ABSTRACT
From their initial identification as ‘nucleolar accessory bodies’ more than a century ago, the relationship between Cajal bodies and nucleoli has been a subject of interest and controversy. In this review, we seek to place recent developments in the understanding of the physical and functional relationships between the 2 structures in the context of historical observations. Biophysical models of nuclear body formation, the molecular nature of CB/nucleolus interactions and the increasing list of joint roles for CBs and nucleoli, predominantly in assembling ribonucleoprotein (RNP) complexes, are discussed.
KEYWORDS: Cajal body, coilin, non-coding RNAs, nuclear structure, nuclear bodies, nucleolus, phase separation, snoRNPs, snRNPs, telomerase
Introduction
The nuclear bodies termed Cajal bodies (CBs, for a review see ref.1) were originally identified as silver-stained spherical bodies in vertebrate neural cells by Ramon Y Cajal in 1903.2 Cajal described the bodies that now bear his name as ‘nucleolar accessory bodies’ due to their frequent close proximity to the nucleolus, the most prominent nuclear substructure. The close physical relationship was confirmed by electron microscopy analyses in the 1990s that revealed close association of CBs with nucleoli in the majority of rat neuronal cells in culture and in situ. CBs found at the nucleolar periphery share the ‘coiled thread’ morphology characteristic of their nucleoplasmic counterparts3 and are clearly structurally distinct from the nucleolus, though it can be difficult to clearly delimit the border between the two structures (Fig. 1).4 Intimate association of the two has also been observed in most plant cells.5 Physical contact between CBs and nucleoli is observed only infrequently in transformed cells, however,6 which has proved a challenge for studying CB/nucleolus interactions in easily-cultured cell lines.
Figure 1.
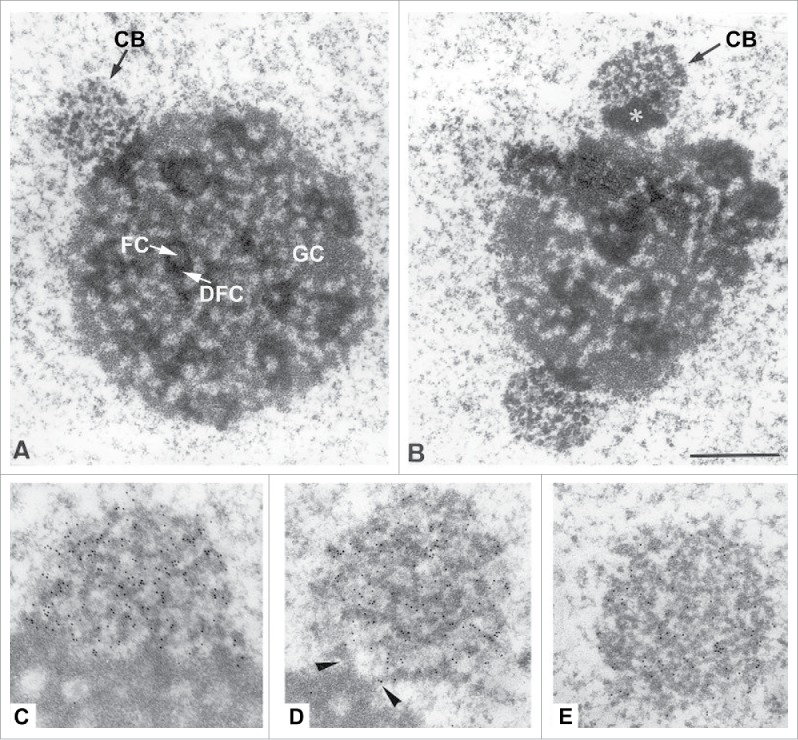
Electron Micrographs showing the relationship between nucleoli and Cajal bodies in rat trigeminal ganglion neurons. A and B) CBs juxtaposed to nucleoli. Note the typical structure of the nucleolus with fibrillar centers (FC), dense fibrillar components (DFC) and granular components (GC). In B, the CB (arrow) appears to be associated with a segregated mass of dense material (asterisk) in close proximity to the nucleolar surface C to D) Immunoelectron localization of coilin. With the anticoilin antibody a high density of immuno-gold particles (small black dots) specifically decorated the dense coiled threads of CBs. In C, an extensive portion of the CB is directly apposed on the dense fibrillar component of the nucleolus, whereas the CB of D maintains a minimal connection with this nucleolar component (arrowheads), and the CB of E appears free into the nucleoplasm. Adapted with permission from.4
In addition to the close physical relationship indicated by these early studies, detailed analyses of these structures have suggested a close functional relationship. This includes the presence of nucleolar proteins such as fibrillarin, nucleolin, Nopp140 and NAP57 in nucleolar-associated and free nucleoplasmic CBs (Fig. 2; for a review see ref.7), and the transient trafficking of CB-resident proteins through nucleoli and nucleolus-resident RNA species through CBs that has been revealed by photokinetic approaches.8,9 Numerous studies have implicated CBs in the modification of non-coding RNAs including those forming the core of snoRNPs (small nucleolar ribonucleoproteins). snoRNPs are required for ribosomal RNA (rRNA) processing by the nucleolus as part of its key role as the site of ribosome biogenesis.10 Evidence also points to the participation of both structures in the sub-cellular localization and function of specific components of the ribonucleoprotein enzyme telomerase, in RNA interference pathways and in the coordinated cellular response to stresses such as DNA damage. This review seeks to place recent developments in our understanding of the relationship between CBs and nucleoli in the context of the emerging appreciation of the biology of non-coding RNAs and the biophysical processes governing nuclear organization.
Figure 2.
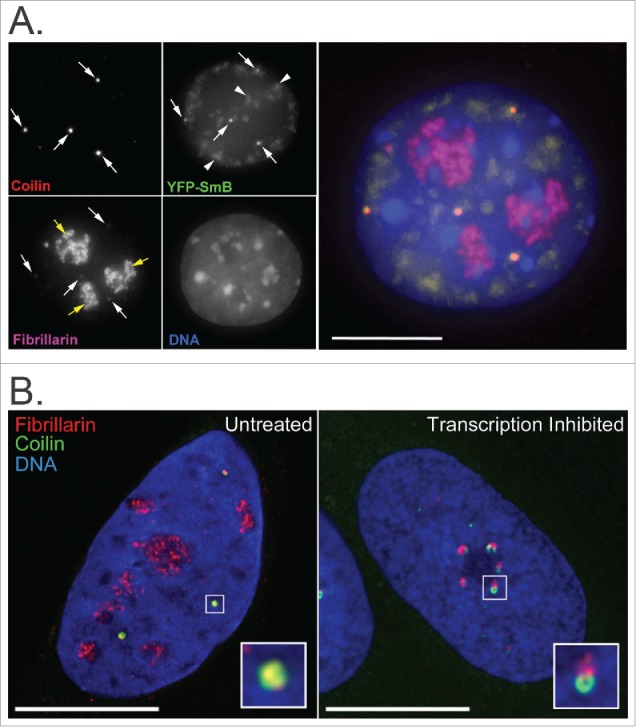
Immunofluorescence images showing co-localization of nucleolar and CB factors. (A) Fibrillarin and SmB co-localize with Coilin at CBs (white arrows) in mouse embryonic fibroblast cells. Coilin (red) was detected using anti-coilin primary and anti-rabbit-Cy5 secondary antibodies, Fibrillarin (magenta) using anti-Fib primary and anti-mouse-TRITC secondary antibodies, SmB (green) by overexpression of YFP-tagged SmB for 48 hours and DNA (blue) by DAPI staining. Fibrillarin is also present in nucleoli (yellow arrows). SmB is also present in speckles (arrowheads). Bar = 10 μm. (B) In untreated U2OS cells, a subset of Fibrillarin (red) accumulates with Coilin (green) in CBs (boxed region, expanded as inset). Upon inhibition of Pol I- and Pol II-mediated RNA transcription by Actinomycin D treatment (2.5 ug/ml for 2 hrs), Fibrillarin and Coilin aggregate in distinct peri-nucleolar caps (boxed region, expanded as inset). Fibrillarin was detected using anti-fibrillarin primary and anti-mouse-Alexa568 secondary antibodies, Coilin using anti-coilin primary and anti-rabbit-Alexa488 secondary antibodies and DNA by DAPI staining. Bar = 10 μm.
Coilin in/at the nucleolus
The protein, coilin was originally identified as an antigen recognized by human autoimmune antibodies that showed accumulation at CBs by light and electron microscopy.11-13 The name ‘coilin’ derives from the term ‘coiled body’ commonly used for CBs at the time due to their ‘coiled thread’ morphology in electron microscopy images.3 Homologs of coilin have since been identified in many species and it is widely used as a marker for CBs, although reliance on this single marker needs to be treated with some caution.11,12,14,15
Nucleolar accumulation of coilin has been observed in a range of primary and cultured cells over the years, as summarized in Table 1.6,16-37 Although this accumulation is often seen in response to a particular cellular perturbation or to mutations induced in coilin, nucleolar coilin has also been observed under normal physiological conditions in certain cell types (Table 1A). Coilin can localize within the nucleolus, or to distinct caps at the edge of the nucleolus. Intranucleolar coilin has been shown shown to colocalize with the rRNA methyltransferase Fibrillarin in the Dense Fibrillar Component (DFC), while coilin in peri-nucleolar caps remains distinct from adjacent Fibrillarin-containing caps (Fig. 2B).
Table 1.
