Abstract
The floral transition is a crucial developmental event, but little is known about the underlying regulatory networks in seasonally and continuously flowering roses. In this study, we compared the genetic basis of flowering in two rose species, Rosa chinensis ‘Old Blush’, which flowers continuously, and R. odorata var. gigantea, which blooms in early spring. Gene ontology (GO) terms related to methylation, light reaction, and starch metabolism were enriched in R. odorata var. gigantea and terms associated with sugar metabolism were enriched in R. chinensis ‘Old Blush’ during the floral transition. A MapMan analysis revealed that genes involved in hormone signaling mediate the floral transition in both taxa. Furthermore, differentially expressed genes (DEGs) involved in vernalization, photoperiod, gibberellin (GA), and starch metabolism pathways converged on integrators, e.g., LFY, AGL24, SOC1, CAL, and COLs, to regulate the floral transition in R. odorata var. gigantea, while DEGs related to photoperiod, sugar metabolism, and GA pathways, including COL16, LFY, AGL11, 6PGDH, GASA4, and BAM, modulated the floral transition in R. chinensis ‘Old Blush.’ Our analysis of the genes underlying the floral transition in roses with different patterns of flowering provides a basis for further functional studies.
Introduction
The floral transition is an important developmental event orchestrated by external and developmental stimuli; for example, photoperiod, vernalization, autonomous, aging, gibberellin (GA), and sugar metabolism pathways1, 2 interact to activate or inhibit the floral transition via a small set of floral integrators, including FLOWERING LOCUS T (FT), CONSTANS-LIKE (COL), SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1), FLOWERING LOCUS C (FLC), and floral meristem identity genes, e.g., LEAFY (LFY), APETALA1 (AP1), AGAMOUS-LIKE (AGL), CAULIFLOWER (CAL), and SEPALLATA (SEP).
Considerable progress has been made toward identifying the molecular networks underlying the floral transition in the annual plant Arabidopsis thaliana 3. However, relatively little is known about the regulation of the floral transition in perennial plants. In perennials, only a portion of buds have competence to complete the phase transition from vegetative to reproductive growth in inductive conditions, while others remain in vegetative growth, indicating the ability to revert back from reproductive to vegetative growth. In temperate regions, most perennials require a period of winter cold (vernalization) to induce the floral transition and show seasonal flowering. However, a small number of perennials, such as Rosa chinensis and Fragaria vesca, can flower continuously in advantageous environmental conditions. The duration of flowering is crucial for the value of ornamental plants, such as modern roses and chrysanthemums.
Some progress has been made in understanding flowering perennials. Vernalization and sugar metabolism, which interacts with ABA signaling and aging pathways, are key factors inducing the floral transition in Malus × domestica 4, 5. The photoperiod-mediated CO/FT modules function as activators in the induction of the floral transition in chrysanthemums and poplars6, 7. Different from other species in the genus Camellia, the floral transition in C. azalea occurs in the spring; it is regulated by low temperatures, photoperiod, and hormone signals8. Jameson and Clemens have preliminarily determined inductive and non-inductive factors that influence the floral transition in New Zealand flora, including Sophora, Clianthus, and Metrosideros 9. The floral transition in Metrosideros is controlled by cool temperatures and photoperiod10, while floral initiation in Sophora does not respond to short day length, but is activated by vernalization11, and inflorescence initiation in Clianthus occurs in late autumn or early winter12.
