Abstract
The exosome is a conserved multiprotein complex essential for RNA processing and degradation. The nuclear exosome is a key factor for pre-rRNA processing through the activity of its catalytic subunits, Rrp6 and Rrp44. In Saccharomyces cerevisiae, Rrp6 is exclusively nuclear and has been shown to interact with exosome cofactors. With the aim of analyzing proteins associated with the nuclear exosome, in this work, we purified the complex with Rrp6-TAP, identified the co-purified proteins by mass spectrometry, and found karyopherins to be one of the major groups of proteins enriched in the samples. By investigating the biological importance of these protein interactions, we identified Srp1, Kap95, and Sxm1 as the most important karyopherins for Rrp6 nuclear import and the nuclear localization signals recognized by them. Based on the results shown here, we propose a model of multiple pathways for the transport of Rrp6 to the nucleus.
Keywords: exosome complex, protein import, protein-nucleic acid interaction, protein-protein interaction, ribosomal RNA processing (rRNA processing), Saccharomyces cerevisiae
Introduction
The RNA exosome is a protein complex involved in processing and degradation of different classes of RNA in the cell. This complex was first identified in the yeast Saccharomyces cerevisiae (1) and later identified in other eukaryotes. The exosome is not present in bacteria but has been identified in archaea, being structurally conserved throughout evolution (2).
In eukaryotes, the exosome is present in both nucleus and cytoplasm, and its nuclease activity is provided by two catalytic subunits, Rrp44/Dis3 and Rrp6 (3). In yeast, the difference between the nuclear and cytoplasmic exosomes is the presence of the subunit Rrp6 in the nucleus. The exosome core is composed of nine subunits: six different subunits each containing an inactive RNase PH domain (Rrp41, Rrp42, Rrp43, Rrp45, Rrp46, and Mtr3) and three RNA-binding subunits (Rrp4, Rrp40, and Csl4). The catalytically active subunits, Rrp44 and Rrp6, bind to opposite sides of the core. Rrp44 is an RNase R-like with both endonucleolytic and processive 3′-5′ exonucleolytic activities (3–6), whereas Rrp6 shows a distributive 3′-5′ exonucleolytic activity (7).
Nuclear exosome function comprises processing of ribosomal RNAs (rRNAs), small nuclear RNAs, and small nucleolar RNAs as well as surveillance and degradation of incorrectly processed RNAs (8). In the pre-rRNA processing pathway, the exosome is directly responsible for the degradation of the 5′-external transcribed spacer sequence after cleavage at site A0 and for trimming of the internal transcribed sequence 2 segment present in the intermediate 7S for the generation of the mature 5.8S rRNA (9). In addition to being important for RNA quality control in the nucleus, the exosome has also been described to be involved in cytoplasmic mRNA degradation (10, 11).
Because the exosome does not show substrate specificity in vitro, the recruitment of the complex in vivo might be performed by its cofactors (12). Interestingly, most of the proteins identified as nuclear exosome cofactors have been shown to interact with Rrp6 (13–15). Due to the low concentration of Rrp6 in the cell or to its restriction to the cell nucleus, however, attempts to isolate nuclear exosome cofactors co-purifying with the exosome when using a core exosome subunit as bait have been shown to be inefficient (16, 17).
A significant amount of information is now available on the structure and function of the exosome, but relatively little is known about the transport of this complex to the nucleus. Protein transport from cytoplasm to nucleus is primarily mediated by specific interactions between karyopherins and signal sequences (nuclear localization signals (NLSs)3) present in the cargo proteins (18, 19). The first identified sequence was the simian virus 40 large tumor antigen-like nuclear localization signal (classical NLS), which is recognized by karyopherin α (Srp1 in yeast) and transported to the nucleus as a trimeric complex with karyopherin β (Kap95 in yeast) and the cargo protein (19, 20). Another nuclear import route occurs through the recognition of a different NLS in the cargo by a karyopherin β and the transport of a heterodimer to the nucleus (21). In S. cerevisiae, there are 14 karyopherins β,10 of which are involved in transport to the nucleus (importins; Kap95, Kap104, Sxm1/Kap108, Mtr10, Kap114, Nmd5, Kap120/Lph2, Pse1/Kap121, Kap122, and Kap123), three that are involved in transport to the cytoplasm (exportins; Cse1, Crm1/Xpo1, and Los1), and one karyopherin involved in transport in both directions, Msn5 (22).
Despite the association described previously of Rrp6 with Srp1 and Kap95 (23–26), there are no conclusive studies describing the nuclear import pathway of yeast Rrp6. In this work, we purified the S. cerevisiae nuclear exosome with Rrp6-TAP for the identification of proteins interacting with this complex. One of the major groups of proteins co-purified with the exosome was that of the karyopherins/importins. The presence of different karyopherins associated with Rrp6/exosome in our purifications raised the possibility of multiple pathways for nuclear import of Rrp6. Here we show the participation of different karyopherins in the transport of the exosome subunit Rrp6 to the nucleus and the sequences that these karyopherins might recognize in Rrp6. The results shown here provide evidence for alternative pathways of Rrp6 transport to the cell nucleus.
Results
Purification of the nuclear exosome with Rrp6-TAP
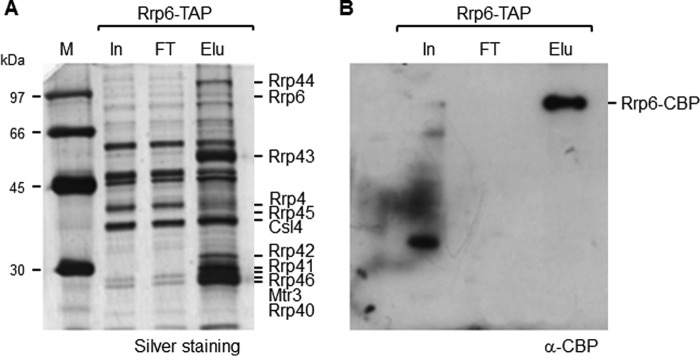
To identify proteins interacting with the yeast nuclear exosome (Exo11), we took advantage of the TAP tag method (27) using an Rrp6-TAP fusion. Because Rrp6 expression in yeast cells is lower than that of the core subunits (16, 17) (Fig. 1), 20 liters of culture were used here for the purification of the exosome with Rrp6-TAP. Despite the difference in expression levels of the bait proteins, the band profile of the proteins co-immunoprecipitated with Rrp6-TAP was similar to that of the exosome co-immunoprecipitated with Rrp43-TAP (16), and all the exosome subunits were identified in the elution fraction (Fig. 1A). Samples from the same Rrp6-TAP elution fraction were analyzed by Western blotting with antibody against the CBP portion of the TAP tag, confirming that Rrp6-TAP bound efficiently to the IgG-Sepharose resin and was eluted after the cleavage reaction with TEV protease (Fig. 1B).
Figure 1.
Coimmunoprecipitation of the exosome with Rrp6-TAP. Total extract was incubated with IgG-Sepharose beads, and proteins were eluted with TEV protease. Samples from total extract (input (In)), flow-through (FT), and elution (Elu) were subjected to SDS-PAGE and silver-stained (A). The exosome subunits were identified by mass spectrometry. B, Western blot of the same samples with anti-CBP. Rrp6-TAP bound efficiently to the resin and was eluted after cleavage with TEV protease.
For the identification of the proteins co-immunoprecipitated with Rrp6-TAP, the eluted fraction was analyzed by mass spectrometry (supplemental Table S1). All the exosome subunits were identified, those that are part of the RNA-binding “cap,” Rrp4, Rrp40, and Csl4; the subunits that form the RNase PH ring, Rrp41, Rrp45, Rrp46, Rrp43, Mtr3, and Rrp42; and the catalytic subunits, Rrp44 and Rrp6 (4, 28–30). Based on the exosome structure described previously, Rrp6 and Rrp44 do not interact directly but rather bind to opposite sides of the exosome core (4, 28). The presence of Rrp44 in all the purifications, therefore, indicates that the exosome obtained here was stable during the co-immunoprecipitation with Rrp6-TAP.
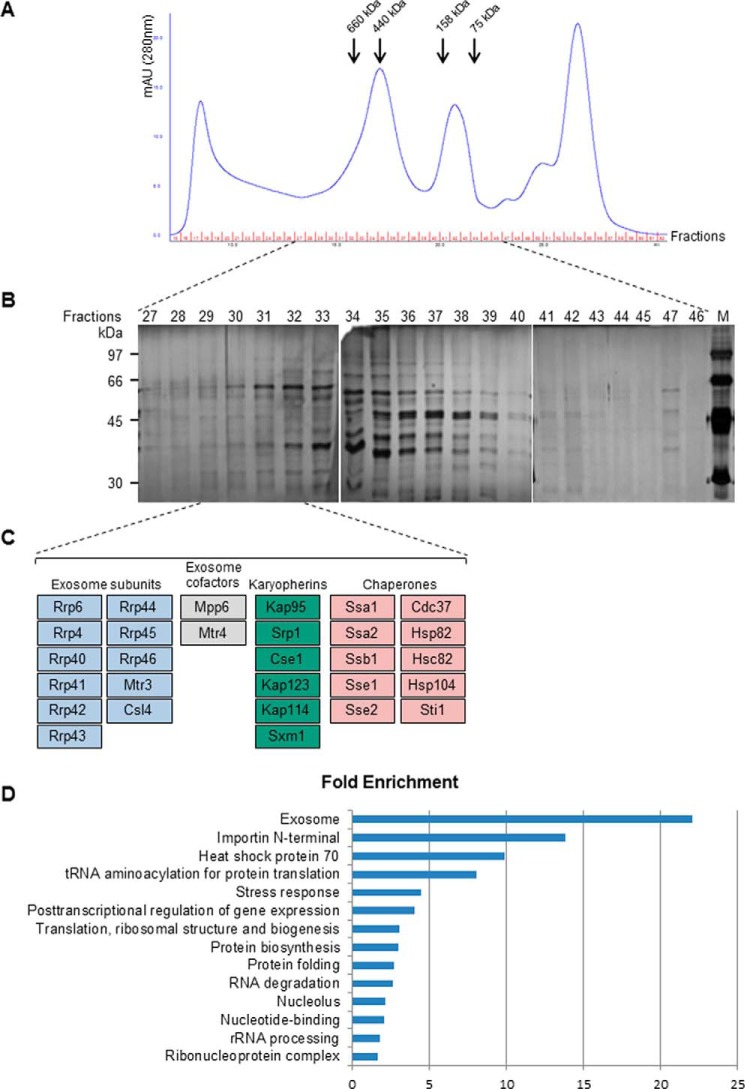
To confirm that the exosome subunits eluted from the column were assembled in the Exo11 complex, proteins co-purified with Rrp6 were subjected to gel filtration for the separation of complexes from free subunits. The collected fractions were subsequently analyzed by SDS-PAGE, showing that the main protein bands were concentrated between fractions 31 and 39 (Fig. 2B). Protein identification of the fractions by mass spectrometry showed that the exosome was eluted mainly in fractions 29 through 32 (supplemental Table S2), corresponding to complexes of 440–660 kDa. These results confirm that the exosome complex was stable under the conditions used here (Fig. 2C and supplemental Fig. S1).
Figure 2.