Studies in which a pool of coilin (in all or a subset of cells) was detected in or surrounding the nucleolus. KO = knockout; KD = knockdown.
| A. Coilin detected in nucleolus | ||
|---|---|---|
| Condition | Cell line(s) | Reference |
| Neuronal cells | Rat primary neurons | 6 |
| Hibernating dormice | Dormouse liver sections | 29 |
| Breast cancer cells | Hep2, MOT4, HBL100, T47D, MCF7 | 30 |
| Inhibition of serine/threonine phosphatase activity | HeLa | 28 |
| Inhibition of CRM1-dependent nuclear export | HeLa | 19 |
| Coilin truncation mutants lacking C-terminus (1–248 and 1–315) | HeLa | 25 |
| High overexpression of exogenous coilin | MEFs (coilin KO), HeLa | 33 |
| Depletion of SMN | HeLa | 22, 27, 32 |
| Depletion of the methyltransferase TGS1 | HeLa | 27 |
| Depletion of the FLICE-associated huge protein FLASH | SAOS2, IMR90, HeLa, MCF7, MEFs | 16 |
| Nonphosphorylatable coilin mutants | HeLa (normal and coilin KD), WI38 | 24 |
| Hypomethylated coilin | MCF7MTAP−/−, HeLa | 34 |
| Depletion of the telomerase complex member WRAP53 | U2OS, HeLa | 36 |
| Pcd (Purkinje cell degeneration) ataxia mouse model | Mouse primary neurons | 37 |
| SMA motor neurons | Human primary motor neurons | 35 |
| Depletion of the SUMO isopeptidase USPL1 |
HeLa, U2OS |
26 |
| B. Coilin detected in perinucleolar caps | ||
| Condition |
Cell line(s) |
Reference |
| Inhibition of RNA transcription | HeLa | 18 |
| Coilin truncation mutant lacking N- and C-termini (94–291) | HeLa | 17 |
| Depletion of SMN | HeLa | 23, 32 |
| Inhibition of RNA transcription | HeLa, U2OS | 31 |
| UVC-induced DNA damage | Various mammalian lines | 20 |
| Depletion of PRMT5 or PRMT7 | HeLa | 23 |
| Cisplatin or gamma-irradiation-induced DNA damage | HeLa, WI38, SAOS2 | 21 |
Analysis of coilin truncation mutants revealed intranucleolar accumulation of the N-terminal 1–248 and 1–315 regions when exogenously over-expressed in cells (Fig. 3; Hebert 2000; Bohmann 1995),25,38 while over-expression of a central fragment (94–291) showed accumulation at nucleolar caps, along with re-organization of nucleolar structure and inhibition of RNA PolI transcription.17 A cryptic nucleolar localization signal (NoLS) similar to that found in the E3 ubiquitin ligase MDM2 was identified at aa160-168 (Fig. 3), and mutation of its basic residues to neutral residues significantly reduced nucleolar accumulation of the N-terminal fragments 1–248 and 1–315.25 Two acidic patches of amino acids downstream of this region (aa242-259 and aa312-325; Fig. 3) were also identified, and mutation of the serine residues within the patches to alanine residues in full-length coilin caused nucleolar accumulation. Interestingly, when all of these regions (the NoLS and two acidic patches) were mutated simultaneously in full-length coilin, there was no effect on its localization. Although the specific mechanism remains unclear, these studies suggest that coilin's nucleolar association may be subject to regulation by post-translational modification.
Figure 3.
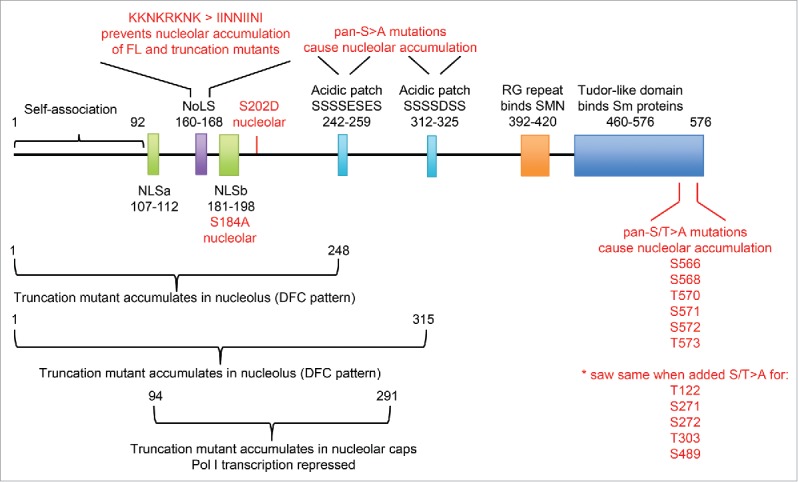
Summary of coilin mutations that affect the protein's association with the nucleolus. Diagram of the protein structure of coilin, highlighting regions that have been linked to regulation of its association with the nucleolus.
Phosphorylation and methylation, both of which have been shown to occur on coilin, have been explored as potential explanations for the occasional presence of a nucleolar pool. An early study using okadaic acid to inhibit serine/threonine protein phosphatase activity showed accumulation of coilin and Sm proteins (normally core components of snRNP splicing factors) in discrete nucleolar regions, consistent with the trafficking of both factors through the nucleolus.28 An exogenously overexpressed phospho-mimic mutant (coilin S202D; Fig. 3) showed a similar accumulation, however phosphorylation at this site was only recently detected in vivo by mass spectrometry (www.proteomicsdb.org) and the functional relevance has not yet been demonstrated. To complicate matters, nucleolar accumulation of coilin is also observed when other serine/threonine residues are mutated to non-phosphorylatable alanine residues (Fig. 3;24,25). With regard to arginine methylation, hypomethylated coilin has been detected in the DFC of the nucleolus in cells that lack a key enzyme in the methionine salvage pathway,34 while depletion of the protein arginine methyl transferases PRMT5 and PRMT7 that methylate coilin induces relocalization to peri-nucleolar caps in HeLa cells23 albeit not in neuroblastoma cells.39 This suggests that regulation of the sub-nuclear localization of coilin is more complex than a simple phosphorylation and/or methylation on/off switch and may vary in different cell types.
Accumulation of coilin in peri-nucleolar caps has also been observed under various other conditions (Table 1B). Formation of these caps, which is accompanied by structural reorganization of the nucleolus, has long been known to occur downstream of the inhibition of rRNA transcription, either directly by drug treatment31,40 or following specific stresses such as DNA damage.21 It is also seen during the transcriptional shut-down and subsequent loss of the nucleus associated with formation of the ocular lens in vivo.41
Overall, these observations suggest that intra-nucleolar coilin accumulation can occur as part of normal transport pathways, while the peri-nucleolar capping phenotype is mainly associated with cellular stress responses that result in shut-down of rRNA transcription.
CBs in the nucleolus
Based on the early ultrastructural analyses of CBs and nucleoli in the rat nervous system that revealed a range of degrees of physical association between the two structures, it was originally suggested that CBs could both fuse with and bud from the nucleolar periphery coupled with the advent of green fluorescent protein technology and its application to nuclear dynamics, allowed this to be tested in live mammalian and plant cells and tissues by time-lapse fluorescence imaging.42-46 In these studies, CBs were observed to fuse and bud within the nucleoplasm, occasionally to form CBs with differing protein compositions44 and to fuse with each other at the nucleolar periphery.42 They were not, however, observed to bud from or fuse with nucleoli.
More recently, a number of other proteins sometimes found in CBs, including the transport factor CRM1 and the small ubiquitin-related modifier SUMO1,47,48 have also been found in discrete structures within nucleoli. These have been described as “intranucleolar bodies” (INBs) that are up-regulated by certain types of DNA damage,49 and as “Crm1 nucleolar bodies”.50 These novel structures, however, contain a relatively low concentration of coilin49 and do not show an ultra-structural appearance typical of CBs,51 so are unlikely to represent intra-nucleolar CBs. The complexity of the nuclear localizations of proteins shared by nucleoli and CBs and of coilin itself, which also shows a substantial diffuse nucleoplasmic pool, continues to make the definite identification of CBs controversial, but current evidence does not appear to support the presence of CBs within nucleoli.
Physical properties of nucleoli and CBs
A longstanding question in the field of sub-nuclear organization has been how non-membranous nuclear bodies such as CBs and nucleoli form and maintain their distinct structures. It is generally accepted that they function to concentrate proteins and RNAs involved in related processes in a confined space, in order to enhance reaction efficiency and facilitate regulation. Models of self-organization, either stochastic or hierarchical, have thus been proposed, with evidence suggesting that a combination of both may drive nuclear body assembly.52 In the case of nucleoli, assembly has been shown to be triggered by activation of rDNA transcription and accumulation of rRNA transcripts at ribosomal gene loci.53 Although the mechanics of CB formation have not yet been studied to the same extent, it has been shown that CBs can be induced by tethering related protein or RNA components to engineered gene loci,54,55 suggesting that a similar activity-driven self-assembly may occur.
The physical step that occurs after an initial “seeding” event to form a gelatinous body from soluble constituents is believed to be a phase transition/separation driven by underlying protein-protein and protein-RNA interactions.56-59 This area of research is providing surprising insight into the potential for these interactions and their consequences to explain the dynamic assembly/disassembly and maintenance of non-membraneous nuclear organelles (for reviews see refs.60-62).
These sol-gel phase transitions are mediated by multiple weak binding interactions between intrinsically disordered low complexity sequences, which are enriched in many RNA- and DNA-binding proteins. Recently, low complexity domains in paraspeckle-localized proteins were implicated in the formation of this sub-nuclear body.63 Although not yet demonstrated, it is likely that CB formation involves similar physical events. The process has been shown to be temperature-dependent,64 for example, while p80-coilin, in common with other proteins shown to drive phase transitions, can self-associate,25 binds RNA,65,66 and contains a central low complexity region.15
Consistent with this general model for nuclear organelle formation, nucleoli in Xenopus oocytes have been demonstrated to behave as viscous fluid droplets that can fuse into larger bodies.67 A more recent study in C.elegans embryos suggested that the concentration of nucleolar components must achieve a certain minimum concentration in order for nucleoli to assemble,68 and that active processes such as rRNA transcription contribute to formation via modulation of effective thermodynamic interaction parameters.69 This, along with the demonstration that purified recombinant nucleolar proteins such as fibrillarin and nucleophosmin can phase separate into liquid droplets mediated by low complexity regions in their amino acid sequences,70 strengthens the argument that nucleolar assembly represents an intracellular phase transition.