Unlike the examples mentioned above, some perennial plants can transform between vegetative and reproductive growth throughout the year. For example, the floral transition in Arabis alpina is regulated by PERPETUAL FLOWERING 1 (PEP1), an ortholog of A. thaliana FLC, which encodes a MADS-box transcription factor that inhibits the floral transition before the winter; the species can return to vegetative growth after flowering. Unexpectedly, mutations in PEP1 enable A. alpina to flower continuously, without vernalization, indicating that the floral transition in A. alpina mainly depends on PEP1 13. It is noteworthy that the repression of vernalization on PEP1 is only transient, and PEP1 expression increases in warm temperatures13. However, the chromatin modification of FLC is stable, even if A. thaliana returns to warm temperatures14. This property distinguishes A. alpina from annual plants. The expression of TFL1 inhibits the conversion from vegetative to reproductive growth in A. thaliana 2. Orthologs of A. thaliana TFL1 have been identified in Fragaria vesca and continuous flowering (CF) roses15. The strawberry genome contains a 2-bp deletion in the first exon of TFL1, which contributes to CF. Likewise, the expression of TFL1 is inhibited by a retrotransposon in CF roses15. GA plays an activating role in regulating the floral transition in A. thaliana, but GAs are maintained at low levels during the floral transition process in both CF and seasonal roses (OF), and increase after flowering16. Spraying application of GA3 could block the floral transition in OF roses, but not in CF roses. Moreover, paclobutrazol treatment does not induce secondary floral transition in OF roses. Thus, the floral transition in OF roses in the spring, but not in CF roses, is influenced by GA3 15. In addition, rose RoKSN acts antagonistically with RoFT, competing with the bZIP transcription factor RoFD to inhibit or activate the floral transition17. Paradoxically, the floral transition in Rosa rugosa ‘Hamanasu’, a CF rose, is not controlled by the KSN copia allele18. Wang et al. proposed that CF is not a strictly qualitative trait, but a qualitative-quantitative trait19. These results indicate that floral genes other than RoKSN regulate the rose floral transition.
R. chinensis ‘Old Blush’ and R. odorata var. gigantea are diploid perennials, belonging to section Chinenses DC, from which modern roses, which share CF and tea scent, are derived. For simplicity, R. chinensis ‘Old Blush’ and R. odorata var. gigantea are abbreviated OB and GIG, respectively, throughout the text. OB flowers continuously in favorable temperature and photoperiod conditions, while GIG only blooms in the early spring. Studies of A. thaliana and other species have demonstrated that most flower-related genes are conserved across plant species2, 20. Thus, based on the complex integrated gene networks underpinning floral transition summarized in previous studies20, 21, a longitudinal comparative transcriptome analysis was performed during the floral transition process, which was divided into vegetative meristem (VM), pre-floral meristem (TM), and floral meristem (FM) stages. Differentially expressed genes (DEGs) between stages in each floral pathway were identified, and further interspecific comparative analyses were performed to investigate key candidate floral genes underlying the floral transition in the two species. This research provides valuable information on the physiology and molecular regulatory mechanisms of the floral transition in two roses that differ in flowering behavior.
Results
GO term enrichment in pairwise comparisons
To discover molecular changes during the floral transition in OB and GIG, floral gene expression was examined at the VM, TM, and FM stages (Supplementary Fig. S1). Moreover, secondary axillary buds that are unable to flower were also examined in GIG (referred to as SVM-GIG). A histological analysis indicated that they were in vegetative growth (Supplementary Fig. S1b).
We investigated 11,692, and 12,665 DGEs in the VM vs. TM (i.e., the difference between the VM and TM stages of development, denoted VM-GIG vs. TM-GIG) and SVM vs. TM comparisons (denoted SVM-GIG vs. TM-GIG) for GIG, respectively, and 790 DEGs in the VM vs. TM comparison for OB (denoted VM-OB vs. TM-OB; Supplementary Table S1). DEGs identified in each comparison were characterized by a GO enrichment analysis to identify the biological functions of these genes. Significantly enriched GO terms belonging to the biological process (BP) and molecular function (MF) categories (p < 0.05) were identified and visualized using the ReviGO tool (Fig. 1 and Supplementary Fig. S2 and Supplementary Table S2). As highlighted in Fig. 1, the GO terms for each comparison were obviously different. In the VM-OB vs. TM-OB comparison, there was enrichment for GO terms associated with sugar metabolism, including the sucrose metabolic process, aminoglycan catabolic process, and carbohydrate metabolic process in the BP category and sugar-phosphatase activity and fructose-bisphosphate aldolase (FBA) activity in the MF category (Fig. 1a and Supplementary Fig. S2a). DNA modification, methylation, sucrose metabolic process, and the regulation of gene expression, epigenetic were enriched in VM-GIG vs. TM-GIG, while DEGs in SVM-GIG vs. TM-GIG were related to many processes, including photosynthesis, light reaction, development process, starch metabolic process, carbohydrate transport, and so on. Additionally, compared with the GO terms in the BP category identified for VM-GIG vs. TM-GIG, terms related to starch metabolism were more highly enriched in the SVM-GIG vs. TM-GIG comparison (Fig. 1b,c). We concluded that sugar metabolism is an important inductive cue affecting the floral transition in both roses, OB and GIG. Additionally, DEGs related to photosynthesis and light reaction were enriched in SVM-GIG vs. TM-GIG, implying that photoperiod plays an important role in regulating the floral transition in GIG. DEGs related to DNA modification and methylation were identified in VM-GIG vs. TM-GIG, and these genes were mainly associated with vernalization and autonomous pathways.
