, see Fig. 6 and Discussion). The majority of the models involve some DNA straightening and partial or complete detachment from core histones. DNA uncoiling is the simplest informative model of nucleosome disassembly. As no high-resolution structure of FACT-rearranged nucleosomes is currently available, we have quantitatively modeled the uncoiling of nucleosomal DNA from the histone octamer for the labeled nucleosomes (N35/112, N13/91 and N57/135) to gain insight into the mechanism of their reorganization (Fig. 4). Low FRET efficiency (<0.333, red area on Fig. 4) is expected for N13/91 and N57/135 if more than ~40 bp of nucleosomal DNA are straightened from either end. In contrast, more than ~60 bp of DNA must be unwrapped from either end of the nucleosome to produce low FRET in the case of N35/112. Thus the observation that low FRET efficiency occurred with all pairs of labels indicates that over ~100 bp of nucleosomal DNA must be reconfigured (red area on Fig. 4, “All labels” panel). This dramatic uncoiling of nucleosomal DNA likely corresponds to a U-shaped DNA that forms less than 1 one superhelical turn of DNA instead of the canonical 1 ¾ DNA turns of nucleosomal DNA. The position of the remaining bent DNA (if any) is unknown, but the ends of nucleosomal DNA are uncoiled at lower forces than are required to affect the central region of nucleosomal DNA 37. Therefore the most likely model involves nearly symmetrical and significant DNA straightening (with or without DNA detachment from (H2A–H2B) dimers, see models in Figs. 6a and b, respectively). Further experiments are needed to reveal the details of nucleosome unfolding.
Fig. 6. Models of FACT-dependent uncoiling of nucleosomal DNA.
a, Previously suggested models of nucleosome unfolding. Unfolding of intact nucleosomes (model 1) could occur via: (2) DNA uncoiling from an intact histone octamer, DNA uncoiling accompanied by octamer disassembly, without (3) or with disruption of the H3:H3 dimer interface (4), (5) opening of the (H3–H4) dimer-dimer interface without further DNA uncoiling. b, A hypothetical integrated model for nucleosome unfolding by yFACT. Nucleosomal DNA and the globular domains of histones and are shown. Expected positions of Nhp6 proteins and various domains of Spt16 and Pob3 are indicated, with connecting lines indicating disordered regions (see text for details). H2A/H2B dimers could be reversibly displaced from DNA by Nhp6; interactions between H3/H4 dimers could be intact. Labels at the positions 35 and 112 are indicated by green and red circles.
Fig. 4. Analysis of FRET efficiency in unfolded nucleosomes by molecular modeling.
On the left: Color-coded maps of predicted FRET efficiencies for molecular models of labeled nucleosomes with different degree of DNA uncoiling (see Materials and Methods). The red and blue areas on the color maps for all labels correspond to the structures where all three label pairs are in low or high FRET conformations, respectively. Shaded rectangles correspond to native structures, in which conformations are close to those expected from the crystal structure. On the right: Models of nucleosome conformations corresponding to positions numbered in cyan on the graphs.
Genetic interactions between mutated FACT and histone H2A are recapitulated in vitro
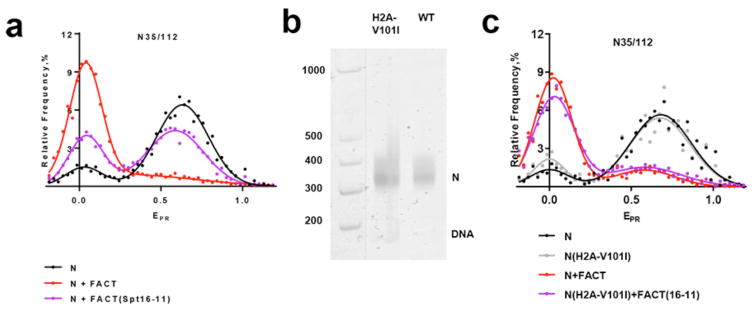
To evaluate physiological relevance and to further study the mechanism of FACT-induced nucleosome unfolding in vitro, we analyzed FACT and histone mutants that interact with one another in vivo. The spt16-11 allele affecting the large subunit of FACT causes phenotypes consistent with transcription and replication defects in vivo 12 as well as a nucleosome reorganization defect detected by restriction endonuclease sensitivity of nucleosomal DNA in vitro 38. Many of the phenotypes and the reorganization defect caused by this mutation were suppressed by an H2A-V101I mutation 12 suggesting that an altered nucleosome is capable of compensating for inefficient FACT activity. We therefore tested these mutant components in our spFRET assay. As expected, FACT(Spt16-11) mutant protein induced less efficient nucleosome unfolding (Fig. 5a). In particular, unfolding was observed in only a subpopulation of nucleosomes, although within this subpolulation the extent of unfolding was the same as in the FACT:nucleosome complexes. The data suggest that nucleosome unfolding is a highly cooperative process that occurs in an “all-or-none” fashion and that the defect caused by FACT(Spt16-11) is in promoting or maintaining the same open state, not in dictating the properties of that state. To test the importance of the properties of the histones core of the nucleosomes in this assay, we assembled N35/112 nucleosomes containing the suppressor mutation H2A-V101I and compared it with wild type (WT) nucleosomes assembled with normal histone octamers. The mutant and WT nucleosomes had similar spFRET profiles, although the H2A-V101I nucleosomes appeared to be less stable as they contained a higher level of free DNA (Fig. 5b, c).