Proteins co-purified with Rrp6-TAP were subjected to gel filtration on a Superose 6 column for the separation of the exosome complex. A, chromatographic profile of the proteins. Arrows indicate the elution volumes of the molecular mass controls run through the same column. B, SDS-PAGE of the fractions obtained. Fractions were also analyzed by mass spectrometry for the identification of the proteins. C, fractions 29–32 contained, in addition to exosome subunits, some exosome cofactors, karyopherins, and chaperones, grouped in different colors. D, proteins co-purifying with Rrp6-TAP were classified by GO using DAVID software. Karyopherins were the second most predominant category of proteins. mAU, milli-absorbance units.
Many fractions in which the exosome was identified corresponded to complexes with masses larger than expected (478 kDa), indicating that the exosome could be associated with additional proteins. Accordingly, some of the previously characterized exosome cofactors that had been shown to bind Rrp6, such as Mpp6 and Mtr4 (26), were identified in the same fractions in which the exosome was concentrated (Fig. 2). Additionally, other proteins were co-purified with the exosome, many of them chaperones (supplemental Table S2), which may interact with the exosome to facilitate the assembly of this large protein complex (31). Interestingly, karyopherins were also identified among the proteins co-purified with the exosome (Fig. 2 and supplemental Table S2).
Karyopherins co-purify with Rrp6-TAP
In nine independent experiments of Rrp6-TAP co-immunoprecipitations, we detected a total of 298 proteins associated with Rrp6 (supplemental Table S1). Gene ontology (GO) analysis of the proteins co-precipitated with Rrp6-TAP showed that the term “importins/karyopherins” was significantly enriched (Fig. 2D). The karyopherins identified co-purifying with Rrp6-TAP were Kap95, Srp1, Kap114, Kap123, Sxm1, and Cse1 (supplemental Table S1). The interaction of Srp1 and Kap95 with Rrp6 had been observed previously in protein purification experiments (23–26). Interestingly, however, interaction of the exosome with the other karyopherins has not been reported previously. Kap114 has been shown to be responsible for nuclear transport of TATA-binding protein; the transcription factor TFIIB; histones H2A and H2B; and the cofactor associated with histone, NAP1 (32–35). Kap123 is involved in nuclear transport of ribosomal proteins before their association with the ribosome subunits and in transport of other proteins associated with ribosomes; of histones H2A, H2B, H3 and H4; the histone acetyltransferase complex; signal recognition particle (SRP) protein; and the endonuclease HO (32, 33, 36–41). Sxm1 is involved in nuclear transport of Lhp1 chaperone (La protein in humans) and complements the absence of Kap123 (42). Finally, Cse1 is an exportin responsible for transporting SRP1 to the cytoplasm (43). Because various nuclear import pathways can be responsible for the transport of proteins to the nucleus, including the recognition of the cargo nuclear localization signals by many karyopherins (32, 44, 45), the identification of different karyopherins complexed with the exosome could suggest multiple nuclear import pathways for the exosome (46).
Depletion of various karyopherins affects Rrp6 localization
Earlier studies showed the presence of a classical NLS at the C-terminal region of Rrp6 that could be recognized by importin α Srp1 (47), suggesting that the transport of Rrp6 to the nucleus would be performed by the heterodimer Kap95/Srp1. To confirm this hypothesis, we first tested the effect of inhibition of Srp1 expression on Rrp6 subcellular localization.
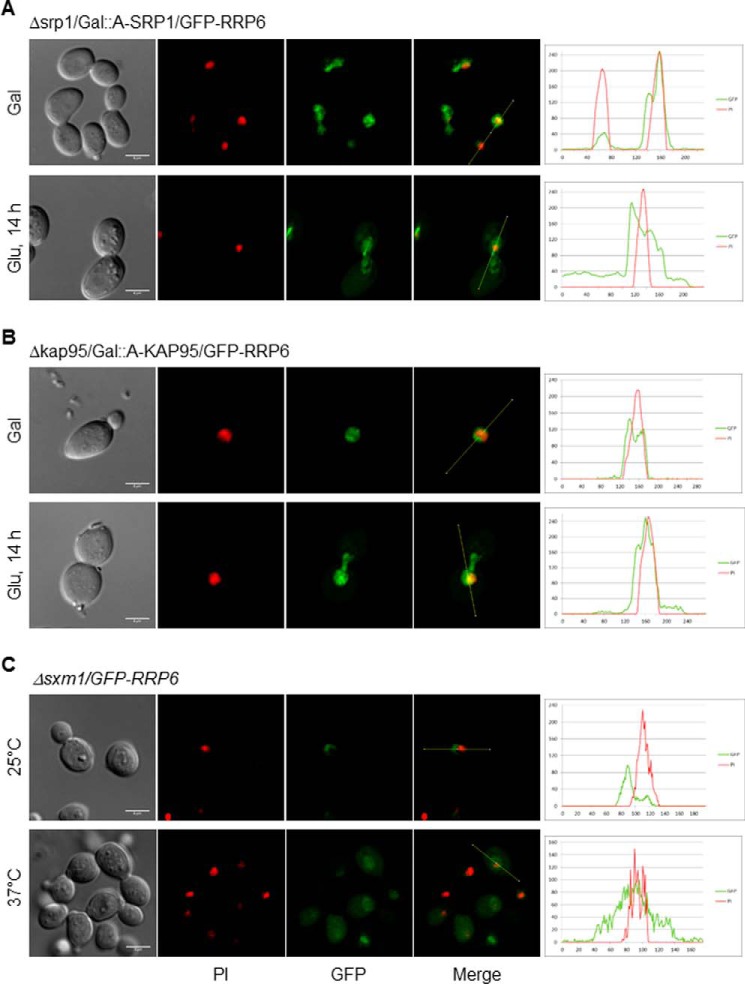
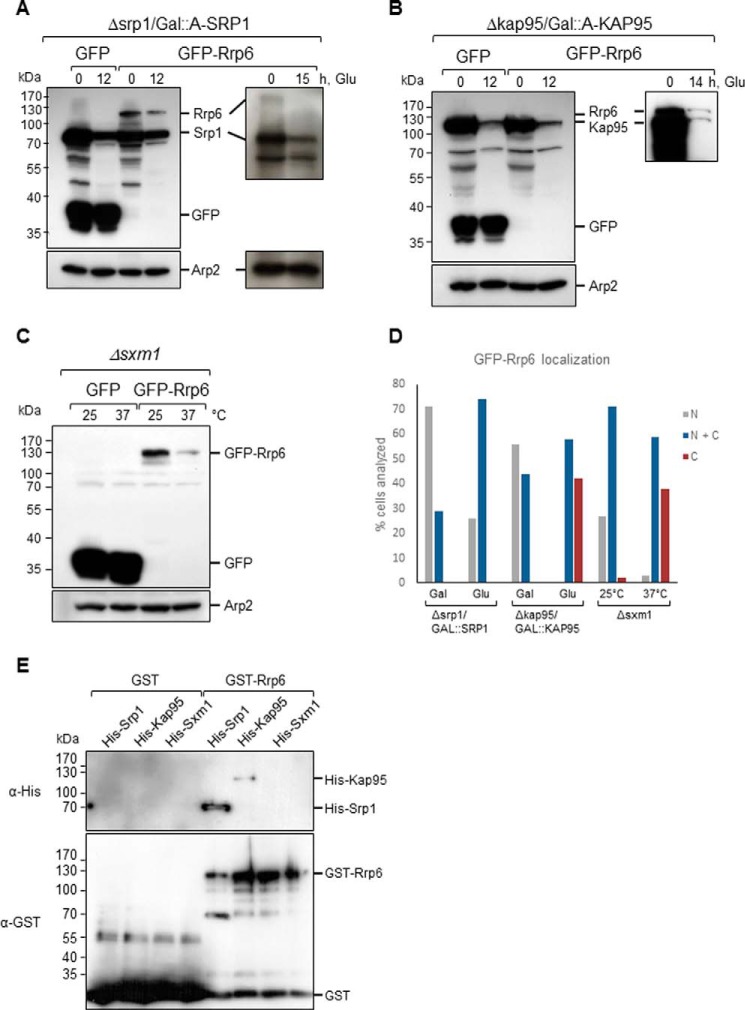
A conditional mutant of SRP1 (Δsrp1/GAL1::A-SRP1) was transformed with plasmids coding for either GFP or GFP-Rrp6 for analysis of Rrp6 localization upon inhibition of expression of Srp1 for 14 h in glucose. The results show that GFP-Rrp6 was localized to the nucleus when cells were incubated in galactose medium, but after incubation for 14 h in glucose medium, a weak protein signal was visualized in the cytoplasm, although GFP-Rrp6 remained concentrated in the nucleus, most probably due to the low levels of Srp1 still present in the cells (Fig. 3A). A control experiment shows GFP in the cytoplasm of these cells (supplemental Fig. S2A). A control Western blot shows that A-Srp1 expression was inhibited after 12 h in glucose, but its levels decreased significantly only after 15 h in glucose (Fig. 4A). Interestingly, the results also show that the levels of Rrp6 decreased upon inhibition of A-Srp1 expression, whereas the levels of GFP alone did not change (Fig. 4A, left panel). These results confirm the involvement of Srp1 in the Rrp6 nuclear import and could suggest that when not efficiently transported to the nucleus Rrp6 may be destabilized.
Figure 3.
Inhibition of karyopherins expression affects the subcellular localization of GFP-Rrp6. A, laser-scanning confocal microscope images show the subcellular localization of GFP-Rrp6 after inhibition of Srp1 expression for 14 h in glucose medium in Δsrp1/GAL::SRP1 cells. Analysis of GFP-Rrp6 relative to PI using ImageJ is shown on the right. Green lines represent GFP, and red lines represent PI. Localization of GFP in Δsrp1/GAL::SRP1 cells is shown in supplemental Fig. S2. B, analysis of the subcellular localization of GFP-Rrp6 after inhibition of Kap95 expression for 14 h in glucose medium in Δkap95/GAL::KAP95 cells. Analysis of GFP-Rrp6 relative to PI using ImageJ is shown on the right. C, analysis of the subcellular localization of GFP-Rrp6 in Δsxm1 cells growing at 25 or 37 °C. Because GFP-Rrp6 expression is lower in this strain growing at 37 °C, gain of the confocal microscope had to be increased for the Rrp6 signal to be detected. Importantly, this signal was above background levels. Scale bars, 4 μm.
Figure 4.
Inhibition of karyopherins expression affects the levels of GFP-Rrp6. Western blotting of total cell extract from karyopherin mutants expressing either GFP or GFP-Rrp6 growing in galactose (Gal)- or glucose (Glu)-containing medium was performed with antibody against GFP, which also allowed the detection of ProtA-Srp1 and ProtA-Kap95. A, Δsrp1/GAL::A-SRP1 growing in glucose shows the lower levels of ProtA-Srp1 after 12 or 15 h in glucose. GFP-Rrp6 also decreases upon inhibition of Srp1 expression. B, Western blot of total cell extract from Δkap95/GAL::A-KAP95 expressing either GFP or GFP-Rrp6 shows the lower levels of ProtA-Kap95 in glucose. GFP-Rrp6 and ProtA-Kap95 have very similar molecular masses. A longer run and exposure for separation and visualization of the bands are shown on the right-hand side. C, Western blot of total cell extract from Δsxm1 expressing either GFP or GFP-Rrp6 growing at 25 or 37 °C shows that the expression levels of GFP-Rrp6 are lower at 37 °C. Western blotting with antibody against Arp2 was used as an internal control. D, quantitative analysis of cells expressing GFP-Rrp6. Approximately 100 cells of each strain were analyzed by fluorescence microscopy, and cells that showed protein localized to the nucleus (N), present both in nucleus and cytoplasm (N + C), or visible mainly in the cytoplasm (C) were counted. Numbers correspond to the percentage of cells showing each phenotype. E, pulldown of Srp1 and Kap95 with Rrp6. GST or GST-Rrp6 bound to glutathione-Sepharose beads was incubated with His-Srp1- or His-Kap95-containing extracts. Elution fractions are shown. The same membrane was incubated with antibody against His tag and subsequently with antibody against GST tag. The figure shown is representative of three independent experiments.