Figure 1.
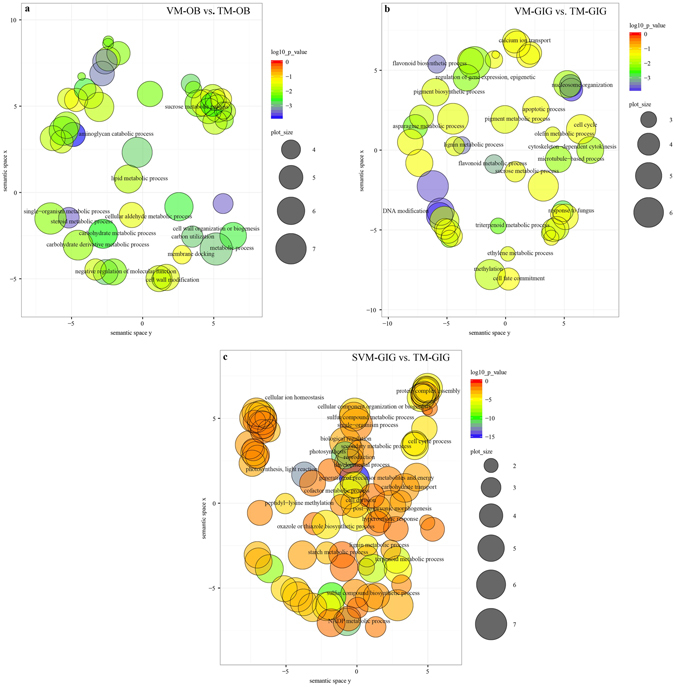
Scatterplot of enriched GO terms in pairwise comparisons. The scatterplot of enriched GO terms (p < 0.05) in biological process in the VM-OB vs. TM-OB comparison (a), in the VM-GIG vs. TM-GIG comparison (b), and in the SVM-GIG vs. TM-GIG comparison (c). Bubble color indicates the p-value of GO term; bubble size indicates the frequency of GO terms in the underlying GOA database.
Functional category enrichment of stage-specific DEGs in GIG and OB
We further investigated transcriptome changes involved in the rose floral transition in response to environmental and developmental cues. We inferred that common DEGs in the VM-GIG vs. TM-GIG and SVM-GIG vs. TM-GIG comparisons were not closely associated with the floral transition. Excluding common DEGs, we identified 7,702 (up-regulated, 3,920; down-regulated, 3,782) and 8,674 (up-regulated, 3,139; down-regulated, 5,536) stage-specific DEGs in VM-GIG vs. TM-GIG and SVM-GIG vs. TM-GIG, respectively (Fig. 2a,b). MapMan was used to map these stage-specific DEGs, identify functional categories, and investigate overrepresented regulation and metabolism terms (Supplementary Table S3). Figure 2c,d shows the comprehensive regulation overview maps of stage-specific pairwise comparisons (VM-GIG vs. TM-GIG and SVM-GIG vs. TM-GIG), including hormones, RNA, protein, and so on. More stage-specific DEGs were detected in the regulation overview in SVM-GIG vs. TM-GIG than in VM-GIG vs. TM-GIG (Fig. 2c,d), revealing that the physiological and molecular properties of SVM-GIG buds were obviously different from those of VM-GIG. Regarding the IAA metabolism subcategory, more DEGs were down-regulated in SVM-GIG vs. TM-GIG than in VM-GIG vs. TM-GIG (Fig. 2c,d and Supplementary Table S3); moreover, auxin contents continued to increase from VM-GIG to SVM-GIG (Supplementary Fig. S3). Accordingly, IAA was not a key cue mediating the floral transition in GIG. DEGs associated with GA metabolism were largely down-regulated in VM-GIG vs. TM-GIG and SVM-GIG vs. TM-GIG (Fig. 2c,d), indicating that GA levels were down-regulated during the floral transition process (Supplementary Fig. S3). Therefore, GA functioned as an inhibitor regulating the rose floral transition.
Figure 2.