Fig. 5. Conditional mutations in FACT(Spt16-11) affect nucleosome unfolding in vitro.
a, c Typical frequency distributions of EPR for nucleosomes N35/112 (N), mutant (H2A-V101I) nucleosomes N35/112 (N(H2A-V101I)), their complexes with FACT and mutant FACT(Spt16-11). Analysis by spFRET microscopy ((4–6)×103 nucleosomes per specimen).
a, Mutations in FACT(Spt16-11) affect nucleosome unfolding. The sample sizes (n, single particle events) were the following: (N) – 3800; (N+FACT(Spt16-11)) – 11642; (N+FACT) – 5958. The mean values of EPR peaks and the standard errors were the following: (N) – 0.02±0.03, 0.645±0.004; (N+FACT(Spt16-11)) – 0.05±0.01, 0.60±0.01; (N+FACT) – 0.033±0.003, 0.51±0.03.
b, Analysis of gel-purified nucleosomes N35/112 (WT) or the nucleosomes containing the suppressor mutation H2A-V101I by native PAGE. Fluorescence of Cy5 (nucleosomes) or FAM (DNA markers, M) was detected using a PhosphorImager.
c, Mutation in the H2A-V101I nucleosome suppresses the defect in FACT(Spt16-11) activity, making the uncoiling of nucleosomal DNA by FACT(Spt16-11) comparable with unwrapping of nucleosomes by FACT. The sample sizes (n, single particle events) were the following: (N) – 562; (N(H2A-V101I)) – 957; (N+FACT)– 4024; (N(H2A-V101I)+FACT(Spt16-11)) – 7245. The mean values of EPR peaks and the standard errors were the following: (N) 0,01±0.02, 0.68±0.01; (N(H2A-V101I) – 0.01±0.02, 0.65±0.01; (N+FACT) – 0.027±0.005, 0.66±0.04; (N(H2A-V101I)+FACT(Spt16-11)) – 0.026±0.003, 0.57±0.02.
Consistent with the in vivo suppression, the diminished ability of FACT(Spt16-11) to promote formation of an unfolded (low-EPR) subpopulation of WT nucleosomes was reversed by combining it with nucleosomes containing H2A-V101I, producing levels of reorganization comparable with those observed with WT FACT on the WT nucleosomes (Fig. 5c).
In summary, the mutations in FACT(Spt16-11) that impair its functions in vivo also result in lower probability of nucleosome unfolding in vitro. A histone point mutation H2A-V101I that suppresses FACT(Spt16-11) defects in vivo also suppressed the uncoiling deficiency in vitro, suggesting that nucleosome uncoiling is an integral part of the mechanism of FACT action in vivo and that our experimental system faithfully recapitulates this essential FACT activity.
Discussion
We have developed an spFRET assay to investigate how FACT alters the structure of DNA within individual nucleosomes during reorganization (Fig. 1). Pairs of fluorescent labels were used to provide information about the proximity of neighboring DNA gyres in three different nucleosome regions during FACT binding. We found that FACT binding induced a dramatic, reversible increase in the distance between labeled sites of nucleosomal DNA in all tested regions, including those near a contact between two H2A–H2B dimers, and the entry/exit points adjacent to and distal from the DNA linker. The uncoiling was ATP-independent, highly cooperative with respect to FACT’s subunits, and extensive. Our modeling suggests that at least 100 bp of DNA is uncoiled during reorganization, with at least 40 bp uncoiled from one entry/exit site and 60 bp uncoiled from the other (Fig. 4). Analysis of uncoiling of nucleosomal DNA by a FACT(Spt16-11) mutant that impairs FACT’s functions and the effect of H2A-V101I that is a suppressor of FACT(Spt 16-11) defects (Fig. 5) suggests that nucleosome uncoiling is an essential feature of FACT action in vivo.
