Abstract
We have determined the kinetics of initiation of transcription of the wild-type ada promoter by abortive initiation assays. In the absence of activation, it is a weak promoter, with an association constant KB and an isomerization rate constant k2 comparable to those obtained under the same conditions for other positively regulated promoters (0.36 x 10(7) M-1 and 1.7 x 10(-2) s-1, respectively, at 37 degrees C and 50 mM NaCl, on a supercoiled template). As already observed for other promoters, these constants are modulated by varying the supercoiling of the plasmid. However, the strength of the promoter (given by the KB.k2 product) remains almost constant, because the maximum value of KB and k2 are obtained for different values of the superhelical density. The ada promoter has a stretch of seven adenosine residues (A7) in its upstream region. We have analysed the effect of this upstream sequence on the efficiency of initiation of the ada promoter by comparing the wild-type sequence with an up-mutant promoter characterized by the inversion of the central base pair in the sequence (A7) leading to the sequence (A3TA3). Although the mutation, which is located outside the promoter consensus regions, has no effect on the isomerization step, it affects the equilibrium constant KB that characterizes the association step. In the mutant promoters, the supercoiling of the plasmid modulates the isomerization and association constants in such a way that both KB and k2 are maximum for the same superhelical density (-0.05), leading to a 12-fold increase of the strength of the promoter, on a supercoiled template.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF
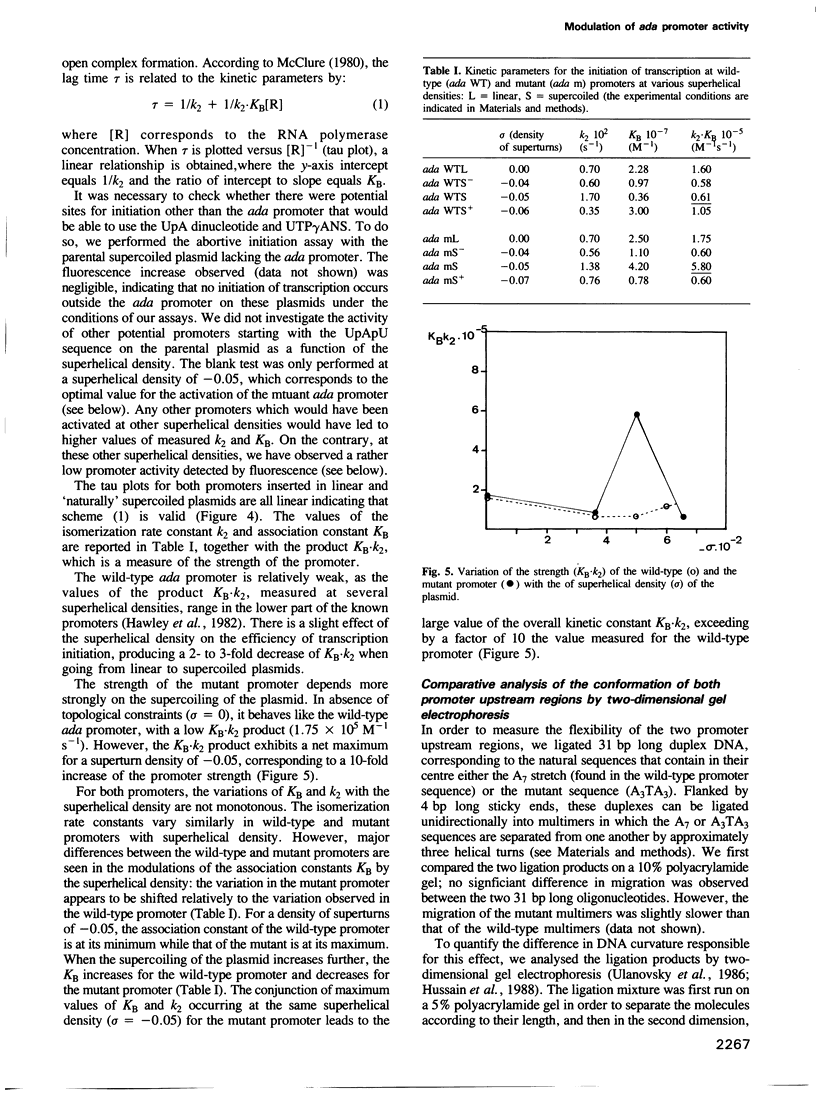



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amouyal M., Buc H. Topological unwinding of strong and weak promoters by RNA polymerase. A comparison between the lac wild-type and the UV5 sites of Escherichia coli. J Mol Biol. 1987 Jun 20;195(4):795–808. doi: 10.1016/0022-2836(87)90485-2. [DOI] [PubMed] [Google Scholar]
- Bertrand-Burggraf E., Lefèvre J. F., Daune M. A new experimental approach for studying the association between RNA polymerase and the tet promoter of pBR322. Nucleic Acids Res. 1984 Feb 10;12(3):1697–1706. doi: 10.1093/nar/12.3.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand-Burggraf E., Schnarr M., Lefevre J. F., Daune M. Effect of superhelicity on the transcription from the tet promoter of pBR322. Abortive initiation and unwinding experiments. Nucleic Acids Res. 1984 Oct 25;12(20):7741–7752. doi: 10.1093/nar/12.20.7741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiec J. A., Gralla J. D. Supercoiling response of the lac ps promoter in vitro. J Mol Biol. 1985 Aug 20;184(4):587–598. doi: 10.1016/0022-2836(85)90305-5. [DOI] [PubMed] [Google Scholar]
- Bracco L., Kotlarz D., Kolb A., Diekmann S., Buc H. Synthetic curved DNA sequences can act as transcriptional activators in Escherichia coli. EMBO J. 1989 Dec 20;8(13):4289–4296. doi: 10.1002/j.1460-2075.1989.tb08615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahms J. G., Dargouge O., Brahms S., Ohara Y., Vagner V. Activation and inhibition of transcription by supercoiling. J Mol Biol. 1985 Feb 20;181(4):455–465. doi: 10.1016/0022-2836(85)90419-x. [DOI] [PubMed] [Google Scholar]
- Burgess R. R., Jendrisak J. J. A procedure for the rapid, large-scall purification of Escherichia coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA-cellulose chromatography. Biochemistry. 1975 Oct 21;14(21):4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- Burkhoff A. M., Tullius T. D. Structural details of an adenine tract that does not cause DNA to bend. Nature. 1988 Feb 4;331(6155):455–457. doi: 10.1038/331455a0. [DOI] [PubMed] [Google Scholar]