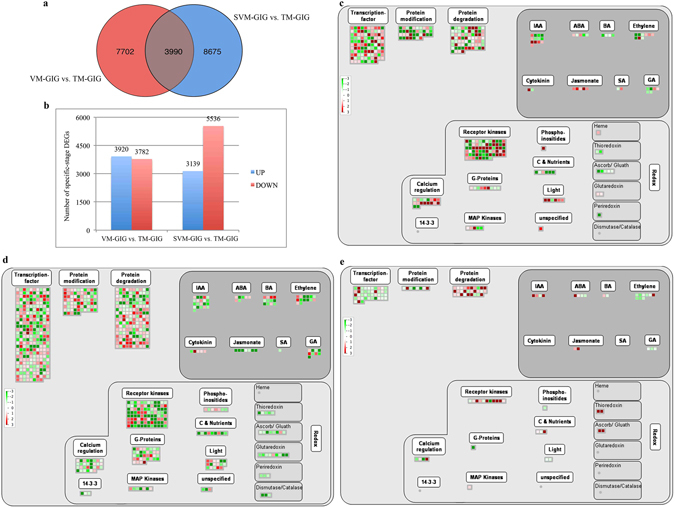
Number of DEGs and MapMan regulation overview maps in each comparison. (a) Number of conserved and stage-specific expression DEGs in pairwise comparisons of GIG. (b) Stage-specific comparison of DEGs in GIG. (c) MapMan regulation overview maps showing the transcript levels of stage-specific DEGs in the VM-GIG vs. TM-GIG comparison. (d) MapMan regulation overview maps showing the transcript levels of stage-specific DEGs in the SVM-GIG vs. TM-GIG comparison. (e) MapMan regulation overview maps showing the transcript levels of DEGs in the VM-OB vs. TM-OB comparison. The color indicates log2 value of fold changes, green color represents down-regulated transcripts, and red color represents up-regulated transcripts.
Regarding RNA categories, zinc finger proteins, IDD2/7 (Unigene0016847 and Unigene0030937) were up-regulated in the SVM-GIG vs. TM-GIG comparison; these regulate the production of floral stimuli or the inhibition of floral repressors22. A cold-inducible zinc finger-containing glycine-rich RNA-binding protein (Unigene0028397) was up-regulated in VM-GIG vs. TM-GIG (Supplementary Table S1). Additionally, Unigene0084554 identified in VM-GIG vs. TM-GIG encodes a small glycine-rich RNA-binding protein, which is responsive to vernalization and interacts with the autonomous pathway. Importantly, two genes, Unigene0046409 and Unigene0018266, which control the vegetative to reproductive phase transition of the meristem, decreased in VM-GIG vs. TM-GIG and SVM-GIG vs. TM-GIG, respectively. It is worth noting that Unigene0001915, Unigene0002888 (DRM1), and Unigene0019386, associated with methylation and epigenetic regulation, were reduced in VM-GIG vs. TM-GIG (Supplementary Table S1). Similarly, Unigene0017617 and Unigene0018266, related to DNA methylation and the silencing of FLOWER LOCUS C (FLC), respectively, were down-regulated in SVM-GIG vs. TM-GIG. These changes demonstrated that vernalization plays a vital role in inducing the floral transition before the TM stage of GIG.
Further comparisons of the metabolism pathways that differ in the VM-GIG vs. TM-GIG and SVM-GIG vs. TM-GIG comparisons were performed. As shown in Fig. S4a,b, more genes in the light reactions subcategory were down-regulated in SVM-GIG vs. TM-GIG, but no DEGs related to light reactions were identified in VM-GIG vs. TM-GIG, suggesting that photoperiod affects the floral transition in GIG. Other photosynthesis genes, involved in the Calvin cycle, photorespiration, and sucrose breakdown subcategories, obviously decreased in SVM-GIG vs. TM-GIG, but did not differ between VM-GIG and TM-GIG. Furthermore, DEGs associated with starch synthesis were up-regulated in SVM-GIG vs. TM-GIG, but were down-regulated in VM-GIG vs. TM-GIG (Supplementary Fig. S4a,b). Combined with changes in the starch contents (Supplementary Fig. S3), these findings suggest that photoperiod affects photosynthesis, sugar metabolism, and further influences the rose floral transition.