The global FACT-dependent uncoiling of nucleosomal DNA has been inferred from population assays as increased sensitivity of nucleosomal DNA to restriction enzymes, DNases and hydroxyl radicals, with tethering of components in an altered configuration suggested by reversibility of the effects and lower sensitivity of the reorganized form to nucleases than that of histone-free DNA 14,28. Previously the structural basis of these changes was unclear 8,28. Our data suggest that dramatic uncoiling of nucleosomal DNA is the principal component of FACT-dependent nucleosome reorganization. Several models for uncoiling of nucleosomal DNA have been proposed previously (Fig. 6): DNA unwrapping from the octamer (Fig. 6a), DNA unwrapping accompanied by opening of the (H2A-H2B) dimer/(H3-H4)2 tetramer interface (Fig. 6b) and DNA unwrapping with complete octamer disassembly 36,39. Uncoiling may also result in opening of the (H3–H4)2 tetramer with minimal DNA unwrapping (Fig. 6d). Each model or a combination suggests a large, FACT-induced change in nucleosome structure, and could explain the increased distance between the labeled DNA sites observed in the presence of FACT.
Fig. 6e shows a likely model that accounts for the following critical features of the FACT-nucleosome complex. (a) dramatic, reversible, Nhp6-dependent uncoiling of nucleosomal DNA (this work), (b) increased accessibility of nucleosomal DNA, as compared with its accessibility in intact nucleosomes 28, (c) the ability of the C-terminal domains of Pob3 and Spt16 to interact with two different H2A-H2B dimers [possibly in the same nucleosome, 26], (d) the ability of human Spt16 to interact with an (H3-H4)2 tetramer 40. Importantly, FACT remains in the reorganized complex and is likely to interact both with the uncoiled nucleosomal DNA and with core histones 26,28, replacing some of DNA-histone and histone-histone interactions.
Our analysis of FACT(Spt16-11) effects suggests that the observed FACT-induced nucleosome reorganization is physiologically relevant. The spt16-11 allele causes sensitivity to elevated temperatures, sensitivity to hydroxyurea, and the Spt− phenotype 12. The last two phenotypes are typically interpreted as disturbances of DNA replication and activation of cryptic transcription initiation start-sites due to incomplete nucleosome assembly during transcript elongation, respectively. Some mutations in core histones can suppress some phenotypes caused by the spt16-11 mutation, in particular the H2A-V101I mutation suppresses temperature- and hydroxyurea sensitivity, but has only a minor effect on the Spt− phenotype 38. The mutations in FACT(Spt16-11) decrease the probability of nucleosomal DNA uncoiling and are suppressed by the H2A-V101I mutation in vitro. Based on these data and the observed phenotypes in vivo we propose that the large-scale FACT-dependent uncoiling of nucleosomal DNA that we detect by spFRET is likely to be critical for replication and possibly for transcription initiation, but not necessarily for FACT function in nucleosome recovery during transcription through chromatin by RNA polymerase II 12.
Materials and Methods
Proteins
Nhp6 was expressed in Escherichia coli and purified as described 41,42. Spt16/Pob3 was purified as the intact heterodimer from yeast cells overexpressing both proteins 43,44. The Spt16-11 (T828I, P859S) 45 mutations were introduced into expression constructs using the Quikchange strategy (Stratagene), and the proteins were purified as described 28,41,46.
DNA templates
Nucleosomal DNA templates were amplified by PCR with the following fluorescently-labeled primers.
For template N35/112, _Fw 5′–ACCCCAGGGACTTGAAGTAATAAGGACGGAGGGCCT#CTTTCAACATCGAT (where T# - is a nucleotide labeled with Cy3), Rev 5′ – CAAGCGACACCGGCACTGGGCCCGGTTCGCGCTCCCT CCTTCCGTGTGTTGTCGTCTCT (where T* - is a nucleotide labeled with Cy5).