That said, refinements to the model would clearly be required in order to explain complex interactions between different sub-nuclear structures such as that observed between the CB and nucleolus. These structures share many protein and RNA components and, under certain conditions, an intimate physical connection, yet remain physically and structurally distinct.
What mediates the physical interaction between nucleoli and CBs?
A variety of nuclear bodies show close interactions with each other, but in many cases it is not yet clear what molecular mechanisms mediate these associations. In the case of nucleoli and CBs, there are many possible factors that may act as the molecular ‘glue’ linking them together. The association of CBs with snRNA gene loci in mammalian cells is known to be dependent on the on-going transcription of snRNA.71-73 Recent work has also implicated the CB-localized SUMO isopeptidase USPL1 as a protein factor mediating the interaction of CBs with snRNA genes. USPL1, though, is thought to promote CB/snRNA gene interactions through an effect on snRNA transcription, resulting in a somewhat circular argument placing RNA ‘in the frame’ for mediating the connection.26 Given that the nucleolus is, essentially, a structure built around transcriptionally active rRNA gene loci,74 there may be similar RNA-dependent mechanisms mediating the association of CBs with nucleoli, but no clear model for this has yet been proposed. Studies using UV cross-linking and immunoprecipitation (CLIP) to identify RNA species able to interact with GFP- coilin in human and mouse cells66 identified rRNAs, including PolI-transcribed 28S and 18S and PolII-transcribed 5S rRNAs. Full validation of these potential interactions was, however, outside the scope of the published study. Interestingly, though, coilin has previously been proposed to interact with 47/45S pre-rRNA, based on immunoprecipitation of endogenous coilin from HeLa cells followed by quantitative real-time reverse transcriptase PCR.65 Taken together, these data suggest that newly transcribed rRNA may indeed be instrumental in tethering CBs to nucleoli.
Electron microscopic examination of nucleoli in plants75 identified CBs in the immediate neighborhood of ‘nucleolar organizer tracks’, which likely represent the rDNA chromatin (for a review see ref.76), although few apparent physical connections were observed. Two recent studies directly probed for interactions between CBs and chromosomal DNA. One used GFP-tagged coilin as the bait protein in a chromatin immunoprecipitation/high-throughput DNA sequencing approach (ChIP-seq;66), while the other used the U1 snRNA genomic regions, widely reported to interact with CBs with high frequency, in a genome-wide chromosome conformation capture analysis (4C-Seq;77). Although the results of both suggested extensive interactions between CBs and loci encoding snRNAs, snoRNAs and histone RNAs, neither explicitly examined rDNA sequences. This leaves the question of whether or not CBs can interact directly with rDNA unanswered.
Functional interactions
snoRNP and snRNP assembly and processing
The most extensively studied functional interaction between CBs and nucleoli is that of RNP (ribonucleoprotein) assembly (Fig. 4A). CBs have long been implicated in the assembly of ribonucleoproteins that play essential house-keeping roles within the nucleus. These include the small nucleolar ribonucleoproteins (snoRNPs) required for rRNA processing within the nucleolus and the small nuclear ribonucleoproteins (snRNPs) required for pre-mRNA splicing. Both are formed from highly structured and modified small RNA cores and specific sub-sets of proteins. As highlighted above (Fig. 2), the snoRNP protein fibrillarin, a methyltransferase essential for the snoRNP-directed modification of rRNA is found in both nucleoli and CBs12,64 and was an early marker for the CB that hinted at a role for this structure in snoRNP biogenesis. Furthermore, transient localization of micro-injected box C/D snoRNAs, which form the RNA core of snoRNPs, to CBs prior to their steady-state accumulation in nucleoli was an early observation in a study in Xenopus oocytes.8 One of the snoRNAs used in this study was U3 snoRNA, vital for rRNA processing. Subsequently, it was demonstrated that key stages of U3 snoRNA processing and addition of proteins to the maturing snoRNP complex take place in the CB.78-80 Both of the two main classes of snoRNAs (box C/D and box H/ACA) have been detected in plant and mammalian CBs,81,82 (for a review see ref.83), strongly suggesting that the CB is involved in the processing and/or assembly of a wide range of snoRNPs.
Figure 4.
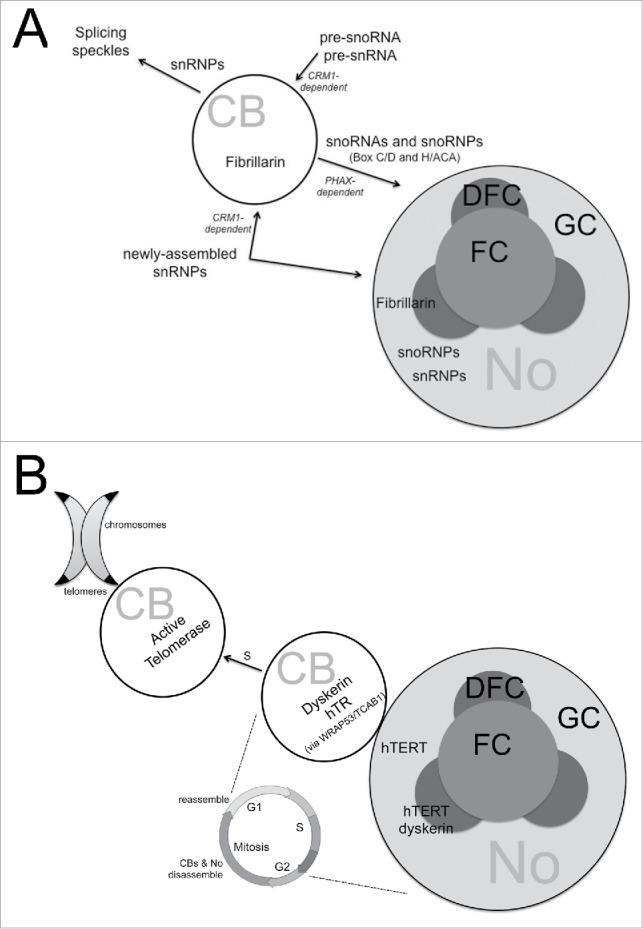
Examples of functional overlap of CBs and nucleoli. (A) RNP biogenesis. CBs have long been implicated in the assembly of both the small nuclear ribonucleoproteins (snRNPs) required for pre-mRNA splicing and the small nucleolar ribonucleoproteins (snoRNPs) required for rRNA processing within the nucleolus. snRNP factors (RNA and protein) have also been shown to shuttle through nucleoli during the maturation process, and to accumulate there in response to certain perturbations. (B) Telomerase assembly and trafficking. In early S-phase, human telomerase reverse transcriptase (hTERT) has ben detected in the granular component (GC) and dense fibrillar component (DFC) of the nucleolus (No), while human telomerase RNA (hTR) is found in nucleolus-associated Cajal bodies (CBs). Dyskerin has been detected in the DFC and CBs. Throughout S-phase, when telomerase is active, CBs containing telomerase (hTERT and hTR) have been seen co-localized with telomeres. The multi-functional protein WRAP53/TCAB1 is required for telomerase retention in CBs.
While the role of CBs in snoRNP assembly is well established, it is not entirely clear whether CBs and nucleoli share roles in splicing snRNP maturation. Splicing snRNPs (U1, U2, U4/5 and U6) comprise a core of snRNA surrounded by a ring of 7 different members of the Sm protein family and a number of additional proteins unique to each snRNP. Splicing snRNPs are found predominantly in CBs and speckles, but accumulate within nucleoli in certain conditions, as discussed in more detail above. In Xenopus oocytes the injection of mutants of U2 snRNA, the RNA core of the U2 snRNP, revealed a correlation between the ability of the snRNA to localize to the nucleolus and the ability of the cell to modify the snRNA.84 In human cells, expression of GFP-tagged members of the Sm protein family results in their accumulation in CBs and nucleoli prior to their accumulation in splicing speckles, suggesting a joint role for the 2 in snRNP assembly.9 However, the localisations of the different Sm proteins within the nucleolus are not identical, so the role of the nucleolus with regard to splicing snRNP maturation and assembly remains unclear.
The traffic of CB-resident snoRNPs and snRNPs between CBs and nucleoli appears to use mechanisms involving the export factors CRM1 and PHAX.48,78,80 A study designed to identify the range of RNA species able to interact with the core CB protein coilin (discussed above),66 revealed that hundreds of non-coding RNAs associate with GFP-coilin. Some are resident in CBs at steady-state while others are not, suggesting that many more small RNAs may traffic through CBs than have previously been identified to reside there. Although small Cajal body RNAs (scaRNAs) and spliceosomal snRNAs, which are well-accepted CB components, were identified in this screen, the most abundant class of RNAs obtained was snoRNAs. Those investigated further by micro-injection of in vitro transcribed RNAs were confirmed to accumulate in CBs as well as in nucleoli, in some cases accumulating in CBs prior to nucleoli. These observations suggest that CBs have a more generalized role in snoRNP assembly and/or maturation than has previously been appreciated.