More DEGs were related to major CHO metabolism, cell wall, and lipid categories in the SVM-GIG vs. TM-GIG comparison than in VM-GIG vs. TM-GIG (Supplementary Fig. S4a,b). Regarding major CHO metabolism categories, sucrose synthase (SUS) and beta-amylase (BAM) were enriched in SVM-GIG vs. TM-GIG, but not in VM-GIG vs. TM-GIG (Supplementary Table S3), indicating that the storage of energy substances, such as sugar, starch, and lipids, was less in SVM-GIG than in VM-GIG. These substances may function as energy sources, osmotic regulators, and molecular regulators of the floral transition in GIG.
To increase our understanding of the expression differences during the floral transition between OB and GIG, we also investigated functional categories that were significantly overrepresented in the VM-OB vs. TM-OB comparison. Based on comparisons of the regulation overview and metabolism overview maps for VM-GIG vs. TM-GIG, SVM-GIG vs. TM-GIG, and VM-OB vs. TM-OB (Fig. 2 and Supplementary Fig. S4), we identified potentially important regulators of the floral transition in the two roses, and the loci explaining the difference between CF and OF phenotypes. Based on the regulation overview maps (Fig. 2c,e), the expression trends of genes related to hormonal regulation was similar to the VM-GIG vs. TM-GIG comparison. For example, IAA genes were up-regulated, whereas genes related to GA decreased from VM to TM; for example, SCL13 (c20369_g1 and Unigene0035053), a negative regulator of GA signal transduction pathway, increased during the floral transition in both roses. The DEGs c37195_g3 in VM-OB vs. TM-OB and Unigene0084554 in VM-GIG vs. TM-GIG, which not only respond to vernalization, but also interact with the autonomous flowering pathway, were identified (Supplementary Table S3). Surprisingly, genes associated with vernalization were also identified in VM-OB vs. TM-OB. For example, c21473_g1 encodes a RNA 3′-end processing factor of FLC silencing, and VERNALIZATION 1 (VRN1), c32035_g1, is essential for the complete repression of FLC in vernalized plants. These results suggested that the floral transition in OB is affected by vernalization. However, OB is characterized by continuous blooming; thus, its floral transition does not appear to be controlled by vernalization. Additionally, PERIANTHIA (PAN) and WUSCHEL (WUS), which control stem cell activity in the determinate floral meristem, were also differentially expressed in OB (Supplementary Table S3).
We further compared the metabolism overviews during the floral transition in the two roses. As shown in Supplementary Fig. S4a,c, more DEGs related to reserve materials (starch and sucrose), light reactions, and glycolysis were up-regulated in OB than in GIG, suggesting that sugar metabolism plays a vital role in regulating the floral transition in OB. Regarding genes in the major CHO metabolism categories, GRANULE-BOUND STARCH SYNTHASE 1 (GBSS1) and SUS2 were identified in VM-OB vs. TM-OB (Supplementary Table S1); these important enzymes related to starch metabolism were also differentially expressed.
Identification of candidate DEGs accounting for the floral transition in OB and GIG
Based on previous studies demonstrating the molecular regulatory networks underlying floral transition2, 21, in the current study, we identified and summarized DEGs significantly involved in the floral transition of OB and GIG, respectively. As shown in Fig. 3, DEGs and stage-specific DEGs belonging to the VM-GIG vs. TM-GIG, SVM-GIG vs. TM-GIG, and VM-OB vs. TM-OB comparisons are displayed in a Venn diagram in Fig. 3a. Notably, sucrose transporter protein (SUT), SUS, alpha-amylase (AMY), and BAM were differentially expressed in the above three comparisons (Fig. 3a). DEGs associated with sucrose synthesis and metabolism as well as starch metabolism were together up-regulated in VM-OB vs. TM-OB, coinciding with changes in the soluble sugar and starch contents during the floral transition (Fig. 3b and Supplementary Fig. S3). Furthermore, FBA, fructose-1,6-bisphosphatase I (FBP), and 6-phosphogluconate dehydrogenase (6PGDH) jointly increased from VM-OB to TM-OB (Fig. 3b). It is noteworthy that an increased soluble sugar content is closely associated with a decreased starch content during the floral transition23. Thus, sugar metabolism plays a vital role in regulating the floral transition in OB. By contrast, BAM3, SUS2, trehalose-phosphate synthase (TPS1), and FBP decreased during the floral transition process in GIG, while SUT, SS1/4, GBSS1, and AMY3 increased. GBSS1, SS1, AMY, and SUS2 were up-regulated from SVM-GIG to VM-GIG (Fig. 3c). In addition, soluble sugar contents did not exhibit obvious changes between developmental stages, whereas the starch content decreased from VM-GIG to TM-GIG, and then increased again at the SVM-GIG stage (Supplementary Fig. S3). Therefore, starch metabolism had a significant influence on the floral transition in GIG. Importantly, the contents of soluble sugar and starch in the buds of OB were higher than those in the buds of GIG (Supplementary Fig. S3), indicating that sugar metabolism plays an inducing role in regulating the floral transition in OB.