Kap95 is the yeast ortholog of human importin β and has been shown to be important for the transport of some transcription factors (48). To analyze the involvement of Kap95 in the nuclear import of Rrp6, a conditional mutant expressing A-Kap95 under control of the GAL1 promoter (Δkap95/GAL1::A-KAP95) was transformed with plasmids expressing either GFP or GFP-Rrp6. Inhibition of Kap95 expression strongly affected GFP-Rrp6 localization, although the latter protein remained concentrated in the nucleus (Fig. 3B). It is noteworthy that some of the Δkap95/GAL1::A-KAP95 cells showed an elongated form when grown in glucose medium (supplemental Fig. S2B), probably due to the role of Kap95 in the transport of proteins involved in cell cycle regulation (48). The analysis of GFP-Rrp6 upon inhibition of Kap95 expression shows that the levels of Rrp6 also decreased (Fig. 4B) as observed after lowering Srp1 levels. The molecular masses of GFP-Rrp6 and ProtA-Kap95 are similar; therefore, to better visualize GFP-Rrp6 band, the gels were also run longer to separate the bands (Fig. 4B, right panel). The results showing that lower levels of Kap95 resulted in a stronger mislocalization of GFP-Rrp6 than lower levels of Srp1 suggest that the complex Srp1–Kap95 is not solely responsible for Rrp6 transport, but rather Kap95 may be involved in the import of Rrp6 to the nucleus, either on its own or associated with other adaptor proteins in addition to Srp1, as shown for other proteins (45).
To test the hypothesis that more than one transport pathway might be involved in the nuclear import of Rrp6, we investigated the subcellular localization of Rrp6 in mutants of other karyopherins found to co-purify with Rrp6-TAP: Kap114, Kap123, and Sxm1. Upon testing deletion of KAP114 or KAP123, however, no effect on Rrp6 localization in the cell was detected (data not shown). These results are interesting because although the karyopherins Kap95, Srp1, Kap114, and Kap123 were co-immunoprecipitated in the same fraction as the exosome in the gel filtration chromatography the individual depletion of Kap95 and Srp1 partially affected Rrp6 localization, whereas deletion of Kap114 and Kap123 did not have any effect, which might suggest that these latter karyopherins interact with other proteins co-purifying with Rrp6-TAP.
Strengthening our hypothesis of alternative pathways for Rrp6 nuclear import, deletion of the β-karyopherin Sxm1/Kap108 gene partially affected the localization of Rrp6. Sxm1 is not essential for growth, and despite not being described as temperature-sensitive (42), the localization of GFP-Rrp6 was tested in the deletion strain Δsxm1 under different temperatures, 25 and 37 °C. The results show that in this strain, although still concentrated in the nucleus, Rrp6 was detected in the cytoplasm, and its mislocalization was stronger at 37 °C (Fig. 3C). Interestingly, when the cells were shifted to 37 °C, the levels of GFP-Rrp6 decreased as visualized by Western blotting (Fig. 4C). Because GFP-Rrp6 expression was lower in this strain growing at 37 °C, gain of the confocal microscope had to be increased so Rrp6 signal could be detected. Importantly, this signal was above background levels. A control experiment shows that GFP-Rrp6 remains nuclear in WT cells incubated at 37 °C (supplemental Fig. S2C). The results of Srp1, Kap95, and Sxm1, an α- and two β-karyopherins, affecting the nuclear localization of Rrp6 further suggest multiple transport pathways for this protein. Quantification of karyopherin mutant cells clearly shows the stronger effect of the deletion of SXM1 on GFP-Rrp6 localization (Fig. 4D).
To determine whether Rrp6 could directly interact with these karyopherins in the absence of other yeast factors, protein pulldown experiments were performed using recombinant epitope-tagged proteins expressed in Escherichia coli. GST or GST-Rrp6 was immobilized on glutathione-Sepharose beads and incubated with extracts of cells expressing either His-Srp1 or His-Kap95. After extensive washing, proteins were eluted and analyzed by Western blotting with antibodies against the tags. The results show that GST-Rrp6 interacts directly with His-Srp1 and with His-Kap95 (Fig. 4E). Rrp6 interacts more strongly with Srp1 than with Kap95 as deduced by the relative amounts of proteins recovered in the elution fractions. Importantly, however, the results shown here confirm that Kap95 can interact with Rrp6 independently of Srp1, strongly indicating alternative pathways for Rrp6 nuclear import.
Because the effect of Sxm1 on Rrp6 localization was stronger than that of Kap95, we also attempted to test Rrp6–Sxm1 interaction. However, despite obtaining satisfactory His-Sxm1 expression levels in E. coli, this protein was mainly present in inclusion bodies, and the little soluble protein was very labile (data not shown), making the performance of pulldown experiments unviable.
We next analyzed the participation of additional karyopherins in Rrp6 nuclear import. Mutants of karyopherins were therefore transformed with the plasmid coding for the GFP-Rrp6 fusion. Deletion of Kap120 had a very small effect on Rrp6 nuclear accumulation when the cells were incubated at 25 °C (supplemental Fig. S3). Deletion of KAP120 did not significantly affect GFP-Rrp6 levels, but when the cells were incubated at 37 °C, GFP-Rrp6 was visualized in an area apparently larger than the nucleus, which could correspond to a perinuclear localization of this protein. This phenotype may be due to a defect in pre-60S maturation caused by the absence of Rpf1, which has been shown to be transported by Kap120 (49), in the nucleus. Additionally, deletion of KAP120 has been shown to cause accumulation of 60S in the nucleus with stronger defects when cells were incubated at 37 °C (50).
Deletion of the karyopherin Msn5 gene resulted in partial mislocalization of Rrp6 (supplemental Fig. S3B). When analyzing GFP-Rrp6 expression in this strain, however, full-length GFP-Rrp6 band was not visualized on Western blots (supplemental Fig. S3D) even though the GFP-Rrp6 signal was visible by fluorescence microscopy. The GFP-Rrp6 degradation product visualized on Western blots cannot correspond to GFP alone because of its mass and its concentration in the nucleus as visualized by fluorescence microscopy (supplemental Fig. S3B). Full-length GFP-Rrp6 might be present in these cells below the levels of detection by Western blotting but at levels sufficient for detection by fluorescence microscopy. These results of GFP-Rrp6 localization in Δmsn5 cells show that despite being concentrated in the nucleus Rrp6 is not stable, suggesting that nuclear accumulation is not the only important factor for maintenance of Rrp6 levels.
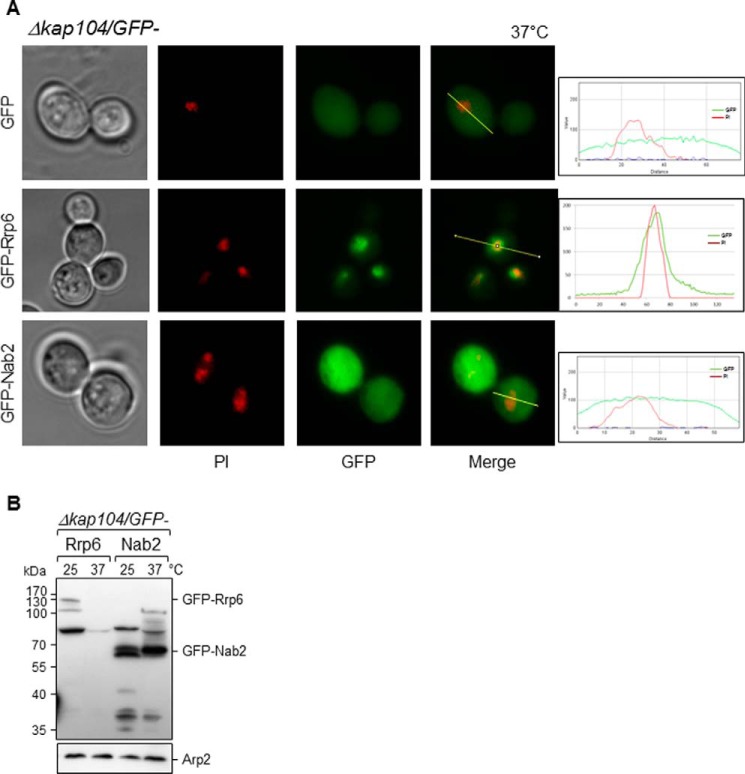
Kap104 (homolog of human karyopherin β2/transportin) has been shown to mediate nuclear import of Nab2 (a nuclear polyadenylated RNA-binding protein), Hrp1 (subunit of cleavage complex required for maturation of pre-mRNA 3′-ends), and the transcription factor Tfg2 (51–54). Kap104 recognizes a multipartite proline-tyrosine nuclear localization signal (PY-NLS) present in its cargos. Multipartite PY-NLSs share a common C-terminal (R/H/K)X2–5PY motif within a positively charged region of ∼30 amino acids. The central region can contain a basic residue-enriched motif or a hydrophobic motif (55). Interestingly, Rrp6 contains a similar PY-NLS motif in its sequence (Fig. 6A, NLS2). To test the involvement of Kap104 in the transport of Rrp6, the Δkap104 strain was transformed with plasmids coding for GFP, GFP-Rrp6, or GFP-Nab2, and the subcellular localization of these proteins was determined by fluorescence microscopy. Absence of Kap104 in cells growing at 37 °C caused some mislocalization of Rrp6, but it remained concentrated in the nucleus (Fig. 5A). Interestingly, from a total of 119 cells observed, 16% showed granules close to the nucleus as well (data not shown). As expected, absence of Kap104 very strongly affected the transport of Nab2, used here as a control (Fig. 5A). Interestingly, upon assessing the levels of expression of GFP-Rrp6 and GFP-Nab2 in Δkap104 cells, Western blotting results show that although GFP-Nab2 levels do not vary when incubating cells at different temperatures the levels of GFP-Rrp6 decreased dramatically at 37 °C (Fig. 5B).
Figure 6.
Expression of Rrp6 deletion mutants in Δrrp6. A, schematic representation of the deletion mutants of Rrp6. Rrp47 and core exosome interaction regions as well as active domains of Rrp6 are highlighted. Positions of putative NLSs are shown in blue. B, Western blot for the determination of the expression of the GFP-Rrp6 mutants in the Δrrp6 strain. Arp2 was used as an internal control. C, analysis of growth of Δrrp6 cells expressing either GFP or the different GFP-Rrp6 constructs at 30 or 37 °C. Mutants lacking the canonical NLS, Rrp6(1–619) and Rrp6(1–693), complement growth of Δrrp6 at 37 °C. HRDC, helicase and RNase D C-terminal domain.
Figure 5.
Deletion of KAP104 does not affect localization of GFP-Rrp6 at 37 °C. A, fluorescence microscopy for the analysis of the subcellular localization of GFP-Rrp6 in Δkap104 cells. GFP-Nab2, a protein affected by the absence of Kap104, was used as a control. Analysis of GFP-Rrp6 and GFP-Nab2 relative to PI in Δkap104 cells growing at 37 °C by is shown on the right. B, Western blotting of total cell extract from Δkap104 cells expressing GFP-Rrp6 or GFP-Nab2 incubated at 25 or 37 °C was performed with antibody against GFP. Expression levels of GFP-Rrp6, but not those of GFP-Nab2, decreased at 37 °C in Δkap120 cells. Antibody against Arp2 was used as an internal control.