Telomerase biogenesis and telomere maintenance
Telomerase is a specialized RNP responsible for the maintenance of telomeric DNA and consequent protection of chromosomal ends. Defects in telomere maintenance, primarily their increased stability, are seen in most cancers while diseases such as aplastic anaemia and dyskeratosis congenita result from deficiencies in telomerase (for a review see ref.85). In humans, the telomerase RNP is comprised of telomerase RNA (hTR or TERC) and the telomerase reverse transcriptase enzyme (hTERT), together with the accessory proteins dyskerin, NOP10, NHP2 and GAR1. The telomerase RNA contains an H/ACA motif, which is also found in snoRNAs and in the small Cajal body RNAs (scaRNAs) that reside in CBs.86 hTERT has been detected in nucleoli using GFP-tagged constructs,87,88 while both hTERT and hTR have been detected in nucleoli and in CBs.89,90 Most of the accessory proteins have also been detected in both structures (for a review see ref.91).
CBs and the signature protein coilin are implicated directly in telomere maintenance through a proposed role in delivering the telomerase RNP to telomeres, where it becomes active (Fig. 4B). A sub-set of CBs co-localize with telomeres during S-phase in the cell cycle, when telomerase is active,92 while depletion of coilin can abolish the accumulation of telomerase at telomeres.93 It has also been suggested that CBs have a role in the maturation of the telomerase RNP, analogous to their role in snRNP and snoRNP maturation, although this is not yet clear (for a review see ref.94). The reason for the localization of telomerase to the nucleolus is still more controversial. Detailed examination of the dynamic nature of hTR and hTERT localizations throughout the cell cycle in HeLa cells95 suggested a role for the nucleolus in telomerase biogenesis, though perhaps not actually in the assembly of the telomerase RNP. Early in S-phase, both hTERT and hTR were localized to the nucleolus. However, while hTERT appeared diffuse throughout the nucleolar interior, hTR was found in CBs associated with the nucleolar periphery, often showing a crescent shape around the nucleolus. Only later in S-phase did the two co-localize in nucleoplasmic Cajal bodies.
More recently, electron microscopy has been used to examine the localization of different components of the telomerase RNP within the nucleolus. The reverse transcriptase enzyme, hTERT, localizes to the dense fibrillar and granular components of the nucleolar interior in S-phase and G1-phase HeLa cells, consistent with previous reports.87,88 The telomerase accessory protein dyskerin, however, was found in nucleolar-associated CBs but also in the dense fibrillar region of the nucleolus.96 This is certainly suggestive of a role for the nucleolus in early telomerase assembly, perhaps passing the complex to CBs for further processing and/or transport to telomeres. In support of a role for the nucleolus in telomerase assembly, in induced pluripotent stem cells from a subset of patients with a telomere homeostasis-related disease called dyskeratosis congenital (DC), telomerase activity was found to be normal, albeit mislocalized from CBs to nucleoli.97 These patients had mutations in a scaffold protein called WRAP53 (also known as WDR79 or TCAB1) that has been shown to bind coilin36 and recruit scaRNA and hTR to CBs.98,99 Defects in telomerase trafficking to telomeres associated with this inability of mutant WRAP53 to target the complex to CBs have been suggested to be part of the molecular pathology of DC.100
Other reports do not, however, support a role for the nucleolus in telomere assembly. In fibroblasts, overexpression of GFP-tagged mutants of TERT that are unable to accumulate in the nucleolus resulted in similar increases in telomere stability and extended replicative life-cycle as were obtained by over-expressing wild-type TERT.101 It should be noted, though, that interpretation of results such as these is complicated by the concept that nuclear bodies such as CBs and nucleoli function to concentrate molecular events that can still occur elsewhere in the nucleus.
Alternative explanations for the presence of telomerase components in nucleoli have also been suggested, such as the stimulation of rDNA transcription in rapidly proliferating cells.102 These ideas, together with the number of telomere-independent roles currently being proposed for TERT, including modulation of gene expression through transcriptional modulation and small interfering RNA (siRNA) processing (for a review see ref.103), make it clear that a dual role for nucleoli and CBs in telomerase biogenesis is far from a certainty.
RNA interference (RNAi) pathways
The fine-tuning of gene regulation by various classes of small RNAs, including micro-RNAs (miRNAs), small interfering RNAs (siRNAs) and PIWI-interacting RNAs (piRNAs) is a hugely complex area. All three of these classes of small RNA can target specific mRNA transcripts for degradation. Modulation of gene expression through DNA methylation can also be mediated by small RNAs in the fission yeast S. pombe104 and in plants. Small RNAs of the piRNA class are able to direct chromatin methylation and gene repression in the germlines of C. elegans, Drosophila and mouse105,106 (for a review see ref107), but DNA methylation targeted by small RNAs has not been documented in mammalian somatic cells.
In plants, nucleoli and CBs have been implicated both in transcriptional gene silencing (TGS) mediated through RNAi-directed epigenetic modification of chromatin and in miRNA directed cleavage of mRNA. RNA-directed DNA methylation (RdDM) in plants requires numerous proteins including the argonaute family member ARGONAUTE 4 (AGO4), RNA polymerase IV (PolIV), RNA-DEPENDENT RNA POLYMERASE 2 (RDR2) and DICER-LIKE 3 (DCL3) together with a 24 nucleotide siRNA. Co-localization of several of these (AGO4, DCL3, RDR2 and NRPD1b (the largest subunit of PolIV)) with siRNAs within Arabidopsis nucleoli has been reported, leading to the proposal of a nucleolar processing center for assembly of silencing complexes within the nucleolus.108 The nucleolar signals for all 4 of these proteins were either at the center or at the periphery of the nucleolus, with clear puncta also seen elsewhere within the nucleus. A partner paper,109 revealed that the nucleolus-associated bodies and nuclear puncta containing the RdDM factors also contain several typical Cajal body markers: tri-methylguanosine-capped snRNAs and the snRNP proteins SmD3 and U2B. This added a further dimension to the model proposed, whereby an siRNA complex containing AGO4 and NRPDb1 is assembled by RDR2 and DCL3 in the nucleolus, Cajal body or both, prior to delivery to target loci where it mediates gene silencing via RNA-directed DNA methylation. These studies did not, however, investigate the presence of the Arabidopsis coilin homolog, Atcoilin, in the nuclear bodies.
Several studies on the localization of machinery required for production of miRNAs targeting mRNAs for cleavage have also implicated the nucleolus and nuclear bodies with miRNA processing in plants. Dicer-like (DCL1), HYPONASTIC LEAVES 1 (HYL1) and SERRATE (SE) have all been identified in structures resembling CBs.110,111 However, while these bodies did contain the CB markers SmD3 and SmB, Atcoilin was absent from them. Furthermore, the putative miRNA processing bodies persisted in mutant plants lacking CBs.110 This complexity makes the potential role for CBs in miRNA processing in plants uncertain and emphasizes the difficulty in positively identifying CBs: can a nuclear body containing a number of CB marker proteins but not coilin still be classified as a CB, or is a new classification of nuclear body required in this instance? Ultrastructural analysis of these mi/siRNA processing bodies is currently lacking, but may provide further clarification.
The miRNA-mediated post-transcription gene silencing pathway in mammalian cells, originally believed to be exclusively cytoplasmic, has also been proposed to function to some extent in the nucleus. Several reports have been made of the presence of essential miRNA factors such as Argonaute-2 (Ago2) and Dicer in the nucleus,112-116 and a recent report suggested they may also be active in the nucleus.117 Analysis of RNAs interacting with Ago1 and Ago2 in human cells identified miRNAs believed to be derived from the snoRNA ACA45.118 Given that the argonaute family, together with the miRNA itself, form the core of gene silencing effector complexes, this is suggestive of a role for the nucleolus in mRNA-targeting RNAi pathways. Further evidence in favor of this idea was provided by the discovery of numerous miRNAs in rat myoblast nucleoli, both in precursor and mature forms.119 While some of these had sequence homology to snoRNAs, others did not, suggesting that they were not processed from snoRNAs and that the nucleolus may have a role in assembling the RNA/protein cores of RNAi silencing complexes in mammalian cells.
Additional roles unrelated to RNA processing
The expanding nucleolar and CB proteomes have highlighted other potentially shared functional roles beyond RNA processing pathways. For example, both structures have been linked to stress response in general, and DNA damage response in particular. It was recognized years ago that in addition to its key role in ribosome biogenesis, the nucleolus also functions as a central hub that senses and mediates the cellular response to stresses that include heat shock, starvation, inhibition of transcription and DNA damage. Interestingly, the structure and composition of Cajal bodies is also altered under most of these conditions (for a review see ref120 ). As discussed above, an early observation was the disruption of CBs and relocalization of p80-coilin to the nucleolar periphery in response to transcriptional inhibition.12,18,31 Recently, peri-nucleolar accumulation of coilin was demonstrated in response to cisplatin- and γ-irradiation-induced DNA damage,21 and correlated with inhibition of RNA Pol I activity. Demonstration of previously unknown interactions between coilin and both the polymerase subunit RPA-194 and the transcription factor UBF suggests a causative role, although further work is needed.
The expanding interactome of coilin has also identified associations with DNA repair factors including the Ku proteins, which associate with a catalytic subunit to form the DNA-dependent protein kinase complex mediating non-homologous DNA end joining121 and the multi-functional scaffold protein WRAP53 (also called WDR79 or TCAB136,122), which is involved in retention of telomerase RNA in CBs and in DNA double strand break repair. As with many aspects of CB biology, the interpretation of these observations is hampered by the pleiotropic functions of many CB-resident proteins.
Do the nucleolus and CBs need each other?