For template N13/91: Fw 5′– AAGCGACACCGGCACTGGGCCCGGTTCGCGCT*CCCGCCTTCCGTGTGTTGTCGT CTCTCGGGCGT (where T* - is a nucleotide labeled with Cy3), _Rev 5′ – ACCCCAGGGACTTGAAGTAAT AAGGACGGAGGGCCTCTTTCAACATCGATGCACGGT#GGTTAG (where T# - is a nucleotide labeled with Cy5).
For template N57/135:Fw 5′–ACACCGGCACTGGGCCCGGTTCGCGCTCCCTCCTTCCGTGTGTTGTCGTCTCTC GGGCGTCTAAGTACGCT*TAGGC (where T* - is a nucleotide labeled with Cy3), Rev 5′ - ACCCCAGGGACTT#GA AGTAATAAG (where T# - is a nucleotide labeled with Cy5).
Plasmid containing the modified 603–42 sequence 30 was used as a template. DNA molecules were purified with a QIAquick PCR Purification Kit (Qiagen).
Nucleosome assembly and purification
Nucleosomes were assembled by octamer transfer from chicken erythrocyte donor -H1 chromatin after dialysis from 1M NaCl as described earlier 47 or with recombinant Xenopus laevis octamers (wild type or H2A-V101I) purified as described 38 Nucleosomes were purified from the octamer exchange reaction components by electrophoretic separation in 4.5% polyacrylamide gel under non-denaturing conditions in buffer HE (10 mM HEPES-NaOH, pH 8.0, 0.2 mM EDTA) at 4°C. Pre-electrophoresis was performed until the current strength decreased and stabilized (about 2 h). Additional pre-electrophoresis was performed with fresh running buffer before loading of samples. Nucleosomes were loaded in 10% sucrose. Detection was performed on a PhosphorImager (General Electric, UK). The gel fragment containing mononucleosomes was crashed and mixed with an equal volume of the HE/BSA buffer (10 mM HEPES-NaOH, pH 8.0, 0.2 mM EDTA, 200 mg/ml BSA), incubated 12 h at 4 °C, then washed with 50 – 100 μL of HE/BSA buffer. Gel fragments were removed by centrifugation and the supernatant containing nucleosomes recovered.
Incubation of the nucleosomes with FACT
Formation of FACT complexes with nucleosomes was evaluated using an electrophoretic mobility shift assay (Fig. 2) under conditions used previously 28 (incubation in 17 mM HEPES pH 7.6, 2 mM Tris-HCl pH 7.5, 0.8 mM Na3EDTA, 0.11 mM 2-mercaptoethanol, 11 mM NaCl, 1.1% glycerin, 12% sucrose).
Incubation of proteins and nucleosomes for spFRET experiments and electrophoretic separation (Fig. 4) was performed in TB (transcription buffer): 20 mM Tris–HCl (pH 8.0), 5 mM MgCl2, 2 mM 2-mercaptoethanol, and 150 mM KCl 33.
Spt16/Pob3 (WT) was used at a final concentration 0.13 μM and Nhp6 at a final concentration of 1.3 μM. Spt16-11/Pob3 was used at a final concentration 0.26 μM (or 0.52 μM, Fig. 4) together with Nhp6 at a final concentration of 2.6 μM. Nucleosomes were added at a final concentration of 0.4 nM.
spFRET measurements
spFRET measurements in solution and calculations were performed as described 33. In-house developed software was used to analyze the data, the code can be provided upon request.
The proximity ratio was calculated as
| (1) |
where IAa is Cy5 fluorescence intensity in the Cy5 detection channel, IDd is Cy3 fluorescence intensity in the Cy3 detection channel (both corrected for background), α is the contribution of Cy3 fluorescence in the Cy5 detection channel (spectral cross-talk) calculated as
| (2) |
where IDa is Cy3 fluorescence intensity in the Cy5 detection channel corrected for background. Proximity ratios EPR were calculated for (0.5–15)×103 single nucleosomes for each experimental sample and presented as a relative frequency distribution plot. This plot was further fitted with a sum of two Gaussians (with a goodness of fit R2 = 0.83–0.96). Mean peak values and standard errors were calculated based on three independent experiments and are described in figure legends. Reproducibility of the results was verified in at least three independent experiments, and typical representative plots are presented in the figures. Over 500 molecules were included in each spFRET experiment to ensure that the experimental margin of error in the mean value of each distinct FRET state across the three experiments is less than 5%.