Combined, the results shown here confirm the hypothesis that Rrp6 can associate with different β-importins for its transport to the nucleus. Accordingly, further analysis of Rrp6 primary sequence using NLS prediction software revealed a third putative nuclear localization signal in its N-terminal region (Fig. 6A, NLS1) in addition to the classical NLS in the C-terminal portion of the protein and the PY-NLS pointed out above.
Deletion mutants of Rrp6 show different subcellular localization profiles
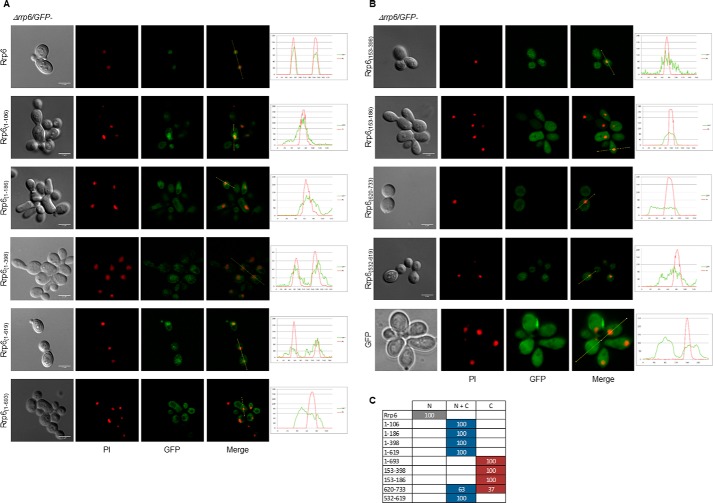
To determine whether the putative NLS at the N-terminal portion of Rrp6 could be involved in its nuclear import, we next constructed GFP-fused Rrp6 deletion mutants lacking one or more putative NLSs (Fig. 6). To analyze the expression levels of the mutants in the Δrrp6 strain, the proteins were first visualized by Western blotting. Although the expression levels of the mutants vary, bands corresponding to the GFP-fused proteins were visualized for all of them with the exception of Rrp6(532–733) (Fig. 6B). Levels of mutants Rrp6(1–106), Rrp6(1–398), and Rrp6(153–398) were similar to that of full-length Rrp6, whereas mutants Rrp6(1–186), Rrp6(1–619), Rrp6(1–693), Rrp6(153–186), Rrp6(620–733), and Rrp6(532–619) showed higher levels than the full-length protein. To determine whether any of the deletion mutants could complement growth of the temperature-sensitive Δrrp6 strain (56), Δrrp6 cells expressing the deletion mutants were incubated at either 30 or 37 °C. Interestingly, mutants Rrp6(1–619) and Rrp6(1–693), which lack the C-terminal region of Rrp6 and therefore the classical NLS (NLS3), did complement growth at 37 °C (Fig. 6C). Remarkably, mutant Rrp6(532–619), containing only the exosome-interacting domain, partially complemented growth of Δrrp6 at 37 °C (Fig. 6C). These findings suggest that the C-terminal NLS is not essential for Rrp6 function and consequently not for its subcellular localization. Surprisingly, mutants Rrp6(1–106), Rrp6(1–186), and Rrp6(1–398), bearing the Rrp47-interacting region, and mutants Rrp6(153–398) and Rrp6(532–733) negatively affected growth of Δrrp6 (Fig. 6C and data not shown). Rrp6(153–398) might interact with exosome subunits or exosome cofactors, sequestering these proteins and causing the negative effects. As indicated above, Rrp6(532–733) could not be detected by Western blotting (Fig. 6B).
The analysis of the subcellular localization of the GFP-Rrp6 deletion mutants in strain Δrrp6 by fluorescence microscopy showed that the full-length GFP-Rrp6 was concentrated in the cell nucleus (Fig. 7) as expected, confirming that the GFP-fused protein was functional as also seen by growth complementation at 37 °C (Fig. 6C). To test the role of the classical NLS in Rrp6 localization, mutants Rrp6(1–106), Rrp6(1–186), Rrp6(1–398), Rrp6(1–619), and Rrp6(1–693), which lack NLS3, were analyzed. Rrp6(1–106), Rrp6(1–186), Rrp6(1–398), and Rrp6(1–619) were concentrated in the nucleus, although significant amounts of the proteins were present in the cytoplasm. Rrp6(1–693), in contrast, was present throughout the cells (Fig. 7). This observation suggests that the putative NLS elements found in the N-terminal portion of Rrp6 could be recognized by karyopherins, allowing the transport of the mutants to the nucleus (Fig. 7). Alternatively, these mutants could interact with proteins containing an NLS and be transported to the nucleus as subcomplexes. This might be the case especially for mutant Rrp6(1–106), which contains only the first three amino acid residues of putative NLS1 (residues 104–132). Because these five mutants contain the Rrp47-interacting domain, these results also suggest that Rrp47 is not sufficient for the nuclear retention of Rrp6.
Figure 7.
Presence of canonical NLS is not the only determinant of Rrp6 nuclear import. Laser-scanning confocal microscope images show the subcellular localization of the Rrp6 deletion mutants expressed in Δrrp6 cells growing at 25 °C. A, GFP-Rrp6, Rrp6(1–106), Rrp6(1–186), Rrp6(1–398), Rrp6(1–619), and Rrp6(1–693). B, Rrp6(153–398), Rrp6(153–186), Rrp6(620–733), Rrp6(532–619), and GFP. Various mutants accumulate in the nucleus although to different levels. Quantification is shown on the right. C, quantitative analysis of cells expressing GFP-fused Rrp6 mutants. N, protein concentrated in the nucleus; N + C, protein present both in nucleus and cytoplasm; C, protein visible mainly in the cytoplasm. Numbers correspond to the percentage of cells showing each phenotype. Scale bars, 4 μm.
Mutants Rrp6(153–398) and Rrp6(153–186), despite containing NLS2 at the N terminus of the exoribonuclease domain, were present throughout the cells (Fig. 7). The signal difference between these mutants is due to their different levels of expression. These results indicate that conformation of the mutant proteins and protein interactions play a significant role in NLS recognition. Mutant Rrp6(620–733), containing only the C-terminal portion of Rrp6 encompassing the classical NLS, was transported to the nucleus albeit with very low efficiency because the protein was mainly visualized in the cytoplasm (Fig. 7). Strikingly, Rrp6(532–619), a mutant that does not contain any NLS but only the Rrp6 portion involved in interaction with the exosome core, although present in the cytoplasm was concentrated in the nucleus (Fig. 7), further confirming that alternative pathways might be used for the transport of Rrp6 through the nuclear pore. These results also suggest that Rrp6 might interact with other proteins containing an NLS sequence and be imported to the nucleus as part of a complex. Based on these results, the NLS in the C-terminal portion of Rrp6 is undoubtedly not the only region of the protein responsible for its nuclear localization. Additionally, there seems to be a correlation between Rrp6 function and cell form. In the case of Rrp6 mutants that did not complement growth and did not localize to the nucleus, the cells seemed more elongated than normal, suggesting some impairment of cell division, as has been described for Drosophila (57). Confirming our observations, changes in Rrp6 expression have been shown to cause filamentous growth (58). Δrrp6 expressing mutants Rrp6(1–106), Rrp6(1–186), Rrp6(1–398), Rrp6(153–186), and Rrp6(532–733) showed the same elongated cell phenotype as the Δrrp6 strain, which correlates with the lack of growth complementation at 37 °C (Fig. 6C and data not shown). Mutants Rrp6(1–619), Rrp6(1–693), and Rrp6(532–619), in contrast, did not show the elongated phenotype, were expressed at high levels, complemented growth, and localized to the nucleus despite lacking the canonical NLS. The results shown here strongly indicate that alternative import pathways are responsible for Rrp6 nuclear localization.
Sxm1 is involved in Rrp6 nuclear import
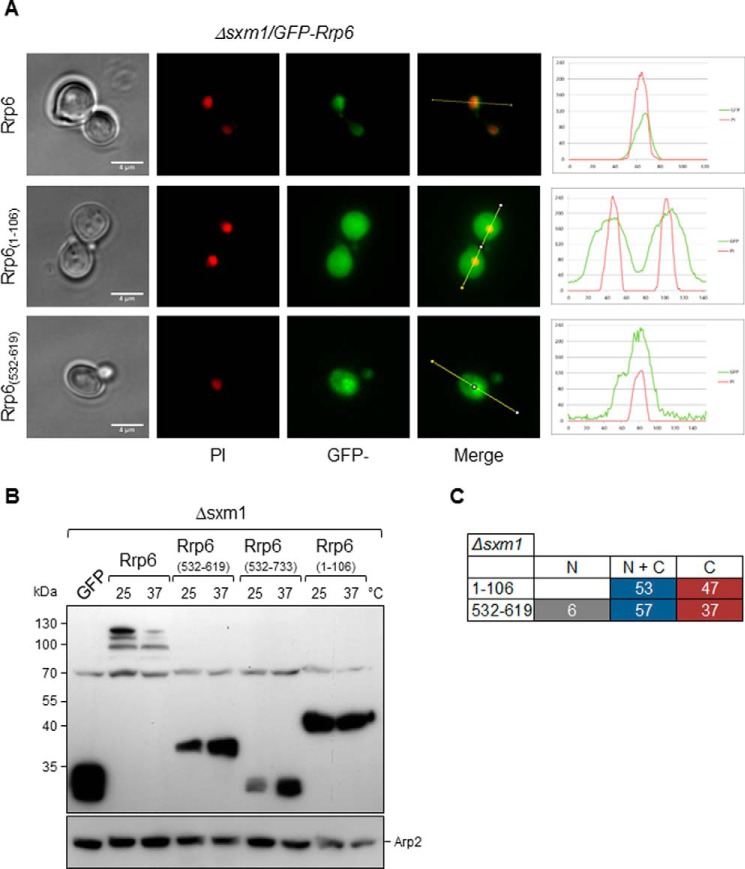
Because Rrp6 mutants lacking its canonical C-terminal NLS were transported to the nucleus and deletion of SXM1 affected Rrp6 localization (Figs. 7 and 3C, respectively), we tested the localization of mutants Rrp6(1–106) and Rrp6(532–619) in the Δsxm1 strain. Rrp6(1–106), containing the Rrp47 interaction domain, was concentrated in the nucleus in Δrrp6 but not in Δsxm1 (Fig. 8A). Rrp6(532–619), containing only the exosome core-interacting domain, localized throughout the cell but was more concentrated in the nucleus in Δsxm1 cells (Fig. 8A). These results strongly suggest that Sxm1 participates in Rrp6 nuclear import either by recognizing the Rrp6 noncanonical NLS or the NLS of a protein complexed with Rrp6. Importantly, as pointed out above, full-length Rrp6 is not stable in Δsxm1 cells at 37 °C, whereas the mutants Rrp6(1–106) and Rrp6(532–619) were expressed at high levels in Δsxm1 cells (Fig. 8B). As in Δrrp6 cells, Rrp6(532–733) was not detected by Western blotting in Δsxm1(Fig. 8B).
Figure 8.