As discussed in the preceding sections, it is clear that CBs and nucleoli have key shared functions, with CBs largely fulfilling the role originally proposed for them as nucleolar accessory bodies. While over-expression of mutants of coilin can clearly disrupt nucleolar structure, a key remaining question is whether the nucleolus actually needs CBs to function correctly. Arabidopsis mutants lacking the plant homolog of coilin and containing no discernable CBs are viable. They show no obvious defects in nucleolar structure using immunohistochemistry to detect fibrillarin or at the EM level.123 Likewise, the coilin knock-out mouse appears essentially normal, though with reduced viability. Again, nucleolar structure appears unaltered as judged by staining with anti-fibrillarin antibodies or by the traditional silver-staining method.124 Nucleoplasmic bodies termed ‘residual CBs’ in this study contain the nucleolar proteins fibrillarin and Nopp140 but fail to recruit the snRNP-associated Sm proteins or the Survival of Motor Neurons (SMN) protein required for snRNP assembly. Taken together, these observations suggest that nucleolar structure is not dependent on the presence of CBs or of coilin, but that the presence of CBs, at least in mammals, is an advantage to the organism most likely by increasing the efficiency of nuclear processes such as snRNP and snoRNP assembly.
Conversely, inhibiting the major function of the nucleolus with transcriptional inhibitors has a profound effect on CB structure. This suggests that CBs need nucleoli for their physical integrity. There are, however, certain unusual situations in which CBs may exist in the absence of functional nucleoli. A study using prolonged nocodazole treatment to induce micronucleation of rat-kangaroo kidney cells reported a number of micronuclei containing CB-like structures but not nucleoli.125 In pigeon oocytes, which lack functional nucleoli, CB-like structures, containing coilin and snRNPs but lacking fibrillarin, have been detected,126 suggesting that CBs may, on occasion, be independent from nucleoli.
Overview
From both a physical and a functional perspective, the relationship between CBs and the nucleolus has always been, and remains, complicated. Although their occasional proximity to the nucleolus has been clear since the earliest discovery of CBs more than a century ago, physical interactions between CBs and nucleoli show species-specific and cell type-specific differences and also vary across the cell cycle in cultured cell lines.
The variations in the physical relationship between CBs and nucleoli have been demonstrated in several species by electron microscopy, but the molecular events and interactions mediating this relationship remain elusive. Tantalizing hints from sequencing studies suggest that RNA or DNA may link the 2 structures, with the ability of newly transcribed rRNA to ‘seed’ assembly of the nucleolus making RNA a clear front-runner. Once formed, the nucleolus and, most likely, CBs behave as viscous fluid droplets, but it remains unclear how simple thermodynamic models of nuclear organization can fully explain the complex relationship between these 2 distinct structures.
Some of the difficulty in elucidating the complex relationship between nucleoli and CBs arises from the continuing ambiguity in positively identifying the latter. While the most commonly used marker is coilin, this protein is not highly conserved throughout evolution and orthologues have not so far been identified in all model organisms. Alternative markers can be used, however the same issue remains: can a nuclear body containing several established CB markers but not coilin itself be classified as a CB? This conundrum is exemplified by the controversy surrounding the nuclear bodies implicated in miRNA processing in plants, which appear to lack the plant coilin homolog Atcoilin. It also bears comparison to the complex relationships between CBs and histone locus bodies and between CBs and gems, both of which are reviewed in detail elsewhere in this issue.
The functional interactions between nucleoli and CBs are more straightforward and revolve predominantly around the assembly and/or processing of ribonucleoprotein complexes (RNPs). Although snoRNPs are the classic example of RNPs required by the nucleolus and assembled with the assistance of CBs, it now seems likely that CBs are a common way-station for small RNAs of different classes within the nucleus, although a substantial amount of further work is required to fully understand the proposed joint roles for CBs and nucleoli in assembling other RNPs such as telomerase and the effectors of RNAi pathways. It is not that far-reaching to suggest that, once the nucleolus evolved as a site enriched in rRNA processing factors, with the CB as an accessory body, the cell subsequently exploited these structures to carry out other functions associated with RNP formation.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Prof. Peter Shaw (John Innes Center, Norfolk, UK), Dr Mirek Dundr (Rosalind Frankin University of Medicine and Science, Chicago), Prof. Karla Neugebauer and Dr Martin Machyna (Yale School of Medicine) for helpful discussions during the writing of this review.
Funding
This work was supported by the Wellcome Trust ISSF award 105621/Z/14/Z.
References
- 1.Gall JG. Cajal bodies: the first 100 years. Ann Rev Cell Dev Biol 2000; 16:273-300; PMID:11031238; https://doi.org/ 10.1146/annurev.cellbio.16.1.273 [DOI] [PubMed] [Google Scholar]
- 2.Cajal S. Un sencillo método de coloración del retículo protoplásmico y sus efectos en los diversos centros nerviosos de vertebrados e invertebrados. Trab Lab Inv Biol Univ Madr 1903; 2:129-221. [Google Scholar]
- 3.Monneron A, Bernhard W. Fine structural organization of the interphase nucleus in some mammalian cells. J Ultrastruct Res 1969; 27:266-88; PMID:5813971; https://doi.org/ 10.1016/S0022-5320(69)80017-1 [DOI] [PubMed] [Google Scholar]
- 4.Pena E, Berciano MT, Fernandez R, Ojeda JL, Lafarga M. Neuronal body size correlates with the number of nucleoli and Cajal bodies, and with the organization of the splicing machinery in rat trigeminal ganglion neurons. J Comp Neurol 2001; 430:250-63; PMID:11135260; https://doi.org/ 10.1002/1096-9861(20010205)430:2%3c250::AID-CNE1029%3e3.0.CO;2-L [DOI] [PubMed] [Google Scholar]
- 5.Brown JW, Shaw PJ, Shaw P, Marshall DF. Arabidopsis nucleolar protein database (AtNoPDB). Nucleic Acids Res 2005; 33:D633-6; PMID:15608277; https://doi.org/ 10.1093/nar/gki052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raska I, Andrade LE, Ochs RL, Chan EK, Chang CM, Roos G, Tan EM. Immunological and ultrastructural studies of the nuclear coiled body with autoimmune antibodies. Exp Cell Res 1991; 195:27-37; PMID:2055273; https://doi.org/ 10.1016/0014-4827(91)90496-H [DOI] [PubMed] [Google Scholar]
- 7.Matera AG. Nuclear bodies: multifaceted subdomains of the interchromatin space. Trends Cell Biol 1999; 9:302-9; PMID:10407409; https://doi.org/ 10.1016/S0962-8924(99)01606-2 [DOI] [PubMed] [Google Scholar]
- 8.Narayanan A, Speckmann W, Terns R, Terns MP. Role of the box C/D motif in localization of small nucleolar RNAs to coiled bodies and nucleoli. Mol Biol cell 1999; 10:2131-47; PMID:10397754; https://doi.org/ 10.1091/mbc.10.7.2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sleeman JE, Lamond AI. Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr Biol 1999; 9:1065-74; PMID:10531003; https://doi.org/ 10.1016/S0960-9822(99)80475-8 [DOI] [PubMed] [Google Scholar]
- 10.Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol 2007; 8:574-85; PMID:17519961; https://doi.org/ 10.1038/nrm2184 [DOI] [PubMed] [Google Scholar]
- 11.Andrade LE, Chan EK, Raska I, Peebles CL, Roos G, Tan EM. Human autoantibody to a novel protein of the nuclear coiled body: immunological characterization and cDNA cloning of p80-coilin. J Exp Med 1991; 173:1407-19; PMID:2033369; https://doi.org/ 10.1084/jem.173.6.1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raska I, Ochs RL, Andrade LE, Chan EK, Burlingame R, Peebles C, Gruol D, Tan EM. Association between the nucleolus and the coiled body. J Structl Biol 1990; 104:120-7; PMID:2088441; https://doi.org/ 10.1016/1047-8477(90)90066-L [DOI] [PubMed] [Google Scholar]
- 13.Raska I, Ochs RL, Salamin-Michel L. Immunocytochemistry of the cell nucleus. Electron Microsc Rev 1990; 3:301-53; PMID:2103346; https://doi.org/ 10.1016/0892-0354(90)90006-E [DOI] [PubMed] [Google Scholar]
- 14.Gall JG. The centennial of the Cajal body. Nat Rev Mol Cell Biol 2003; 4:975-80; PMID:14685175; https://doi.org/ 10.1038/nrm1262 [DOI] [PubMed] [Google Scholar]