Modeling of DNA uncoiling from the histone octamer
To estimate the FRET efficiencies for all pairs of dyes in different stages of DNA uncoiling, a set of static molecular models was built based on the nucleosome structure 1KX5 48 Uncoiled models were created with the 3DNA software package 49 by straightening the DNA coils by several bp from one or the other end of the nucleosome (a total of 10658 structures in different uncoiling states were analyzed). FRET efficiencies were calculated from the distances between the fluorescent labels which were modeled as described 50 In short, labels were placed on short DNA oligomers and modeled by molecular dynamics simulations, then the average position of chromophores with respect to the labeled nucleotide was calculated. Average positions of the labels were superimposed on the models of DNA uncoiling, FRET efficiencies were calculated from the distances between the average positions. The Förster radius for Cy3-Cy5 label pair was assumed to be 5.6 nm.
Supplementary Material
Acknowledgments
We thank Daria Gaykalova for help with designing the fluorescent probes. This work was supported by NIH grants GM58650 to V.M.S. and R01GM064649 to T.F., and by the Program of the Presidium of the Russian Academy of Sciences “Basic Research for the Development of Biomedical Technologies” (FIMT-2014-011). Part of the work was performed using the equipment of the Center for Collective Use “Genom” of the Institute of Molecular Biology, Russian Academy of Sciences (http://www.eimb.ru/RUSSIAN_NEW/INSTITUTE/ccu_genome_c.php) supported by the Ministry of Education and Science of the Russian Federation (Agreement no. 14.621.21.0001, unique project identification number RFMEFI62114X0001). Development and applications of spFRET were supported by the Russian Science Foundation grant 14-24-00031. Facilities of the Supercomputing Center of Lomonosov Moscow State University were used for the modeling of FRET in nucleosomes.
Footnotes
Supplementary Materials
Movies S1–S4
Author contributions: M.E.V. constructed templates, designed and performed spFRET experiments, analyzed spFRET data, performed gel-shift and native gel experiments, wrote the manuscript; G.A.A. designed and performed computer modelling, developed a program for spFRET raw data analyzes, contributed to writing the manuscript; K.S.K. designed and managed spFRET experiments; N.S.G. designed and performed fluorescent marker for gel-shift experiments, interpreted results, designed some experiments; A.K.S. designed computer modelling, wrote the manuscript; O.I.K. designed templates and analyzed the data; L.L.McC. purified yFACT and some histones; T.F. wrote the manuscript, interpreted data; P.G.G. interpreted data; M.P.K. interpreted data; V.M.S. purified donor chromatin, designed experiments, interpreted results and wrote the manuscript; A.V.F. designed spFRET experiments, interpreted results and wrote the manuscript.
References and Notes
- 1.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Shaytan AK, Landsman D, Panchenko AR. Nucleosome adaptability conferred by sequence and structural variations in histone H2A-H2B dimers. Curr Opin Struct Biol. 2015;32:48–57. doi: 10.1016/j.sbi.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kulaeva OI, Hsieh FK, Chang HW, Luse DS, Studitsky VM. Mechanism of transcription through a nucleosome by RNA polymerase II. Biochim Biophys Acta. 2013;1829:76–83. doi: 10.1016/j.bbagrm.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gurard-Levin ZA, Quivy JP, Almouzni G. Histone chaperones: assisting histone traffic and nucleosome dynamics. Annu Rev Biochem. 2014;83:487–517. doi: 10.1146/annurev-biochem-060713-035536. [DOI] [PubMed] [Google Scholar]
- 5.Park YJ, Luger K. Histone chaperones in nucleosome eviction and histone exchange. Curr Opin Struct Biol. 2008;18:282–289. doi: 10.1016/j.sbi.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ransom M, Dennehey BK, Tyler JK. Chaperoning histones during DNA replication and repair. Cell. 2010;140:183–195. doi: 10.1016/j.cell.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belotserkovskaya R, Saunders A, Lis JT, Reinberg D. Transcription through chromatin: understanding a complex FACT. Biochim Biophys Acta. 2004;1677:87–99. doi: 10.1016/j.bbaexp.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 8.Formosa T. The role of FACT in making and breaking nucleosomes. Biochim Biophys Acta. 2012;1819:247–255. doi: 10.1016/j.bbagrm.