- DiGabriele A. D., Sanderson M. R., Steitz T. A. Crystal lattice packing is important in determining the bend of a DNA dodecamer containing an adenine tract. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1816–1820. doi: 10.1073/pnas.86.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval-Valentin G., Ehrlich R. Far upstream sequences of the bla promoter from TN3 are involved in complexation with E. coli RNA-polymerase. Nucleic Acids Res. 1988 Mar 25;16(5):2031–2044. doi: 10.1093/nar/16.5.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartenberg M. R., Crothers D. M. DNA sequence determinants of CAP-induced bending and protein binding affinity. Nature. 1988 Jun 30;333(6176):824–829. doi: 10.1038/333824a0. [DOI] [PubMed] [Google Scholar]
- Greaves D. R., Patient R. K., Lilley D. M. Facile cruciform formation by an (A-T)34 sequence from a Xenopus globin gene. J Mol Biol. 1985 Oct 5;185(3):461–478. doi: 10.1016/0022-2836(85)90064-6. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J. Sequence dependence of the curvature of DNA: a test of the phasing hypothesis. Biochemistry. 1985 Dec 3;24(25):7033–7037. doi: 10.1021/bi00346a001. [DOI] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain I., Griffith J., Sancar A. Thymine dimers bend DNA. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2558–2562. doi: 10.1073/pnas.85.8.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo Y. L., Nara F., Ichihara S., Mizuno T., Mizushima S. Purification and characterization of the OmpR protein, a positive regulator involved in osmoregulatory expression of the ompF and ompC genes in Escherichia coli. J Biol Chem. 1986 Nov 15;261(32):15252–15256. [PubMed] [Google Scholar]
- Lane A. N., Lefèvre J. F., Jardetzky O. The effect of the trp repressor from Escherichia coli on the structure and dynamics of dA20dT20. Biochim Biophys Acta. 1986 May 27;867(1-2):45–56. doi: 10.1016/0167-4781(86)90028-x. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Sedgwick B., Sekiguchi M., Nakabeppu Y. Regulation and expression of the adaptive response to alkylating agents. Annu Rev Biochem. 1988;57:133–157. doi: 10.1146/annurev.bi.57.070188.001025. [DOI] [PubMed] [Google Scholar]
- Lodge J. K., Kazic T., Berg D. E. Formation of supercoiling domains in plasmid pBR322. J Bacteriol. 1989 Apr;171(4):2181–2187. doi: 10.1128/jb.171.4.2181-2187.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman T. M., deHaseth P. L., Record M. T., Jr Pentalysine-deoxyribonucleic acid interactions: a model for the general effects of ion concentrations on the interactions of proteins with nucleic acids. Biochemistry. 1980 Jul 22;19(15):3522–3530. doi: 10.1021/bi00556a017. [DOI] [PubMed] [Google Scholar]
- Malan T. P., Kolb A., Buc H., McClure W. R. Mechanism of CRP-cAMP activation of lac operon transcription initiation activation of the P1 promoter. J Mol Biol. 1984 Dec 25;180(4):881–909. doi: 10.1016/0022-2836(84)90262-6. [DOI] [PubMed] [Google Scholar]
- Malan T. P., McClure W. R. Dual promoter control of the Escherichia coli lactose operon. Cell. 1984 Nov;39(1):173–180. doi: 10.1016/0092-8674(84)90203-4. [DOI] [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure W. R. Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- McClure W. R. Rate-limiting steps in RNA chain initiation. Proc Natl Acad Sci U S A. 1980 Oct;77(10):5634–5638. doi: 10.1073/pnas.77.10.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel R., Gellert M. Fusions of the Escherichia coli gyrA and gyrB control regions to the galactokinase gene are inducible by coumermycin treatment. J Bacteriol. 1987 Mar;169(3):1272–1278. doi: 10.1128/jb.169.3.1272-1278.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau J. G., Crothers D. M. Structural basis for DNA bending. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2622–2626. doi: 10.1073/pnas.86.8.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Tokumoto Y., Sakumi K., Koike G., Nakabeppu Y., Sekiguchi M. Expression of the ada gene of Escherichia coli in response to alkylating agents. Identification of transcriptional regulatory elements. J Mol Biol. 1988 Aug 5;202(3):483–494. doi: 10.1016/0022-2836(88)90280-x. [DOI] [PubMed] [Google Scholar]