- 15.Machyna M, Neugebauer KM, Stanek D. Coilin: The first 25 years. RNA biology 2015; 12:590-6; PMID:25970135; https://doi.org/ 10.1080/15476286.2015.1034923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barcaroli D, Dinsdale D, Neale MH, Bongiorno-Borbone L, Ranalli M, Munarriz E, Sayan AE, McWilliam JM, Smith TM, Fava E, et al.. FLASH is an essential component of Cajal bodies. Proc Natl Acad Sci U S A 2006; 103:14802-7; PMID:17003126; https://doi.org/ 10.1073/pnas.0604225103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bohmann K, Ferreira JA, Lamond AI. Mutational analysis of p80 coilin indicates a functional interaction between coiled bodies and the nucleolus. J Cell Biol 1995; 131:817-31; PMID:7490287; https://doi.org/ 10.1083/jcb.131.4.817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carmo-Fonseca M, Pepperkok R, Carvalho MT, Lamond AI. Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J Cell Biol 1992; 117:1-14; PMID:1532583; https://doi.org/ 10.1083/jcb.117.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carvalho T, Almeida F, Calapez A, Lafarga M, Berciano MT, Carmo-Fonseca M. The spinal muscular atrophy disease gene product, SMN: A link between snRNP biogenesis and the Cajal (coiled) body. J Cell Biol 1999; 147:715-28; PMID:10562276; https://doi.org/ 10.1083/jcb.147.4.715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cioce M, Lamond AI. Cajal bodies: a long history of discovery. Ann Rev Cell Dev Biol 2005; 21:105-31; PMID:16212489; https://doi.org/ 10.1146/annurev.cellbio.20.010403.103738 [DOI] [PubMed] [Google Scholar]
- 21.Gilder AS, Do PM, Carrero ZI, Cosman AM, Broome HJ, Velma V, Martinez LA, Hebert MD. Coilin participates in the suppression of RNA polymerase I in response to cisplatin-induced DNA damage. Mol Biol cell 2011; 22:1070-9; PMID:21289084; https://doi.org/ 10.1091/mbc.E10-08-0731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Girard C, Neel H, Bertrand E, Bordonne R. Depletion of SMN by RNA interference in HeLa cells induces defects in Cajal body formation. Nucleic Acids Res 2006; 34:2925-32; PMID:16738131; https://doi.org/ 10.1093/nar/gkl374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonsalvez GB, Tian L, Ospina JK, Boisvert FM, Lamond AI, Matera AG. Two distinct arginine methyltransferases are required for biogenesis of Sm-class ribonucleoproteins. J Cell Biol 2007; 178:733-40; PMID:17709427; https://doi.org/ 10.1083/jcb.200702147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hearst SM, Gilder AS, Negi SS, Davis MD, George EM, Whittom AA, Toyota CG, Husedzinovic A, Gruss OJ, Hebert MD. Cajal-body formation correlates with differential coilin phosphorylation in primary and transformed cell lines. J Cell Sci 2009; 122:1872-81; PMID:19435804; https://doi.org/ 10.1242/jcs.044040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hebert MD, Matera AG. Self-association of coilin reveals a common theme in nuclear body localization. Mol Biol cell 2000; 11:4159-71; PMID:11102515; https://doi.org/ 10.1091/mbc.11.12.4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hutten S, Chachami G, Winter U, Melchior F, Lamond AI. A role for the Cajal-body-associated SUMO isopeptidase USPL1 in snRNA transcription mediated by RNA polymerase II. J Cell Sci 2014; 127:1065-78; PMID:24413172; https://doi.org/ 10.1242/jcs.141788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lemm I, Girard C, Kuhn AN, Watkins NJ, Schneider M, Bordonne R, Luhrmann R. Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies. Mol Biol cell 2006; 17:3221-31; PMID:16687569; https://doi.org/ 10.1091/mbc.E06-03-0247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lyon CE, Bohmann K, Sleeman J, Lamond AI. Inhibition of protein dephosphorylation results in the accumulation of splicing snRNPs and coiled bodies within the nucleolus. Exp Cell Res 1997; 230:84-93; PMID:9013710; https://doi.org/ 10.1006/excr.1996.3380 [DOI] [PubMed] [Google Scholar]
- 29.Malatesta M, Zancanaro C, Martin TE, Chan EK, Amalric F, Luhrmann R, Vogel P, Fakan S. Is the coiled body involved in nucleolar functions? Exp Cell Res 1994; 211:415-9; PMID:8143790; https://doi.org/ 10.1006/excr.1994.1106 [DOI] [PubMed] [Google Scholar]
- 30.Ochs RL, Stein TW Jr., Tan EM. Coiled bodies in the nucleolus of breast cancer cells. J Cell Sci 1994; 107(Pt 2):385-99; PMID:8207070 [DOI] [PubMed] [Google Scholar]
- 31.Shav-Tal Y, Blechman J, Darzacq X, Montagna C, Dye BT, Patton JG, Singer RH, Zipori D. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol Biol cell 2005; 16:2395-413; PMID:15758027; https://doi.org/ 10.1091/mbc.E04-11-0992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shpargel KB, Matera AG. Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins. Proc Natl Acad Sci U S A 2005; 102:17372-7; PMID:16301532; https://doi.org/ 10.1073/pnas.0508947102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shpargel KB, Ospina JK, Tucker KE, Matera AG, Hebert MD. Control of Cajal body number is mediated by the coilin C-terminus. J Cell Sci 2003; 116:303-12; PMID:12482916; https://doi.org/ 10.1242/jcs.00211 [DOI] [PubMed] [Google Scholar]
- 34.Tapia O, Bengoechea R, Berciano MT, Lafarga M. Nucleolar targeting of coilin is regulated by its hypomethylation state. Chromosoma 2010; 119:527-40; PMID:20449600; https://doi.org/ 10.1007/s00412-010-0276-7 [DOI] [PubMed] [Google Scholar]
- 35.Tapia O, Bengoechea R, Palanca A, Arteaga R, Val-Bernal JF, Tizzano EF, Berciano MT, Lafarga M. Reorganization of Cajal bodies and nucleolar targeting of coilin in motor neurons of type I spinal muscular atrophy. Histochem Cell Biol 2012; 137:657-67; PMID:22302308; https://doi.org/ 10.1007/s00418-012-0921-8 [DOI] [PubMed] [Google Scholar]
- 36.Mahmoudi S, Henriksson S, Weibrecht I, Smith S, Soderberg O, Stromblad S, Wiman KG, Farnebo M. WRAP53 is essential for Cajal body formation and for targeting the survival of motor neuron complex to Cajal bodies. PLoS Biol 2010; 8:e1000521; PMID:21072240; https://doi.org/ 10.1371/journal.pbio.10000521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baltanas FC, Casafont I, Weruaga E, Alonso JR, Berciano MT, Lafarga M. Nucleolar disruption and cajal body disassembly are nuclear hallmarks of DNA damage-induced neurodegeneration in purkinje cells. Brain Pathol 2011; 21:374-88; PMID:21054627; https://doi.org/ 10.1111/j.1750-3639.2010.00461.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bohmann K, Ferreira J, Santama N, Weis K, Lamond AI. Molecular analysis of the coiled body. J Cell Sci Suppl 1995; 19:107-13; PMID:8655641; https://doi.org/ 10.1242/jcs.1995.Supplement_19.16 [DOI] [PubMed] [Google Scholar]
- 39.Clelland AK, Kinnear NP, Oram L, Burza J, Sleeman JE. The SMN protein is a key regulator of nuclear architecture in differentiating neuroblastoma cells. Traffic 2009; 10:1585-98; PMID:19735367; https://doi.org/ 10.1111/j.1600-0854.2009.00972.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carmo-Fonseca M. New clues to the function of the Cajal body. EMBO Rep 2002; 3:726-7; PMID:12151329; https://doi.org/ 10.1093/embo-reports/kvf154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dahm R, Gribbon C, Quinlan RA, Prescott AR. Changes in the nucleolar and coiled body compartments precede lamina and chromatin reorganization during fibre cell denucleation in the bovine lens. Eur J Cell Biol 1998; 75:237-46; PMID:9587055; https://doi.org/ 10.1016/S0171-9335(98)80118-0 [DOI] [PubMed] [Google Scholar]
- 42.Boudonck K, Dolan L, Shaw PJ. The movement of coiled bodies visualized in living plant cells by the green fluorescent protein. Mol Biol cell 1999; 10:2297-307; PMID:10397766; https://doi.org/ 10.1091/mbc.10.7.2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Misteli T, Caceres JF, Spector DL. The dynamics of a pre-mRNA splicing factor in living cells. Nature 1997; 387:523-7; PMID:9168118; https://doi.org/ 10.1038/387523a0 [DOI] [PubMed] [Google Scholar]
- 44.Platani M, Goldberg I, Swedlow JR, Lamond AI. In vivo analysis of Cajal body movement, separation, and joining in live human cells. J Cell Biol 2000; 151:1561-74; PMID:11134083; https://doi.org/ 10.1083/jcb.151.7.1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sleeman JE, Trinkle-Mulcahy L, Prescott AR, Ogg SC, Lamond AI. Cajal body proteins SMN and Coilin show differential dynamic behaviour in vivo. J Cell Sci 2003; 116:2039-50; PMID:12679382; https://doi.org/ 10.1242/jcs.00400 [DOI] [PubMed] [Google Scholar]
- 46.Snaar S, Wiesmeijer K, Jochemsen AG, Tanke HJ, Dirks RW. Mutational analysis of fibrillarin and its mobility in living human cells. J Cell Biol 2000; 151:653-62; PMID:11062265; https://doi.org/ 10.1083/jcb.151.3.653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Navascues J, Bengoechea R, Tapia O, Casafont I, Berciano MT, Lafarga M. SUMO-1 transiently localizes to Cajal bodies in mammalian neurons. J Structl Biol 2008; 163:137-46; PMID:18571432; https://doi.org/ 10.1016/j.jsb.2008.04.013 [DOI] [PubMed] [Google Scholar]
- 48.Sleeman J. A regulatory role for CRM1 in the multi-directional trafficking of splicing snRNPs in the mammalian nucleus. J Cell Sci 2007; 120:1540-50; PMID:17405816; https://doi.org/ 10.1242/jcs.001529 [DOI] [PubMed] [Google Scholar]
- 49.Hutten S, Prescott A, James J, Riesenberg S, Boulon S, Lam YW, Lamond AI. An intranucleolar body associated with rDNA. Chromosoma 2011; 120:481-99; PMID:21698343; https://doi.org/ 10.1007/s00412-011-0327-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ernoult-Lange M, Wilczynska A, Harper M, Aigueperse C, Dautry F, Kress M, Weil D. Nucleocytoplasmic traffic of CPEB1 and accumulation in Crm1 nucleolar bodies. Mol Biol cell 2009; 20:176-87; PMID:18923137; https://doi.org/ 10.1091/mbc.E08-09-0904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Souquere S, Weil D, Pierron G. Comparative ultrastructure of CRM1-Nucleolar bodies (CNoBs), Intranucleolar bodies (INBs) and hybrid PML/p62 bodies uncovers new facets of nuclear body dynamic and diversity. Nucleus 2015; 6:326-38; PMID:26275159; https://doi.org/ 10.1080/19491034.2015.1082695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dundr M. Nuclear bodies: multifunctional companions of the genome. Curr Opin Cell Biol 2012; 24:415-22; PMID:22541757; https://doi.org/ 10.1016/j.ceb.2012.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karpen GH, Schaefer JE, Laird CD. A Drosophila rRNA gene located in euchromatin is active in transcription and nucleolus formation. Genes Dev 1988; 2:1745-63; PMID:3149250; https://doi.org/ 10.1101/gad.2.12b.1745 [DOI] [PubMed] [Google Scholar]