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hondele M, Ladurner AG. Catch me if you can: how the histone chaperone FACT capitalizes on nucleosome breathing. Nucleus. 2013;4:443–449. doi: 10.4161/nucl.27235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winkler DD, Luger K. The histone chaperone FACT: structural insights and mechanisms for nucleosome reorganization. J Biol Chem. 2011;286:18369–18374. doi: 10.1074/jbc.R110.180778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brewster NK, Johnston GC, Singer RA. A bipartite yeast SSRP1 analog comprised of Pob3 and Nhp6 proteins modulates transcription. Mol Cell Biol. 2001;21:3491–3502. doi: 10.1128/MCB.21.10.3491-3502.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Formosa T, et al. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 2001;20:3506–3517. doi: 10.1093/emboj/20.13.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reinberg D, Sims RJ., 3rd de FACTo nucleosome dynamics. The Journal of biological chemistry. 2006;281:23297–23301. doi: 10.1074/jbc.R600007200. [DOI] [PubMed] [Google Scholar]
- 14.Formosa T. FACT and the reorganized nucleosome. Mol Biosyst. 2008;4:1085–1093. doi: 10.1039/b812136b. [DOI] [PubMed] [Google Scholar]
- 15.Mandemaker IK, Vermeulen W, Marteijn JA. Gearing up chromatin: A role for chromatin remodeling during the transcriptional restart upon DNA damage. Nucleus. 2014;5:203–210. doi: 10.4161/nucl.29085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erkina TY, Erkine A. ASF1 and the SWI/SNF complex interact functionally during nucleosome displacement, while FACT is required for nucleosome reassembly at yeast heat shock gene promoters during sustained stress. Cell Stress Chaperones. 2015;20:355–369. doi: 10.1007/s12192-014-0556-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahata S, Yu Y, Stillman DJ. FACT and Asf1 regulate nucleosome dynamics and coactivator binding at the HO promoter. Mol Cell. 2009;34:405–415. doi: 10.1016/j.molcel.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheung V, et al. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 2008;6:e277. doi: 10.1371/journal.pbio.0060277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jamai A, Imoberdorf RM, Strubin M. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol Cell. 2007;25:345–355. doi: 10.1016/j.molcel.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 20.Voth WP, et al. A role for FACT in repopulation of nucleosomes at inducible genes. PLoS ONE. 2014;9:e84092. doi: 10.1371/journal.pone.0084092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsieh FK, et al. Histone chaperone FACT action during transcription through chromatin by RNA polymerase II. Proc Natl Acad Sci USA. 2013;110:7654–7659. doi: 10.1073/pnas.1222198110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belotserkovskaya R, et al. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 23.Winkler DD, Muthurajan UM, Hieb AR, Luger K. Histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events. J Biol Chem. 2011;286:41883–41892. doi: 10.1074/jbc.M111.301465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stuwe T, et al. The FACT Spt16 ‘peptidase’ domain is a histone H3-H4 binding module. Proc Natl Acad Sci USA. 2008;105:8884–8889. doi: 10.1073/pnas.0712293105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hondele M, et al. Structural basis of histone H2A-H2B recognition by the essential chaperone FACT. Nature. 2013;499:111–114. doi: 10.1038/nature12242. [DOI] [PubMed] [Google Scholar]
- 26.Kemble DJ, McCullough LL, Whitby FG, Formosa T, Hill CP. FACT Disrupts Nucleosome Structure by Binding H2A–H2B with Conserved Peptide Motifs. Mol Cell. 2015;60:294–306. doi: 10.1016/j.molcel.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jamai A, Puglisi A, Strubin M. Histone chaperone spt16 promotes redeposition of the original h3–h4 histones evicted by elongating RNA polymerase. Mol Cell. 2009;35:377–383. doi: 10.1016/j.molcel.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 28.Xin H, et al. yFACT induces global accessibility of nucleosomal DNA without H2A-H2B displacement. Mol Cell. 2009;35:365–376. doi: 10.1016/j.molcel.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaykalova DA, et al. Structural analysis of nucleosomal barrier to transcription. Proc Natl Acad Sci USA. 2015;112:E5787–5795. doi: 10.1073/pnas.1508371112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kulaeva OI, et al. Mechanism of chromatin remodeling and recovery during passage of RNA polymerase II. Nat Struct Mol Biol. 2009;16:1272–1278. doi: 10.1038/nsmb.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vasudevan D, Chua EYD, Davey CA. Crystal structures of nucleosome core particles containing the ‘601’ strong positioning sequence. J Mol Biol. 2010;403:1–10. doi: 10.1016/j.jmb.2010.08.039. [DOI] [PubMed] [Google Scholar]