Deletion of SXM1 strongly affects the subcellular localization of Rrp6 mutants. A, fluorescence microscopy for the analysis of the subcellular localization of GFP-Rrp6 mutants in Δsxm1 cells growing at 25 °C. Mutants Rrp61–106 and Rrp6(532–619) were analyzed in this strain. Quantification is shown on the right. B, Western blot for the determination of the expression of the GFP-Rrp6 mutants in the Δsxm1 strain. Arp2 was used as an internal control. Mutants are expressed at higher levels than full-length Rrp6, and their levels do not decrease at 37 °C. C, quantitative analysis of Δsxm1 cells expressing GFP-fused Rrp6 mutants. N, protein concentrated in the nucleus; N + C, protein present both in nucleus and cytoplasm; C, protein visible mainly in the cytoplasm. Numbers correspond to the percentage of cells showing each phenotype. Scale bars, 4 μm.
Rrp47 is not the only factor affecting Rrp6 nuclear retention
Rrp6 has been shown to interact with Rrp47 through its N-terminal domain (59). To determine whether Rrp47 could influence Rrp6 mutant localization, full-length Rrp6 and deletion mutants containing the Rrp47-interacting region, Rrp6 (1–106), Rrp6(1–398), Rrp6(1–619), and Rrp6(1–693), were transformed into Δrrp47 cells for analysis. The results show that localization of full-length GFP-Rrp6, which remained exclusively nuclear (Fig. 9), was not affected by the absence of Rrp47. Rrp6 deletion mutants, in contrast, were mainly visualized in the cytoplasm (Fig. 9). These results are very interesting because although mutants Rrp6(1–106) and Rrp6(1–398) do not contain the canonical C-terminal NLS they were present in the nucleus in strain Δrrp6, but remarkably, significantly lower amounts of these proteins were visible in the nucleus in Δrrp47 cells (Figs. 7 and 9A). Mutants Rrp6(1–619) and Rrp6(1–693) showed similar localization in Δrrp6 and Δrrp47 cells (Figs. 7 and 9).
Figure 9.
Analysis of the subcellular localization of Rrp6 or Rrp6 mutants in Δrrp47 cells. A, fluorescence microscopy for the analysis of the subcellular localization of GFP-Rrp6 or the mutants containing the Rrp47-interacting domain, Rrp6(1–106), Rrp6(1–398), Rrp6(1–619), andRrp6(1–693), in Δrrp47 cells. Absence of Rrp47 affects localization of the Rrp6(1–106) and Rrp6(1–398) mutants. B, analysis of the expression levels of Rrp6 deletion mutants in Δrrp47 cells by Western blotting. Mutants are expressed at higher levels than full-length Rrp6. Arp2 was used as an internal control. C, analysis of growth of Δrrp47 cells expressing either GFP or the different GFP-Rrp6 constructs at 25 or 37 °C. Full-length Rrp6 and mutants Rrp6(1–619) and Rrp6(1–693) complement growth of Δrrp47 at 37 °C.
Similar to what was seen in the Δrrp6 strain, Rrp6 mutants showed different levels of expression in Δrrp47 cells. Most of the mutants were expressed at higher levels than full-length Rrp6 (Fig. 9B). Interestingly, overexpression of Rrp6 and the mutants Rrp6(1–619) and Rrp6(1–693) complemented growth of Δrrp47 cells at 37 °C (Fig. 9C). Combined, these results suggest that Rrp47 plays a role in, but is not the only factor, regulating the nuclear retention of Rrp6.
Discussion
Previous attempts to purify the exosome from yeast cells with tagged core subunits resulted in very little recovery of Rrp6 (16). By using Rrp6 as bait, however, Exo11 complex was purified in a stable form that could be separated by gel filtration. In addition, exosome cofactors and karyopherins were identified that were not present in the complex isolated with TAP-Rrp43 (16). These results show that different proteins remain associated with the exosome depending on the bait used for the purification, probably because the core exosome subunits are present both in nucleus and cytoplasm, whereas Rrp6 is exclusively nuclear.
Among the karyopherins identified with Exo11 were the yeast α-importin Srp1/Kap60 and the β-importins Kap95, Sxm1, Kap114, and Kap123. As shown here, lower levels of Srp1 or of Kap95 partially affect the localization of Rrp6 because despite being concentrated in the nucleus Rrp6 can also be visualized in the cytoplasm upon inhibition of expression of these karyopherins. Kap114 and Kap123, in contrast, do not affect Rrp6 nuclear localization. Co-purification of Kap114 and Kap123 with Rrp6-TAP may therefore be due to the interaction of these karyopherins with other proteins co-purifying with Rrp6.
It has been proposed that the complex Srp1–Kap95 is responsible for the nuclear import of Rrp6 based on pulldown of Srp1 with ProtA-Rrp6 (59); however, as shown here, despite the co-purification of these proteins from yeast cells, inhibition of expression of Srp1 or Kap95 has different effects on the localization of Rrp6. Srp1 possibly recognizes the classical NLS sequence at the Rrp6 C terminus, but in its absence, alternative NLS sequences in the N-terminal portion of Rrp6 may be recognized by other karyopherins because Rrp6 partially localizes to the nucleus in Δsrp1/GAL::SRP1 and Δkap95/GAL::KAP95 strains growing in glucose medium. Importantly, Rrp6 mutants lacking the canonical C-terminal NLS, Rrp6(1–619) and Rrp6(1–693) are not concentrated but are transported to the nucleus and complement growth of Δrrp6 cells at the nonpermissive temperature. Overlapping and redundant import pathways have been reported for other proteins and may also occur in the case of Rrp6 (60).
Confirming this hypothesis, by performing protein pulldown experiments, we show here that Rrp6 not only interacts directly with Srp1 but also with Kap95 in the absence of any other yeast proteins. To the best of our knowledge, this is the first time such Rrp6 direct interactions have been shown. Importantly, by binding directly to the β-karyopherin Kap95, Rrp6 may be transported to the nucleus independently of the α-karyopherin Srp1.
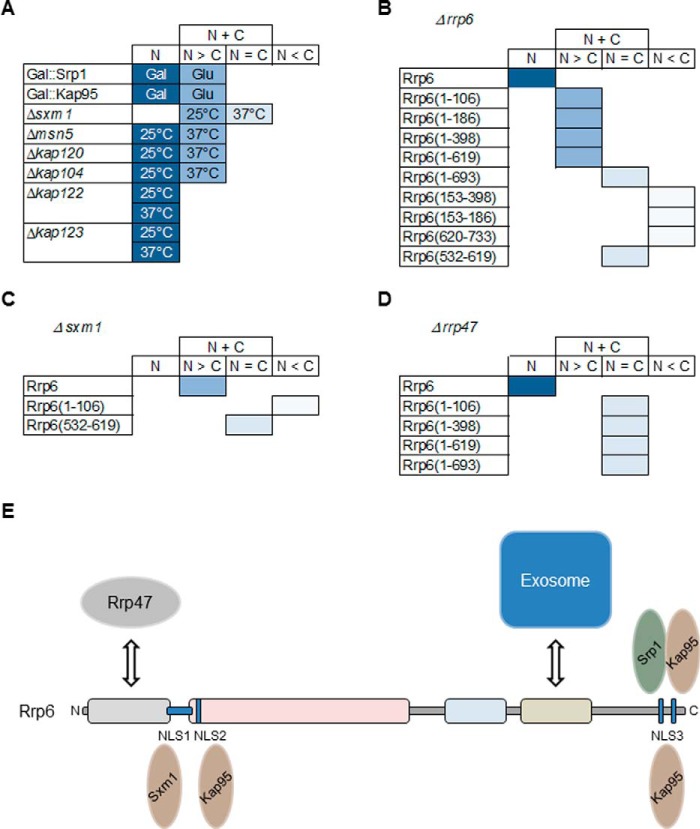
Upon testing other karyopherins, we identified the involvement of Sxm1 in the transport of Rrp6. The deletion of Sxm1 gene strongly affects Rrp6 localization in the cells, suggesting that alternative pathways might be used for the transport of Rrp6 to the nucleus. Confirming that hypothesis, Rrp6(1–106) mislocalized in Δsxm1 cells, a very different phenotype from that observed in the Δrrp6 strain. Sxm1 might recognize an alternative NLS in the Rrp6 sequence or in proteins interacting with Rrp6. This hypothesis is supported by the observation that Rrp6 mutants lacking the canonical NLS still localize to the nucleus but not in the absence of Sxm1. In all karyopherin mutants tested here, despite the presence of Rrp6 in the cytoplasm in some cases, the full-length protein was concentrated in the nucleus. Srp1, Kap95, and Sxm1 were the karyopherins that affected Rrp6 more strongly. Deletion of Msn5 and Kap104 mildly affected Rrp6 localization, whereas Kap120, Kap114, Kap122, and Kap123 had little or no effect (Fig. 10). These results strongly support the idea of overlapping mechanisms for Rrp6 nuclear import.
Figure 10.
A, score chart to summarize the effects of karyopherin depletions (Srp1 and Kap95; galactose (Gal) or glucose (Glu)) or deletions (Sxm1, Msn5, Kap104, Nmd5, Kap120, Kap114, Kap122, and Kap123; 25 or 37 °C) on Rrp6 localization. B, localization of Rrp6 deletion mutants in the Δrrp6 strain. C, localization of Rrp6 deletion mutants in the Δsxm1 strain. D, localization of Rrp6 deletion mutants in the Δrrp47 strain. N, nuclear localization; N > C, mainly nuclear but also present in cytoplasm; N = C, protein visualized both in nucleus and cytoplasm; N < C, protein mainly present in cytoplasm. E, model of the alternative and overlapping pathways for the nuclear import of Rrp6. Rrp6 transport to the nucleus can be facilitated by the α/β dimer Srp1–Kap95 recognizing the canonical NLS3 at the C-terminal portion of Rrp6 or by the β-importins Kap95 and Sxm1 recognizing one of its nuclear localization signals. Alternatively, Rrp6 could be transported in a subcomplex with the exosome complex or other exosome-interacting proteins.
Rrp6 structure has been determined in the context of the exosome complex (4, 28, 61), but the cell compartment in which its association with the exosome occurs has not been described. Considering the structure of Rrp6 when bound to the exosome core, its N-terminal portion is free to interact with Rrp47 (61) and possibly with karyopherins that might recognize the putative N-terminal NLS (Fig. 6A). The classical NLS is positioned in the C-terminal portion of Rrp6, which is also exposed in the exosome structure (61). Alternatively, these NLSs could be recognized in Rrp6 molecules not complexed with the exosome core.
One possible nuclear import pathway for Rrp6 is through the recognition of its classical C-terminal NLS by Srp1–Kap95 heterodimer. However, as shown here, another pathway involves the recognition of an additional N-terminal NLS in Rrp6. We have identified one additional β-karyopherin involved in Rrp6 nuclear import, Sxm1. In the absence of this protein, transport of full-length Rrp6 is less efficient, whereas mutant Rrp6(1–106) is no longer transported to the nucleus. Because of Rrp6 interactions with other exosome subunits and with exosome cofactors, it may also be transported in the form of protein subcomplexes.
As shown here, inhibition of Kap95 expression leads to the appearance of abnormal elongated cells in the culture, similar to the Δrrp6 strain or Δrrp6 expressing deletion mutants of Rrp6. Rrp6 has been shown to be involved in processing of histone mRNAs in yeast, indirectly affecting cell cycle regulation (62, 63). Taken together, these results suggest that there may be a cell division impairment in the absence of functional Rrp6 in the cells or when Rrp6 is mislocalized.