- 54.Dundr M. Seed and grow: a two-step model for nuclear body biogenesis. J Cell Biol 2011; 193:605-6; PMID:21576389; https://doi.org/ 10.1083/jcb.201104087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaiser TE, Intine RV, Dundr M. De novo formation of a subnuclear body. Science 2008; 322:1713-7; PMID:18948503; https://doi.org/ 10.1126/science.1165216 [DOI] [PubMed] [Google Scholar]
- 56.Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G, et al.. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 2012; 149:768-79; PMID:22579282; https://doi.org/ 10.1016/j.cell.2012.04.016 [DOI] [PubMed] [Google Scholar]
- 57.Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al.. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 2012; 149:753-67; PMID:22579281; https://doi.org/ 10.1016/j.cell.2012.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwartz JC, Wang X, Podell ER, Cech TR. RNA seeds higher-order assembly of FUS protein. Cell Rep 2013; 5:918-25; PMID:24268778; https://doi.org/ 10.1016/j.celrep.2013.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sleeman JE, Trinkle-Mulcahy L. Nuclear bodies: new insights into assembly/dynamics and disease relevance. Curr Opin Cell Biol 2014; 28:76-83; PMID:24704702; https://doi.org/ 10.1016/j.ceb.2014.03.004 [DOI] [PubMed] [Google Scholar]
- 60.Courchaine EM, Lu A, Neugebauer KM. Droplet organelles? EMBO J 2016; 35:1603-12; PMID:27357569; https://doi.org/ 10.15252/embj.201593517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu B, Buxbaum AR, Katz ZB, Yoon YJ, Singer RH. Quantifying protein-mRNA interactions in single live cells. Cell 2015; 162:211-20; PMID:26140598; https://doi.org/ 10.1016/j.cell.2015.05.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu L, Brangwynne CP. Nuclear bodies: the emerging biophysics of nucleoplasmic phases. Curr Opin cell biol 2015; 34:23-30; PMID:25942753; https://doi.org/ 10.1016/j.ceb.2015.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hennig S, Kong G, Mannen T, Sadowska A, Kobelke S, Blythe A, Knott GJ, Iyer KS, Ho D, Newcombe EA, et al.. Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles. J Cell Biol 2015; 210:529-39; PMID:26283796; https://doi.org/ 10.1083/jcb.201504117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carmo-Fonseca M, Ferreira J, Lamond AI. Assembly of snRNP-containing coiled bodies is regulated in interphase and mitosis–evidence that the coiled body is a kinetic nuclear structure. J Cell Biol 1993; 120:841-52; PMID:7679389; https://doi.org/ 10.1083/jcb.120.4.841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Broome HJ, Hebert MD. Coilin displays differential affinity for specific RNAs in vivo and is linked to telomerase RNA biogenesis. J Mol Biol 2013; 425:713-24; PMID:23274112; https://doi.org/ 10.1016/j.jmb.2012.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Machyna M, Kehr S, Straube K, Kappei D, Buchholz F, Butter F, Ule J, Hertel J, Stadler PF, Neugebauer KM. The coilin interactome identifies hundreds of small noncoding RNAs that traffic through Cajal bodies. Mol Cell 2014; 56:389-99; PMID:25514182; https://doi.org/ 10.1016/j.molcel.2014.10.004 [DOI] [PubMed] [Google Scholar]
- 67.Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci U S A 2011; 108:4334-9; PMID:21368180; https://doi.org/ 10.1073/pnas.1017150108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weber SC, Brangwynne CP. Inverse size scaling of the nucleolus by a concentration-dependent phase transition. Curr Biol 2015; 25:641-6; PMID:25702583; https://doi.org/ 10.1016/j.cub.2015.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Berry J, Weber SC, Vaidya N, Haataja M, Brangwynne CP. RNA transcription modulates phase transition-driven nuclear body assembly. Proc Natl Acad Sci U S A 2015; 112:E5237-45; PMID:26351690; https://doi.org/ 10.1073/pnas.1509317112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, Brangwynne CP. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell 2016; 165:1686-97; PMID:27212236; https://doi.org/ 10.1016/j.cell.2016.04.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Frey MR, Matera AG. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc Natl Acad Sci U S A 1995; 92:5915-9; PMID:7597053; https://doi.org/ 10.1073/pnas.92.13.5915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Frey MR, Matera AG. RNA-mediated interaction of Cajal bodies and U2 snRNA genes. J Cell Biol 2001; 154:499-509; PMID:11489914; https://doi.org/ 10.1083/jcb.200105084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smith KP, Carter KC, Johnson CV, Lawrence JB. U2 and U1 snRNA gene loci associate with coiled bodies. J Cell Biochem 1995; 59:473-85; PMID:8749717; https://doi.org/ 10.1002/jcb.240590408 [DOI] [PubMed] [Google Scholar]
- 74.Lam YW, Trinkle-Mulcahy L. New insights into nucleolar structure and function. F1000prime Rep 2015; 7:48; PMID:26097721; https://doi.org/ 10.12703/P7-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lafontaine JG, Chamberland H. Relationship of nucleolus-associated bodies with the nucleolar organizer tracks in plant interphase nuclei (Pisum sativum). Chromosoma 1995; 103:545-53; PMID:7621704 [DOI] [PubMed] [Google Scholar]
- 76.Shaw PJ, Jordan EG. The nucleolus. Ann Rev Cell Dev Biol 1995; 11:93-121; PMID:8689574; https://doi.org/ 10.1146/annurev.cb.11.110195.000521 [DOI] [PubMed] [Google Scholar]
- 77.Wang Q, Sawyer IA, Sung MH, Sturgill D, Shevtsov SP, Pegoraro G, Hakim O, Baek S, Hager GL, Dundr M. Cajal bodies are linked to genome conformation. Nat Commun 2016; 7:10966; PMID:26997247; https://doi.org/ 10.1038/ncomms10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Boulon S, Verheggen C, Jady BE, Girard C, Pescia C, Paul C, Ospina JK, Kiss T, Matera AG, Bordonne R, et al.. PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol Cell 2004; 16:777-87; PMID:15574332; https://doi.org/ 10.1016/j.molcel.2004.11.013 [DOI] [PubMed] [Google Scholar]
- 79.Verheggen C, Lafontaine DL, Samarsky D, Mouaikel J, Blanchard JM, Bordonne R, Bertrand E. Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J 2002; 21:2736-45; PMID:12032086; https://doi.org/ 10.1093/emboj/21.11.2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Watkins NJ, Lemm I, Ingelfinger D, Schneider C, Hossbach M, Urlaub H, Luhrmann R. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol Cell 2004; 16:789-98; PMID:15574333; https://doi.org/ 10.1016/j.molcel.2004.11.012 [DOI] [PubMed] [Google Scholar]
- 81.Richard P, Darzacq X, Bertrand E, Jady BE, Verheggen C, Kiss T. A common sequence motif determines the Cajal body-specific localization of box H/ACA scaRNAs. EMBO J 2003; 22:4283-93; PMID:12912925; https://doi.org/ 10.1093/emboj/cdg394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shaw PJ, Beven AF, Leader DJ, Brown JW. Localization and processing from a polycistronic precursor of novel snoRNAs in maize. J Cell Sci 1998; 111 (Pt 15):2121-8; PMID:9664033 [DOI] [PubMed] [Google Scholar]
- 83.Kiss T, Fayet E, Jady BE, Richard P, Weber M. Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb Symp Quant Biol 2006; 71:407-17; PMID:17381323; https://doi.org/ 10.1101/sqb.2006.71.025 [DOI] [PubMed] [Google Scholar]
- 84.Yu YT, Shu MD, Narayanan A, Terns RM, Terns MP, Steitz JA. Internal modification of U2 small nuclear (sn)RNA occurs in nucleoli of Xenopus oocytes. J Cell Biol 2001; 152:1279-88; PMID:11257127; https://doi.org/ 10.1083/jcb.152.6.1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Armanios M, Blackburn EH. The telomere syndromes. Nat Rev Genet 2012; 13:693-704; PMID:22965356; https://doi.org/ 10.1038/nrg3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kiss T, Fayet-Lebaron E, Jady BE. Box H/ACA small ribonucleoproteins. Mol Cell 2010; 37:597-606; PMID:20227365; https://doi.org/ 10.1016/j.molcel.2010.01.032 [DOI] [PubMed] [Google Scholar]
- 87.Etheridge KT, Banik SS, Armbruster BN, Zhu Y, Terns RM, Terns MP, Counter CM. The nucleolar localization domain of the catalytic subunit of human telomerase. J Biol Chem 2002; 277:24764-70; PMID:11956201; https://doi.org/ 10.1074/jbc.M201227200 [DOI] [PubMed] [Google Scholar]
- 88.Yang Y, Chen Y, Zhang C, Huang H, Weissman SM. Nucleolar localization of hTERT protein is associated with telomerase function. Exp Cell Res 2002; 277:201-9; PMID:12083802; https://doi.org/ 10.1006/excr.2002.5541 [DOI] [PubMed] [Google Scholar]
- 89.Jady BE, Bertrand E, Kiss T. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J Cell Biol 2004; 164:647-52; PMID:14981093; https://doi.org/ 10.1083/jcb.200310138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhu Y, Tomlinson RL, Lukowiak AA, Terns RM, Terns MP. Telomerase RNA accumulates in Cajal bodies in human cancer cells. Mol Biol cell 2004; 15:81-90; PMID:14528011; https://doi.org/ 10.1091/mbc.E03-07-0525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Machyna M, Heyn P, Neugebauer KM. Cajal bodies: where form meets function. Wiley Interdiscip Rev RNA 2013; 4:17-34; PMID:23042601; https://doi.org/ 10.1002/wrna.1139 [DOI] [PubMed] [Google Scholar]