Figure 3.
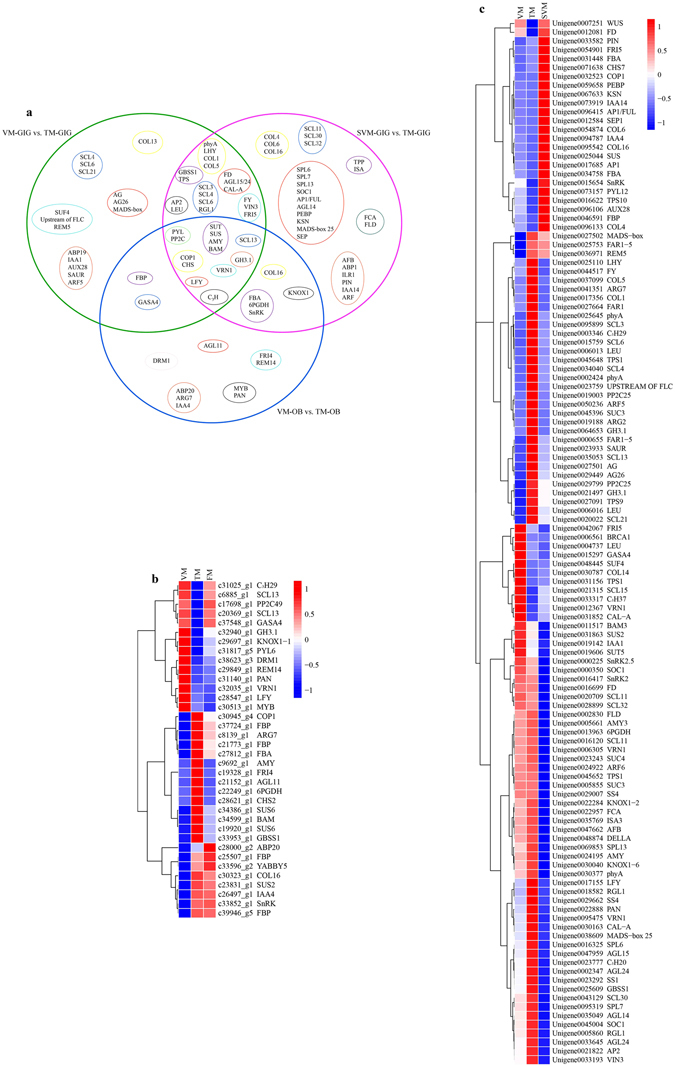
The overview of DEGs was identified between different comparisons of OB and GIG, respectively. (a) A Venn diagram showing the candidate floral DEGs commonly or individually belonging to each comparison of GIG and OB. Big green circle represents DEGs in the VM-GIG vs. TM-GIG comparison; big rose-red circle represents DEGs in the SVM-GIG vs. TM-GIG comparison; big blue circle stands for DEGs in the VM-OB vs. TM-OB comparison. Small yellow, cyan-blue, purple, light blue, orange-red, black, red circles indicate DEGs involved in photoperiod, vernalization, sugar metabolism, GA, auxin pathways, TFs, floral integrators, respectively. (b) Hierarchical clustering analysis of key floral candidate DEGs during floral transition process in OB. (c) Hierarchical clustering analysis of key floral candidate DEGs in different developmental stages of GIG. Data for gene expression levels were normalized by Z-score. Red and blue colors represent up- and down- regulated genes, respectively.