Another important observation described here was that Rrp6 levels decreased in most of the karyopherin mutants, suggesting that when not efficiently transported to the nucleus Rrp6 may be directed for degradation. Incubation of the deletion strains Δmsn5 and Δkap104 at 37 °C led to a very strong decrease in Rrp6 levels so that it could no longer be detected by Western blotting. These results suggest that not only the subcellular localization of Rrp6 affects its levels but additional factors are important as well. It is tempting to hypothesize that Msn5 may be involved in the transport of other proteins that are important for maintaining Rrp6 levels.
Based on the data shown here, the direct interactions between Rrp6 and karyopherins, and the sequences of the putative NLS sequences present in Rrp6, we propose a model according to which Rrp6 transport to the nucleus can be facilitated by the α/β dimer Srp1–Kap95 recognizing the canonical NLS3 at the C-terminal portion of Rrp6 or by the β-karyopherins Kap95 and Sxm1 recognizing one of the nuclear localization signals. In addition to these pathways, Rrp6 may be transported to the nucleus complexed with other exosome subunits, which could be recognized by Sxm1 and other β-karyopherin (Fig. 10E). Importantly, interaction of Rrp6 with the exosome core or with Rrp47 may be necessary for nuclear retention and maintenance of Rrp6 stability.
Experimental procedures
DNA manipulation and plasmid construction
Plasmids used in this study, described in Table 1, were constructed according to cloning techniques described previously (64) and sequenced by the Big Dye method (PerkinElmer Life Sciences). Cloning strategies are briefly described below.
Table 1.
Plasmids used in this work
| Name | Description | Ref. |
|---|---|---|
| pGADC-RRP6 | AD::RRP6; LEU2; 2mm | 11 |
| pUG34 | MET25::GFP, HIS3, CEN6 | 65 |
| pUG36 | URA3, CEN6 | 65 |
| pUG34-ASR1 | MET25::GFP-ASR1, HIS3, CEN6 | This study |
| pUG34-H2A | MET25::GFP-H2A, HIS3, CEN6 | This study |
| pUG34-GCN4 | MET25::GFP-GCN4, HIS3, CEN6 | This study |
| pUG34-NAB2 | MET25::GFP-NAB2, HIS3, CEN6 | This study |
| pUG34-RRP6 | MET25::GFP-RRP6, HIS3, CEN6 | This study |
| pUG34-rrp6(1–106) | MET25::GFP-rrp6 1–106, HIS3, CEN6 | This study |
| pUG34-rrp6(1–398) | MET25::GFP-rrp6 1–398, HIS3, CEN6 | This study |
| pUG34-rrp6(1–619) | MET25::GFP-rrp6 1–619, HIS3, CEN6 | This study |
| pUG34-rrp6(1–693) | MET25::GFP-rrp6 1–693, HIS3, CEN6 | This study |
| pUG34-rrp6(153–398) | MET25::GFP-rrp6 153–398, HIS3, CEN6 | This study |
| pUG34-rrp6(532–733) | MET25::GFP-rrp6 532–733, HIS3, CEN6 | This study |
| pUG34-rrp6(620–733) | MET25::GFP-rrp6 620–733, HIS3, CEN6 | This study |
| pUG34-rrp6(532–619) | MET25::GFP-rrp6 532–619, HIS3, CEN6 | This study |
| pUG36-DsRed-NOP1 | MET25::DsRED-NOP1, URA3, CEN6 | 66 |
| YCplac33 | URA3; CEN4 | 67 |
| YCplac111 | LEU2; CEN4 | 67 |
| YCplac33-GAL-A-RRP43 | GAL1::ProtA-RRP43, URA3, CEN4 | 68 |
| YCplac33-GAL-A-SRP1 | GAL1::ProtA-SRP1, URA3, CEN4 | This study |
| YCplac33-GAL-A-KAP95 | GAL1::ProtA-KAP95, URA3, CEN4 | This study |
| YCplac111-RFP-NOP1 | MET25::DsRED-NOP1, LEU2, CEN4 | This study |
| YCplac111-GFP-RRP6 | MET25::GFP-RRP6, LEU2, CEN4 | This study |
| pGEX-RRP6 | GST::RRP6, AmpR | This study |
| pET28-KAP95 | His::KAP95, KanR | This study |
| pET29-SRP1 | His::SRP1, KanR | This study |
| pET28-SXM1 | His::SXM1, KanR | This study |
Plasmids expressing Srp1 and Kap95 fused to Protein A were constructed by inserting the PCR-amplified open reading frames into YCplac33-GAL-A-RRP43 (68), which was previously digested to remove the RRP43 coding sequence. SRP1 fragment was cloned using BamHI and SalI restriction sites, and KAP95 fragment was cloned using BamHI and PstI restriction sites. In both constructs, the expression of the fusion proteins was regulated by the GAL1 promoter.
Plasmids expressing the GFP fusions in yeast were constructed by inserting DNA fragments into pUG34 plasmid (65) using oligonucleotides with specific restriction sites. An RRP6 fragment was extracted from pGADC2-RRP6 (11) and inserted into pUG34 using EcoRI and SalI restriction sites. Constructs expressing Rrp6 truncated mutants 1–106, 1–186, 1–398, 1–619, 1–693, 153–398, 153–186, 532–733, 620–733, and 532–619 (numbers denote amino acid positions in full-length Rrp6) were amplified by PCR using specific oligonucleotides and inserted into pUG34 with the same restriction enzymes. NAB2 gene was inserted into pUG34 vector using SpeI and SalI restriction enzymes. Expression of these GFP fusions was regulated by MET25 promoter. MET-RFP-NOP1 fragment was extracted from pUG36-DsRed-NOP1 (66) and inserted into YCplac111 SacI and HindIII restriction sites. To construct the plasmid YCplac111-GFP-RRP6, fragment MET25-GFP-RRP6 from pUG34-RRP6 was extracted after digestion with SacI and SalI restrictions enzymes and inserted into YCplac111 plasmid digested with the same enzymes.
Yeast maintenance, transformation, and sporulation
Yeast genetic techniques were conducted as described (69). Yeast strains (Table 2) were maintained in synthetic dropout medium complemented with the appropriate amino acids or nitrogenous base mixture or synthetic complete medium. Glucose or galactose was added as carbon source to a final concentration of 2% (w/v) as indicated. Yeast cells were transformed using the lithium acetate method (70).
Table 2.
Strains used in this work
| Strain | Name | Genotype | Ref. |
|---|---|---|---|
| FGY-25 | KAP95/Δkap95 | BY4743; Mat a/α; his3Δ1/his3Δ1; leu2Δ0/leu2Δ0; lys2Δ0/LYS2; MET15/met15Δ0; ura3Δ0/ura3Δ0; YLR347c::kanMX4/YLR347c | EUROSCARF |
| FGY-53 | Δkap95 | YLR347c::kanMX4; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; Ycplac33-GAL-A-KAP95 | This study |
| FGY-97 | KAP104/Δkap104 | BY4743; Mat a/a; his3Δ1/his3Δ1; leu2Δ0/leu2Δ0; lys2D0/LYS2; MET15/met15Δ0; ura3Δ0/ura3Δ0; YBR017c::kanMX4/YBR017c | EUROSCARF |
| FGY-112 | Δkap104 | CEN.ZI5-3B; CEN.PK; Mat a; ura3–52; his3Δ1; leu2-3_112; trp1–289; YBR017c::URA3 | EUROSCARF |
| FGY-60 | Δkap114 | BY4742; Mat a; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; YGL241w::kanMX4 | EUROSCARF |
| FGY-100 | Δkap120 | BY4742; Mat α; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; YPL125w::kanMX4 | EUROSCARF |
| FGY-101 | Δkap122 | BY4742; Mat a; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; YGL016w::kanMX4 | EUROSCARF |
| FGY-61 | Δkap123 | BY4742; Mat a; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; YER110C::kanMX4 | EUROSCARF |
| FGY-105 | Δmsn5 | BY4742; Mat a; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; YDR335w::kanMX4 | EUROSCARF |
| FGY-87 | Δnmd5 | CEN.RO22–4B; CEN.PK; Mat a; ura3–52; his3Δ1; leu2-3_112; trp1–289; YJR132w::HIS3 | EUROSCARF |
| FGY-5 | RRP6-TAP | MAT a; ade2; arg4; leu2–3,112; trp1–289; ura3–52; YOR001w::TAP | EUROSCARF |
| FGY-88 | Δrrp6 | BY4742; Mat a; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; YOR001w::kanMX4 | EUROSCARF |
| FGY-209 | Δrrp47 | BY4742; Mat α; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; YOR001w::kanMX4 | EUROSCARF |
| FGY-26 | SRP1/Δsrp1 | BY4743; Mat a/α; his3Δ1/his3Δ1; leu2Δ0/leu2Δ0; lys2Δ0/LYS2; MET15/met15Δ0; ura3Δ0/ura3Δ0; YNL189w::kanMX4/YNL189w | EUROSCARF |
| FGY-41 | Δsrp1 | YNL189w::kanMX4; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; Ycplac33-GAL-A-SRP1 | This study |
| FGY-86 | Δsxm1 | BY4742; Mat a; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; YDR395w::kanMX4 | EUROSCARF |
Tandem affinity purification
Rrp6-TAP and empty-TAP purifications were performed as described previously (16). Briefly, 20 liters of yeast cells expressing TAP-tagged Rrp6 were grown to an A600 of 1.0–1.2 in synthetic complete medium containing glucose as a carbon source. Cells were harvested by centrifugation; resuspended in 50 mm Tris-HCl, pH8.0, 150 mm NaCl, 10% glycerol, and 1 mm phenylmethylsulfonyl fluoride (PMSF); immediately frozen in liquid N2; and stored at −80 °C. Cells were lysed by grinding using a ball mill device (Retsch, Mixer Mill MM 200 or Planetary Ball Mill PM 100) and centrifuged at 40,000 rpm for 1 h at 4 °C in a P5OAT2-716 rotor (Hitachi). The supernatant was incubated for 2 h at 4 °C with IgG-Sepharose beads (GE Healthcare) followed by extensive washing with the same buffer. Proteins were eluted from beads by incubating the resin with 20 units of tobacco etch virus protease (Invitrogen) for 16 h at 4 °C in the presence of 1 mm DTT and 0.5 mm EDTA, pH 8.0. The elution fraction from IgG-Sepharose chromatography was subjected to gel filtration using a prepacked glass column, SuperoseTM 6 10/300 GL (GE Healthcare, catalog number 17-5172-01) connected to an AKTA-FPLC (GE Healthcare, catalog number 18-1900-26) at 0.5 ml/min flow rate.