- 92.Jady BE, Richard P, Bertrand E, Kiss T. Cell cycle-dependent recruitment of telomerase RNA and Cajal bodies to human telomeres. Mol Biol cell 2006; 17:944-54; PMID:16319170; https://doi.org/ 10.1091/mbc.E05-09-0904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stern JL, Zyner KG, Pickett HA, Cohen SB, Bryan TM. Telomerase recruitment requires both TCAB1 and Cajal bodies independently. Mol Cell Biol 2012; 32:2384-95; PMID:22547674; https://doi.org/ 10.1128/MCB.00379-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schmidt JC, Cech TR. Human telomerase: biogenesis, trafficking, recruitment, and activation. Genes Dev 2015; 29:1095-105; PMID:26063571; https://doi.org/ 10.1101/gad.263863.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tomlinson RL, Ziegler TD, Supakorndej T, Terns RM, Terns MP. Cell cycle-regulated trafficking of human telomerase to telomeres. Mol Biol cell 2006; 17:955-65; PMID:16339074; https://doi.org/ 10.1091/mbc.E05-09-0903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lee JH, Lee YS, Jeong SA, Khadka P, Roth J, Chung IK. Catalytically active telomerase holoenzyme is assembled in the dense fibrillar component of the nucleolus during S phase. Histochem Cell Biol 2014; 141:137-52; PMID:24318571; https://doi.org/ 10.1007/s00418-013-1166-x [DOI] [PubMed] [Google Scholar]
- 97.Batista LF, Pech MF, Zhong FL, Nguyen HN, Xie KT, Zaug AJ, Crary SM, Choi J, Sebastiano V, Cherry A, et al.. Telomere shortening and loss of self-renewal in dyskeratosis congenita induced pluripotent stem cells. Nature 2011; 474:399-402; PMID:21602826; https://doi.org/ 10.1038/nature10084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tycowski KT, Shu MD, Kukoyi A, Steitz JA. A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol Cell 2009; 34:47-57; PMID:19285445; https://doi.org/ 10.1016/j.molcel.2009.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, Veenstra TD, Terns MP, Artandi SE. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 2009; 323:644-8; PMID:19179534; https://doi.org/ 10.1126/science.1165357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhong F, Savage SA, Shkreli M, Giri N, Jessop L, Myers T, Chen R, Alter BP, Artandi SE. Disruption of telomerase trafficking by TCAB1 mutation causes dyskeratosis congenita. Genes Dev 2011; 25:11-6; PMID:21205863; https://doi.org/ 10.1101/gad.2006411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lin J, Jin R, Zhang B, Chen H, Bai YX, Yang PX, Han SW, Xie YH, Huang PT, Huang C, et al.. Nucleolar localization of TERT is unrelated to telomerase function in human cells. J Cell Sci 2008; 121:2169-76; PMID:18522991; https://doi.org/ 10.1242/jcs.024091; https://doi.org/ [DOI] [PubMed] [Google Scholar]
- 102.Gonzalez OG, Assfalg R, Koch S, Schelling A, Meena JK, Kraus J, Lechel A, Katz SF, Benes V, Scharffetter-Kochanek K, et al.. Telomerase stimulates ribosomal DNA transcription under hyperproliferative conditions. Nat Commun 2014; 5:4599; PMID:25118183; https://doi.org/ 10.1038/ncomms5599 [DOI] [PubMed] [Google Scholar]
- 103.Maida Y, Masutomi K. Telomerase reverse transcriptase moonlights: Therapeutic targets beyond telomerase. Cancer Sci 2015; 106:1486-92; PMID:26331588; https://doi.org/ 10.1111/cas.12806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 2002; 297:1833-7; PMID:12193640; https://doi.org/ 10.1126/science.1074973 [DOI] [PubMed] [Google Scholar]
- 105.Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 2008; 31:785-99; PMID:18922463; https://doi.org/ 10.1016/j.molcel.2008.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Watanabe T, Tomizawa S, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S, Iida N, Hoki Y, Murphy PJ, Toyoda A, et al.. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science 2011; 332:848-52; PMID:21566194; https://doi.org/ 10.1126/science.1203919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet 2013; 14:100-12; PMID:23329111; https://doi.org/ 10.1038/nrg3355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pontes O, Li CF, Costa Nunes P, Haag J, Ream T, Vitins A, Jacobsen SE, Pikaard CS. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell 2006; 126:79-92; PMID:16839878; https://doi.org/ 10.1016/j.cell.2006.05.031 [DOI] [PubMed] [Google Scholar]
- 109.Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, Chan SW, Lagrange T, Pikaard CS, Jacobsen SE. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell 2006; 126:93-106; PMID:16839879; https://doi.org/ 10.1016/j.cell.2006.05.032 [DOI] [PubMed] [Google Scholar]
- 110.Fang Y, Spector DL. Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr Biol 2007; 17:818-23; PMID:17442570; https://doi.org/ 10.1016/j.cub.2007.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fujioka Y, Utsumi M, Ohba Y, Watanabe Y. Location of a possible miRNA processing site in SmD3/SmB nuclear bodies in Arabidopsis. Plant Cell Physiol 2007; 48:1243-53; PMID:17675322; https://doi.org/ 10.1093/pcp/pcm099 [DOI] [PubMed] [Google Scholar]
- 112.Chu Y, Yue X, Younger ST, Janowski BA, Corey DR. Involvement of argonaute proteins in gene silencing and activation by RNAs complementary to a non-coding transcript at the progesterone receptor promoter. Nucleic Acids Res 2010; 38:7736-48; PMID:20675357; https://doi.org/ 10.1093/nar/gkq648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Doyle M, Badertscher L, Jaskiewicz L, Guttinger S, Jurado S, Hugenschmidt T, Kutay U, Filipowicz W. The double-stranded RNA binding domain of human Dicer functions as a nuclear localization signal. Rna 2013; 19:1238-52; PMID:23882114; https://doi.org/ 10.1261/rna.039255.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ohrt T, Mutze J, Staroske W, Weinmann L, Hock J, Crell K, Meister G, Schwille P. Fluorescence correlation spectroscopy and fluorescence cross-correlation spectroscopy reveal the cytoplasmic origination of loaded nuclear RISC in vivo in human cells. Nucleic Acids Res 2008; 36:6439-49; PMID:18842624; https://doi.org/ 10.1093/nar/gkn693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rudel S, Flatley A, Weinmann L, Kremmer E, Meister G. A multifunctional human Argonaute2-specific monoclonal antibody. Rna 2008; 14:1244-53; PMID:18430891; https://doi.org/ 10.1261/rna.973808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Weinmann L, Hock J, Ivacevic T, Ohrt T, Mutze J, Schwille P, Kremmer E, Benes V, Urlaub H, Meister G. Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell 2009; 136:496-507; PMID:19167051; https://doi.org/ 10.1016/j.cell.2008.12.023 [DOI] [PubMed] [Google Scholar]
- 117.Gagnon KT, Li L, Chu Y, Janowski BA, Corey DR. RNAi factors are present and active in human cell nuclei. Cell Rep 2014; 6:211-21; PMID:24388755; https://doi.org/ 10.1016/j.celrep.2013.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol Cell 2008; 32:519-28; PMID:19026782; https://doi.org/ 10.1016/j.molcel.2008.10.017 [DOI] [PubMed] [Google Scholar]
- 119.Politz JC, Hogan EM, Pederson T. MicroRNAs with a nucleolar location. Rna 2009; 15:1705-15; PMID:19628621; https://doi.org/ 10.1261/rna.1470409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. The nucleolus under stress. Mol Cell 2010; 40:216-27; PMID:20965417; https://doi.org/ 10.1016/j.molcel.2010.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Velma V, Carrero ZI, Cosman AM, Hebert MD. Coilin interacts with Ku proteins and inhibits in vitro non-homologous DNA end joining. FEBS Lett 2010; 584:4735-9; PMID:21070772; https://doi.org/ 10.1016/j.febslet.2010.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Henriksson S, Rassoolzadeh H, Hedstrom E, Coucoravas C, Julner A, Goldstein M, Imreh G, Zhivotovsky B, Kastan MB, Helleday T, et al.. The scaffold protein WRAP53beta orchestrates the ubiquitin response critical for DNA double-strand break repair. Genes Dev 2014; 28:2726-38; PMID:25512560; https://doi.org/ 10.1101/gad.246546.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Collier S, Pendle A, Boudonck K, van Rij T, Dolan L, Shaw P. A distant coilin homologue is required for the formation of cajal bodies in Arabidopsis. Mol Biol cell 2006; 17:2942-51; PMID:16624863; https://doi.org/ 10.1091/mbc.E05-12-1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tucker KE, Berciano MT, Jacobs EY, LePage DF, Shpargel KB, Rossire JJ, Chan EK, Lafarga M, Conlon RA, Matera AG. Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J Cell Biol 2001; 154:293-307; PMID:11470819; https://doi.org/ 10.1083/jcb.200104083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Silva NP, Christofolini DM, Mortara RA, Andrade LE. Colocalization of coilin and nucleolar proteins in Cajal body-like structures of micronucleated PtK2 cells. Braz J Med Biol Res 2004; 37:997-1003; PMID:15264006; https://doi.org/ 10.1590/S0100-879X2004000700008 [DOI] [PubMed] [Google Scholar]
- 126.Khodyuchenko T, Gaginskaya E, Krasikova A. Non-canonical Cajal bodies form in the nucleus of late stage avian oocytes lacking functional nucleolus. Histochem Cell Biol 2012; 138:57-73; PMID:22382586; https://doi.org/ 10.1007/s00418-012-0938-z [DOI] [PubMed] [Google Scholar]