- Nelson H. C., Finch J. T., Luisi B. F., Klug A. The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature. 1987 Nov 19;330(6145):221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Energetics of B-to-Z transition in DNA. Proc Natl Acad Sci U S A. 1983 Oct;80(20):6206–6210. doi: 10.1073/pnas.80.20.6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Transcriptional block caused by a negative supercoiling induced structural change in an alternating CG sequence. Cell. 1985 Jan;40(1):129–137. doi: 10.1016/0092-8674(85)90316-2. [DOI] [PubMed] [Google Scholar]
- Pruss G. J., Drlica K. DNA supercoiling and prokaryotic transcription. Cell. 1989 Feb 24;56(4):521–523. doi: 10.1016/0092-8674(89)90574-6. [DOI] [PubMed] [Google Scholar]
- Ptashne M. Gene regulation by proteins acting nearby and at a distance. Nature. 1986 Aug 21;322(6081):697–701. doi: 10.1038/322697a0. [DOI] [PubMed] [Google Scholar]
- Rahmouni A. R., Wells R. D. Stabilization of Z DNA in vivo by localized supercoiling. Science. 1989 Oct 20;246(4928):358–363. doi: 10.1126/science.2678475. [DOI] [PubMed] [Google Scholar]
- Raibaud O., Schwartz M. Positive control of transcription initiation in bacteria. Annu Rev Genet. 1984;18:173–206. doi: 10.1146/annurev.ge.18.120184.001133. [DOI] [PubMed] [Google Scholar]
- Sakumi K., Sekiguchi M. Regulation of expression of the ada gene controlling the adaptive response. Interactions with the ada promoter of the Ada protein and RNA polymerase. J Mol Biol. 1989 Jan 20;205(2):373–385. doi: 10.1016/0022-2836(89)90348-3. [DOI] [PubMed] [Google Scholar]
- Travers A. A. DNA conformation and protein binding. Annu Rev Biochem. 1989;58:427–452. doi: 10.1146/annurev.bi.58.070189.002235. [DOI] [PubMed] [Google Scholar]
- Travers A. A., Lamond A. I., Mace H. A., Berman M. L. RNA polymerase interactions with the upstream region of the E. coli tyrT promoter. Cell. 1983 Nov;35(1):265–273. doi: 10.1016/0092-8674(83)90229-5. [DOI] [PubMed] [Google Scholar]
- Travers A. A. Structure and function of E. coli promoter DNA. CRC Crit Rev Biochem. 1987;22(3):181–219. doi: 10.3109/10409238709101483. [DOI] [PubMed] [Google Scholar]
- Tsao Y. P., Wu H. Y., Liu L. F. Transcription-driven supercoiling of DNA: direct biochemical evidence from in vitro studies. Cell. 1989 Jan 13;56(1):111–118. doi: 10.1016/0092-8674(89)90989-6. [DOI] [PubMed] [Google Scholar]
- Tse-Dinh Y. C., Beran R. K. Multiple promoters for transcription of the Escherichia coli DNA topoisomerase I gene and their regulation by DNA supercoiling. J Mol Biol. 1988 Aug 20;202(4):735–742. doi: 10.1016/0022-2836(88)90554-2. [DOI] [PubMed] [Google Scholar]
- Ulanovsky L., Bodner M., Trifonov E. N., Choder M. Curved DNA: design, synthesis, and circularization. Proc Natl Acad Sci U S A. 1986 Feb;83(4):862–866. doi: 10.1073/pnas.83.4.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. Y., Shyy S. H., Wang J. C., Liu L. F. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988 May 6;53(3):433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- Yarbrough L. R., Schlageck J. G., Baughman M. Synthesis and properties of fluorescent nucleotide substrates for DNA-dependent RNA polymerases. J Biol Chem. 1979 Dec 10;254(23):12069–12073. [PubMed] [Google Scholar]
- Zahn K., Blattner F. R. Sequence-induced DNA curvature at the bacteriophage lambda origin of replication. Nature. 1985 Oct 3;317(6036):451–453. doi: 10.1038/317451a0. [DOI] [PubMed] [Google Scholar]