Protein digestion and mass spectrometric identification by LC-MS/MS
For identification of proteins obtained from TAP, the elution fraction was resolved by SDS-PAGE, and bands were removed by cutting gels in slices, reduced, alkylated, and digested with trypsin (71). The digested samples were desalted using a Sep-Pak C18 Plus Short Cartridge (Waters) or EMD Millipore ZipTipTM pipette tips (Millipore) according to the manufacturers' instructions. Tryptic peptides were resuspended in 20 μl of formic acid (0.1%), and an aliquot (4.5 μl) was injected onto a Q-Tof Ultima mass spectrometer (Waters) through a coupled nanoUPLC system (Acquity, Waters). The peptide mixture was first desalted into a C18 trap column (180-μm inner diameter × 20 mm; Waters) with 100% solvent A (0.1% formic acid) at 5 μl/min for 3 min. Peptides were fractionated onto an analytical C18 column (75-μm inner diameter × 100 mm; Waters) in a 20-min gradient (5–40% acetonitrile in 0.1% formic acid) at a flow rate of 600 nl/min. Spray voltage was set at 3.2 kV, and the instrument was operated in data-dependent mode in which one full MS scan was acquired in the m/z range of 200–2000 followed by MS/MS acquisition using collision-induced dissociation of the three most intense ions from the MS scan. Phosphoric acid (0.05% in acetonitrile) was used as a lock mass and therefore was continuously sprayed into the electrospray ionization source and detected every 15 s. Alternatively, peptides originating from in-gel digestion were also analyzed by LC-MS/MS using a Q-Tof Premier mass spectrometer (Waters) as described (72). The raw data were processed and transformed into pkl format using ProteinLynx Global Server (Waters) after lock mass (at m/z 784,823) correction. In-solution digestion was also performed for proteins both directly eluted from TAP constructs (including empty vector) and from those eluted from TAP constructs followed by gel filtration chromatography (73). Glycerol was previously removed from protein mixtures through acetone precipitation. Peptide mixtures originating from total TAP eluates were analyzed by LC-MS/MS using a Q-Tof Premier mass spectrometer as described (16). Tryptic peptides from gel filtration chromatography eluates were analyzed by LC-MS/MS using an LTQ-Velos Orbitrap (Thermo Fisher) as described (16). Proteins were identified by searching against a database sequence of S. cerevisiae (S288c strain, downloaded at UniProt). Carbamidomethylation (Cys) was set as a fixed modification, and oxidation (Met) was set as a variable modification. For Q-Tof and LTQ-Velos Orbitrap mass spectrometry, MS1 tolerance was set to 0.1–0.5 Da and 10 ppm, respectively, and MS2 was set to 0.1–0.5 Da and 0.5 Da, respectively. High resolution data were also analyzed at Proteome Discover 1.4 (Thermo) where the false discovery rate was set to 1%. Proteins present in the empty-TAP (negative control) were excluded from the final list of Rrp6-TAP (interactors detected in all nine purifications). Functional annotation and GO enrichment analysis were performed by DAVID (74, 75) with the parameter Ease = 0.01.
Protein pulldown and immunoblot analysis
In the pulldown assay, cellular extracts (generated in 20 mm Tris, 150 mm NaCl, 1 mm EDTA, 0.8% Nonidet, 1 mm DTT) of E. coli cells expressing either GST or GST-Rrp6 were incubated for 1 h at 4 °C with 250 μl of glutathione-Sepharose beads (GE Healthcare), and the unbound material was washed. Beads were then incubated with cellular extract containing His-Srp1 or His-Kap95, flow-through was collected, and beads were washed with the same buffer followed by washing with buffer containing 250 mm NaCl. Bound proteins were eluted with 50 mm Tris, pH 8.0, 10 mm reduced glutathione.
Immunoblotting experiments
Protein samples were resolved by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (GE Healthcare). Membranes were incubated with primary antibodies against CBP, GFP (Sigma-Aldrich), or Arp2 (Santa Cruz Biotechnology) in phosphate-buffered saline (PBS)/Tween 20/nonfat milk. Secondary antibodies used were anti-rabbit or anti-goat IgG conjugated to peroxidase (Sigma-Aldrich). Western blots were developed using Immobilon Western Chemiluminescent HRP Substrate (Millipore).
Fluorescence microscopy
Cells were fixed in 70% methanol for 15 min, rinsed with cold PBS, and then treated with 1 mg/ml RNase for 30 min. Nuclei were counterstained in a dye solution containing 3 mg/ml propidium iodide (PI) for 15 min. Cells were observed using a Nikon Eclipse Ti microscope equipped with filters for green fluorescence (GFP-3035B-000-ZERO, Semrock) and red fluorescence (Texas Red BrightLine set, TXRED4040-B, Semrock). The exposure times varied from 1 to 3 s. Images were processed and analyzed using the programs Nis Elements (version 3.07; Nikon) and ImageJ (National Institutes of Health, Bethesda, MD). Confocal images were captured in a 1024 × 1024-pixel format using a Zeiss LSM 780 confocal laser-scanning inverted microscope (Carl Zeiss, Germany) at Centro de Facilidades para a Pesquisa (CEFAP-USP). Image stacks comprised eight images captured with an alpha Plan-Apochromat 100×/1.46 oil differential interference contrast M27 objective (Carl Zeiss), applying a zoom factor of 1.5. Step intervals along the z axis ranged from 200 to 250 nm. Image processing was performed using Zen 2011 software (version 11.00.190; Carl Zeiss).
Identification of putative NLS elements
PSORT II prediction software (GenScript), NLS Mapper (76), and NLStradamus (77) were used for the identification of putative NLS elements. Examples of monopartite, bipartite, and basic residue-enriched NLSs can be found in Refs. 19, 52, and 53. Sequences of NLSs are as follows: NLS1, 104NSKSRGSDLQYLGEFSGKNFSPTKRVEKP132; NLS2, 153KEKPNALKPLSESLRLVDDDENNPSHYPHPY183; NLS3, 697RQQKKRRFDPSSSDSNGPRAAKKRRPA723.
Author contributions
C. C. O. conceived and coordinated the study. F. A. G.-Z. designed, performed, and analyzed the experiments. E. K. O. contributed to confocal analysis, protein pulldown experiments, and preparation of the figures. F. A. G.-Z. and J. P. C. D. C. analyzed the mass spectrometry results. F. A. G.-Z., J. P. C. D. C., and C. C. O. wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank all the members of the Oliveira laboratory, especially Bruna F. Rech, for help with cloning and Western blotting and Fiorella Orellana-Peralta, Felipe F. M. Bagatelli, and M. Griselda Perona for reagents and discussions. We thank Frederico Gueiros Filho, members of his laboratory, and Glaucia M. Machado-Santelli for the use of fluorescence microscopes, reagents, and helpful suggestions. We also thank the Brazilian Biosciences National Laboratory (LNBio) for some of the analysis of proteins by mass spectrometry.
This work was supported in part by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) Grants 10/51842-3 and 15/06477-9 (to C. C. O.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S3 and Tables S1 and S2.
- NLS
- nuclear localization signal
- TAP
- tandem affinity purification
- CBP
- calmodulin-binding peptide
- TEV
- tobacco etch virus
- GO
- gene ontology
- SRP
- signal recognition particle
- ProtA
- Protein A
- PI
- propidium iodide.
References
- 1. Mitchell P., Petfalski E., Shevchenko A., Mann M., and Tollervey D. (1997) The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell 91, 457–466 [DOI] [PubMed] [Google Scholar]
- 2. Schmid M., and Jensen T. H. (2008) The exosome: a multipurpose RNA-decay machine. Trends Biochem. Sci. 33, 501–510 [DOI] [PubMed] [Google Scholar]
- 3. Liu Q., Greimann J. C., and Lima C. D. (2006) Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell 127, 1223–1237 [DOI] [PubMed] [Google Scholar]
- 4. Makino D. L., Baumgärtner M., and Conti E. (2013) Crystal structure of an RNA-bound 11-subunit eukaryotic exosome complex. Nature 495, 70–75 [DOI] [PubMed] [Google Scholar]
- 5. Dziembowski A., Lorentzen E., Conti E., and Séraphin B. (2007) A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol. 14, 15–22 [DOI] [PubMed] [Google Scholar]
- 6. Schaeffer D., Tsanova B., Barbas A., Reis F. P., Dastidar E. G., Sanchez-Rotunno M., Arraiano C. M., and van Hoof A. (2009) The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat. Struct. Mol. Biol. 16, 56–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burkard K. T., and Butler J. S. (2000) A nuclear 3′-5′ exonuclease involved in mRNA degradation interacts with poly(A) polymerase and the hnRNA protein Npl3p. Mol. Cell. Biol. 20, 604–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Houseley J., LaCava J., and Tollervey D. (2006) RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 7, 529–539 [DOI] [PubMed] [Google Scholar]
- 9. Allmang C., Petfalski E., Podtelejnikov A., Mann M., Tollervey D., and Mitchell P. (1999) The yeast exosome and human PM-Scl are related complexes of 3′ → 5′ exonucleases. Genes Dev. 13, 2148–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Anderson J. S., and Parker R. P. (1998) The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 17, 1497–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oliveira C. C., Gonzales F. A., and Zanchin N. I. (2002) Temperature-sensitive mutants of the exosome subunit Rrp43p show a deficiency in mRNA degradation and no longer interact with the exosome. Nucleic Acids Res. 30, 4186–4198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schneider C., Kudla G., Wlotzka W., Tuck A., and Tollervey D. (2012) Transcriptome-wide analysis of exosome targets. Mol. Cell 48, 422–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mitchell P., Petfalski E., Houalla R., Podtelejnikov A., Mann M., and Tollervey D. (2003) Rrp47p is an exosome-associated protein required for the 3′ processing of stable RNAs. Mol. Cell. Biol. 23, 6982–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schilders G., Raijmakers R., Raats J. M., and Pruijn G. J. (2005) MPP6 is an exosome-associated RNA-binding protein involved in 5.8S rRNA maturation. Nucleic Acids Res. 33, 6795–6804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Granato D. C., Machado-Santelli G. M., and Oliveira C. C. (2008) Nop53p interacts with 5.8S rRNA co-transcriptionally, and regulates processing of pre-rRNA by the exosome. FEBS J. 275, 4164–4178 [DOI] [PubMed] [Google Scholar]
- 16. Lourenço R. F., Leme A. F., and Oliveira C. C. (2013) Proteomic analysis of yeast mutant RNA exosome complexes. J. Proteome Res. 12, 5912–5922 [DOI] [PubMed] [Google Scholar]
- 17. Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., and Weissman J. S. (2003) Global analysis of protein expression in yeast. Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
- 18. Xu D., Farmer A., and Chook Y. M. (2010) Recognition of nuclear targeting signals by karyopherin-β proteins. Curr. Opin. Struct. Biol. 20, 782–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lange A., Mills R. E., Lange C. J., Stewart M., Devine S. E., and Corbett A. H. (2007) Classical nuclear localization signals: definition, function, and interaction with importin α. J. Biol. Chem. 282, 5101–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Görlich D., Kostka S., Kraft R., Dingwall C., Laskey R. A., Hartmann E., and Prehn S. (1995) Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr. Biol. 5, 383–392 [DOI] [PubMed] [Google Scholar]
- 21. Lusk C. P., Blobel G., and King M. C. (2007) Highway to the inner nuclear membrane: rules for the road. Nat. Rev. Mol. Cell Biol. 8, 414–420 [DOI] [PubMed] [Google Scholar]
- 22. Mosammaparast N., and Pemberton L. F. (2004) Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 14, 547–556 [DOI] [PubMed] [Google Scholar]
- 23. Ho Y., Gruhler A., Heilbut A., Bader G. D., Moore L., Adams S. L., Millar A., Taylor P., Bennett K., Boutilier K., Yang L, Wolting C., Donaldson I., Schandorff S., Shewnarane J., et al. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415, 180–183 [DOI] [PubMed] [Google Scholar]