- 32.Morozov AV, et al. Using DNA mechanics to predict in vitro nucleosome positions and formation energies. Nucleic Acids Res. 2009;37:4707–4722. doi: 10.1093/nar/gkp475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kudryashova KS, et al. Preparation of mononucleosomal templates for analysis of transcription with RNA polymerase using spFRET. Methods Mol Biol. 2015;1288:395–412. doi: 10.1007/978-1-4939-2474-5_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gurunathan K, Levitus M. Single-molecule fluorescence studies of nucleosome dynamics. Curr Pharm Biotechnol. 2009;10:559–568. doi: 10.2174/138920109788922074. [DOI] [PubMed] [Google Scholar]
- 35.Koopmans WJA, Brehm A, Logie C, Schmidt T, van Noort J. Single-pair FRET microscopy reveals mononucleosome dynamics. J Fluoresc. 2007;17:785–795. doi: 10.1007/s10895-007-0218-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Böhm V, et al. Nucleosome accessibility governed by the dimer/tetramer interface. Nucleic Acids Res. 2011;39:3093–3102. doi: 10.1093/nar/gkq1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mihardja S, Spakowitz AJ, Zhang Y, Bustamante C. Effect of force on mononucleosomal dynamics. Proc Natl Acad Sci USA. 2006;103:15871–15876. doi: 10.1073/pnas.0607526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCullough L, et al. Insight Into the Mechanism of Nucleosome Reorganization From Histone Mutants That Suppress Defects in the FACT Histone Chaperone. Genetics. 2011;188:835–846. doi: 10.1534/genetics.111.128769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zlatanova J, Bishop TC, Victor JM, Jackson V, van Holde K. The nucleosome family: dynamic and growing. Structure. 2009;17:160–171. doi: 10.1016/j.str.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 40.Tsunaka Y, Fujiwara Y, Oyama T, Hirose S, Morikawa K. Integrated molecular mechanism directing nucleosome reorganization by human FACT. Genes Dev. 2016;30:673–686. doi: 10.1101/gad.274183.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruone S, Rhoades AR, Formosa T. Multiple Nhp6 molecules are required to recruit Spt16-Pob3 to form yFACT complexes and to reorganize nucleosomes. J Biol Chem. 2003;278:45288–45295. doi: 10.1074/jbc.M307291200. [DOI] [PubMed] [Google Scholar]
- 42.Paull TT, Johnson RC. DNA looping by Saccharomyces cerevisiae high mobility group proteins NHP6A/B. Consequences for nucleoprotein complex assembly and chromatin condensation. J Biol Chem. 1995;270:8744–8754. doi: 10.1074/jbc.270.15.8744. [DOI] [PubMed] [Google Scholar]
- 43.Biswas D, Yu Y, Prall M, Formosa T, Stillman DJ. The yeast FACT complex has a role in transcriptional initiation. Mol Cell Biol. 2005;25:5812–5822. doi: 10.1128/MCB.25.14.5812-5822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wittmeyer J, Joss L, Formosa T. Spt16 and Pob3 of Saccharomyces cerevisiae form an essential, abundant heterodimer that is nuclear, chromatin-associated, and copurifies with DNA polymerase alpha. Biochemistry. 1999;38:8961–8971. doi: 10.1021/bi982851d. [DOI] [PubMed] [Google Scholar]
- 45.Formosa T, et al. Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway: polymerase passage may degrade chromatin structure. Genetics. 2002;162:1557–1571. doi: 10.1093/genetics/162.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rhoades AR, Ruone S, Formosa T. Structural features of nucleosomes reorganized by yeast FACT and its HMG box component, Nhp6. Mol Cell Biol. 2004;24:3907–3917. doi: 10.1128/MCB.24.9.3907-3917.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kireeva ML, et al. Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol Cell. 2002;9:541–552. doi: 10.1016/s1097-2765(02)00472-0. [DOI] [PubMed] [Google Scholar]
- 48.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 A resolution. J Mol Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 49.Lu XJ, Olson WK. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat Protoc. 2008;3:1213–1227. doi: 10.1038/nprot.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klose D, et al. Simulation vs. reality: a comparison of in silico distance predictions with DEER and FRET measurements. PLoS ONE. 2012;7:e39492. doi: 10.1371/journal.pone.0039492. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.