- 24. Gavin A. C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., Remor M., Höfert C., Schelder M., Brajenovic M., Ruffner H., et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147 [DOI] [PubMed] [Google Scholar]
- 25. Gavin A. C., Aloy P., Grandi P., Krause R., Boesche M., Marzioch M., Rau C., Jensen L. J., Bastuck S., Dümpelfeld B., Edelmann A., Heurtier M. A., Hoffman V., Hoefert C., Klein K., et al. (2006) Proteome survey reveals modularity of the yeast cell machinery. Nature 440, 631–636 [DOI] [PubMed] [Google Scholar]
- 26. Synowsky S. A., van Wijk M., Raijmakers R., and Heck A. J. (2009) Comparative multiplexed mass spectrometric analyses of endogenously expressed yeast nuclear and cytoplasmic exosomes. J. Mol. Biol. 385, 1300–1313 [DOI] [PubMed] [Google Scholar]
- 27. Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., and Séraphin B. (2001) The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24, 218–229 [DOI] [PubMed] [Google Scholar]
- 28. Wasmuth E. V., Januszyk K., and Lima C. D. (2014) Structure of an Rrp6-RNA exosome complex bound to poly(A) RNA. Nature 511, 435–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hernández H., Dziembowski A., Taverner T., Séraphin B., and Robinson C. V. (2006) Subunit architecture of multimeric complexes isolated directly from cells. EMBO Rep. 7, 605–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Luz J. S., Tavares J. R., Gonzales F. A., Santos M. C., and Oliveira C. C. (2007) Analysis of the Saccharomyces cerevisiae exosome architecture and of the RNA binding activity of Rrp40p. Biochimie 89, 686–691 [DOI] [PubMed] [Google Scholar]
- 31. Macario A. J., and Conway de Macario E. (2005) Sick chaperones, cellular stress, and disease. N. Engl. J. Med. 353, 1489–1501 [DOI] [PubMed] [Google Scholar]
- 32. Mosammaparast N., Jackson K. R., Guo Y., Brame C. J., Shabanowitz J., Hunt D. F., and Pemberton L. F. (2001) Nuclear import of histone H2A and H2B is mediated by a network of karyopherins. J. Cell Biol. 153, 251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mosammaparast N., Ewart C. S., and Pemberton L. F. (2002) A role for nucleosome assembly protein 1 in the nuclear transport of histones H2A and H2B. EMBO J. 21, 6527–6538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hodges J. L., Leslie J. H., Mosammaparast N., Guo Y., Shabanowitz J., Hunt D. F., and Pemberton L. F. (2005) Nuclear import of TFIIB is mediated by Kap114p, a karyopherin with multiple cargo-binding domains. Mol. Biol. Cell 16, 3200–3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Morehouse H., Buratowski R. M., Silver P. A., and Buratowski S. (1999) The importin/karyopherin Kap114 mediates the nuclear import of TATA-binding protein. Proc. Natl. Acad. Sci. U.S.A. 96, 12542–12547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rout M. P., Blobel G., and Aitchison J. D. (1997) A distinct nuclear import pathway used by ribosomal proteins. Cell 89, 715–725 [DOI] [PubMed] [Google Scholar]
- 37. Schlenstedt G., Smirnova E., Deane R., Solsbacher J., Kutay U., Görlich D., Ponstingl H., and Bischoff F. R. (1997) Yrb4p, a yeast ran-GTP-binding protein involved in import of ribosomal protein L25 into the nucleus. EMBO J. 16, 6237–6249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Franke J., Reimann B., Hartmann E., Köhlerl M., and Wiedmann B. (2001) Evidence for a nuclear passage of nascent polypeptide-associated complex subunits in yeast. J. Cell Sci. 114, 2641–2648 [DOI] [PubMed] [Google Scholar]
- 39. Schaper S., Franke J., Meijsing S. H., and Ehrenhofer-Murray A. E. (2005) Nuclear import of the histone acetyltransferase complex SAS-I in Saccharomyces cerevisiae. J. Cell Sci. 118, 1473–1484 [DOI] [PubMed] [Google Scholar]
- 40. Grosshans H., Deinert K., Hurt E., and Simos G. (2001) Biogenesis of the signal recognition particle (SRP) involves import of SRP proteins into the nucleolus, assembly with the SRP-RNA, and Xpo1p-mediated export. J. Cell Biol. 153, 745–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bakhrat A., Baranes K., Krichevsky O., Rom I., Schlenstedt G., Pietrokovski S., and Raveh D. (2006) Nuclear import of Ho endonuclease utilizes two nuclear localization signals and four importins of the ribosomal import system. J. Biol. Chem. 281, 12218–12226 [DOI] [PubMed] [Google Scholar]
- 42. Sydorskyy Y., Dilworth D. J., Yi E. C., Goodlett D. R., Wozniak R. W., and Aitchison J. D. (2003) Intersection of the Kap123p-mediated nuclear import and ribosome export pathways. Mol. Cell. Biol. 23, 2042–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hood J. K., and Silver P. A. (1998) Cse1p is required for export of Srp1p/importin-alpha from the nucleus in Saccharomyces cerevisiae. J. Biol. Chem. 273, 35142–35146 [DOI] [PubMed] [Google Scholar]
- 44. Marfori M., Mynott A., Ellis J. J., Mehdi A. M., Saunders N. F., Curmi P. M., Forwood J. K., Bodén M., and Kobe B. (2011) Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim. Biophys. Acta 1813, 1562–1577 [DOI] [PubMed] [Google Scholar]
- 45. Fries T., Betz C., Sohn K., Caesar S., Schlenstedt G., and Bailer S. M. (2007) A novel conserved nuclear localization signal is recognized by a group of yeast importins. J. Biol. Chem. 282, 19292–19301 [DOI] [PubMed] [Google Scholar]
- 46. Tran E. J., King M. C., and Corbett A. H. (2014) Macromolecular transport between the nucleus and the cytoplasm: advances in mechanism and emerging links to disease. Biochim. Biophys. Acta 1843, 2784–2795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Phillips S., and Butler J. S. (2003) Contribution of domain structure to the RNA 3′ end processing and degradation functions of the nuclear exosome subunit Rrp6p. RNA 9, 1098–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kimura M., and Imamoto N. (2014) Biological significance of the importin-β family-dependent nucleocytoplasmic transport pathways. Traffic 15, 727–748 [DOI] [PubMed] [Google Scholar]
- 49. Caesar S., Greiner M., and Schlenstedt G. (2006) Kap120 functions as a nuclear import receptor for ribosome assembly factor Rpf1 in yeast. Mol. Cell. Biol. 26, 3170–3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stage-Zimmermann T., Schmidt U., and Silver P. A. (2000) Factors affecting nuclear export of the 60S ribosomal subunit in vivo. Mol. Biol. Cell 11, 3777–3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lee D. C., and Aitchison J. D. (1999) Kap104p-mediated nuclear import. Nuclear localization signals in mRNA-binding proteins and the role of Ran and RNA. J. Biol. Chem. 274, 29031–29037 [DOI] [PubMed] [Google Scholar]
- 52. Kim B. J., and Lee H. (2006) Importin-β mediates Cdc7 nuclear import by binding to the kinase insert II domain, which can be antagonized by importin-α. J. Biol. Chem. 281, 12041–12049 [DOI] [PubMed] [Google Scholar]
- 53. Lange A., Mills R. E., Devine S. E., and Corbett A. H. (2008) A PY-NLS nuclear targeting signal is required for nuclear localization and function of the Saccharomyces cerevisiae mRNA-binding protein Hrp1. J. Biol. Chem. 283, 12926–12934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Süel K. E., and Chook Y. M. (2009) Kap104p imports the PY-NLS-containing transcription factor Tfg2p into the nucleus. J. Biol. Chem. 284, 15416–15424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lee B. J., Cansizoglu A. E., Süel K. E., Louis T. H., Zhang Z., and Chook Y. M. (2006) Rules for nuclear localization sequence recognition by karyopherin β2. Cell 126, 543–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Briggs M. W., Burkard K. T., and Butler J. S. (1998) Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J. Biol. Chem. 273, 13255–13263 [DOI] [PubMed] [Google Scholar]
- 57. Graham A. C., Kiss D. L., and Andrulis E. D. (2009) Core exosome-independent roles for Rrp6 in cell cycle progression. Mol. Biol. Cell 20, 2242–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jin R., Dobry C. J., McCown P. J., and Kumar A. (2008) Large-scale analysis of yeast filamentous growth by systematic gene disruption and overexpression. Mol. Biol. Cell 19, 284–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Feigenbutz M., Jones R., Besong T. M., Harding S. E., and Mitchell P. (2013) Assembly of the yeast exoribonuclease Rrp6 with its associated cofactor Rrp47 occurs in the nucleus and is critical for the controlled expression of Rrp47. J. Biol. Chem. 288, 15959–15970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chook Y. M., and Süel K. E. (2011) Nuclear import by karyopherin-βs: recognition and inhibition. Biochim. Biophys. Acta 1813, 1593–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Makino D. L., Schuch B., Stegmann E., Baumgärtner M., Basquin C., and Conti E. (2015) RNA degradation paths in a 12-subunit nuclear exosome complex. Nature 524, 54–58 [DOI] [PubMed] [Google Scholar]
- 62. Canavan R., and Bond U. (2007) Deletion of the nuclear exosome component RRP6 leads to continued accumulation of the histone mRNA HTB1 in S-phase of the cell cycle in Saccharomyces cerevisiae. Nucleic Acids Res. 35, 6268–6279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Reis C. C., and Campbell J. L. (2007) Contribution of Trf4/5 and the nuclear exosome to genome stability through regulation of histone mRNA levels in Saccharomyces cerevisiae. Genetics 175, 993–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sambrook J., and Russell D. W. (2001) Molecular Cloning: a Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 65. Niedenthal R. K., Riles L., Johnston M., and Hegemann J. H. (1996) Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast 12, 773–786 [DOI] [PubMed] [Google Scholar]
- 66. Goldfeder M. B., and Oliveira C. C. (2010) Utp25p, a nucleolar Saccharomyces cerevisiae protein, interacts with U3 snoRNP subunits and affects processing of the 35S pre-rRNA. FEBS J. 277, 2838–2852 [DOI] [PubMed] [Google Scholar]
- 67. Gietz R. D., and Prakash S. (1988) Cloning and nucleotide sequence analysis of the Saccharomyces cerevisiae RAD4 gene required for excision repair of UV-damaged DNA. Gene 74, 535–541 [DOI] [PubMed] [Google Scholar]
- 68. Zanchin N. I., and Goldfarb D. S. (1999) The exosome subunit Rrp43p is required for the efficient maturation of 5.8S, 18S and 25S rRNA. Nucleic Acids Res. 27, 1283–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Guthrie C. and Fink G. R. (eds) (1991) Guide to Yeast Genetics and Molecular Biology, Volume 194, Academic Press, Inc., San Diego, CA [Google Scholar]
- 70. Chen D. C., Yang B. C., and Kuo T. T. (1992) One-step transformation of yeast in stationary phase. Curr. Genet. 21, 83–84 [DOI] [PubMed] [Google Scholar]
- 71. Aragão A. Z., Belloni M., Simabuco F. M., Zanetti M. R., Yokoo S., Domingues R. R., Kawahara R., Pauletti B. A., Gonçalves A., Agostini M., Graner E., Coletta R. D., Fox J. W., and Paes Leme A. F. (2012) Novel processed form of syndecan-1 shed from SCC-9 cells plays a role in cell migration. PLoS One 7, e43521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Shevchenko A., Wilm M., Vorm O., and Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 73. Villén J., and Gygi S. P. (2008) The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat. Protoc. 3, 1630–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Huang da W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 75. Huang da W., Sherman B. T., and Lempicki R. A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kosugi S., Hasebe M., Tomita M., and Yanagawa H. (2009) Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. U.S.A. 106, 10171–10176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Nguyen Ba A. N., Pogoutse A., Provart N., and Moses A. M. (2009) NLStradamus: a simple hidden Markov model for nuclear localization signal prediction. BMC Bioinformatics 10, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.