DELLA proteins are central nodes that function as negative regulators in GA signaling24. In the study, RGL1, SCL3, SCL4, SCL6, SCL13, and SCL21 were increased in VM-GIG vs. TM-GIG and SVM-GIG vs. TM-GIG, and SCL11, SCL30, and SCL32 were only up-regulated in the SVM-GIG vs. TM-GIG comparison (Fig. 3c). Additionally, Gibberellic Acid-stimulated in Arabidopsis 4 (GASA4) was down-regulated in the VM-GIG vs. TM-GIG and VM-OB vs. TM-OB comparisons (Fig. 3b,c). Simultaneously, the contents of GA1, GA3, and GA4 were all down-regulated during the floral transition in the two roses (Supplementary Fig. S3). Paradoxically, SCL13 decreased from VM-OB to TM-OB (Fig. 3b). In this scenario, we speculated that RGL1 and SCLs functioned as activators to induce the floral transition in GIG, and GASA4 played a negative role in regulating the floral transition in OB.
Regarding auxin signals, auxin-binding proteins, ABP19 and ABP20, and auxin-induced proteins, ARG2 and ARG7, were up-regulated during the floral transition in the two roses (Fig. 3b,c), while auxin-responsive proteins, such as IAA1, IAA4, and IAA14 decreased. Indole-3-acetic acid-amido synthetase (GH3) facilitates the conversion of auxin to amino acids, maintaining auxin homoeostasis. GH3.1 expression increased from VM-GIG to TM-GIG, but was down-regulated from VM-OB to TM-OB; in addition, auxin contents continued to increase during the floral transition in the two roses (Supplementary Fig. S3). In that respect, auxin may induce the floral transition in the two roses. However, it is worth noting that auxin contents were highest at the SVM stage of GIG (Supplementary Fig. S3), indicating that auxin was not a determinant of the regulation of the floral transition in GIG.
In the ABA signaling pathway, Pyrabactin Resistance 1-like (PYL) is an ABA receptor that directly inhibits protein phosphatase 2C (PP2C), which in turn directly represses SNF1-related protein kinases (SnRK)25. In GIG, PYL12 and SnRK2/2.5 decreased, but PP2C25 increased from VM-GIG to TM-GIG (Fig. 3c). Likewise, DEGs involved in ABA signaling were down-regulated from VM-OB to TM-OB (Fig. 3b). Meanwhile, the ABA content decreased during the floral transition in the two roses (Supplementary Fig. S3), indicating that ABA functioned as an inhibitor in inducing the floral transition in the two roses.
The floral transition of temperate plants is usually mediated by seasonal cues, including photoperiod (circadian clock) and vernalization. Phytochrome A (phyA), LATE ELONGATED HYPOCOTYL (LHY), and CONSTANS-LIKE (COL1, COL3, and COL5), involved in photoperiod and circadian clock pathways, were up-regulated in VM-GIG vs. TM-GIG, and phyA, LHY, and COL5 also increased from SVM-GIG to TM-GIG (Fig. 3b,c). Unexpectedly, COL4, COL6, and COL16 were down-regulated from SVM-GIG vs. TM-GIG (Fig. 3c). During the floral transition of OB, CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1), CHALCONE SYNTHASE 2 (CHS2), and COL16 were up-regulated (Fig. 3b), indicating that photoperiod plays an inducing role. While COP1 decreased in VM-GIG vs. TM-GIG and SVM-GIG vs. TM-GIG, CHS7 increased in VM-GIG vs. TM-GIG, but decreased in SVM-GIG vs. TM-GIG, indicating that photoperiod functions as an activator, and there was potential functional diversification (Fig. 3b,c).
Seasonal perennials require vernalization to obtain the competence to complete floral transition, even if plants return to warmer temperatures26. SUPPRESSOR OF FRI4 (SUF4) decreased, but REPRODUCTIVE MERISTEM 5 (REM5) increased from VM-GIG to TM-GIG (Fig. 3c). VERNALIZATION INSENSITIVE 3 (VIN3) and FY were up-regulated, and FRIGIDA 5 (FRI5) was down-regulated in VM-GIG vs. TM-GIG and SVM-GIG vs. TM-GIG comparisons. As shown in Fig. 3, VERNALIZATION 1 (VRN1) was up-regulated in VM-GIG vs. TM-GIG and SVM-GIG vs. TM-GIG, but decreased from VM-OB to TM-OB (Fig. 3b,c). In addition, FCA, FLOWERING LOCUS D (FLD), and SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL6/7/13) were up-regulated in SVM-GIG vs. TM-GIG (Fig. 3c). Unexpectedly, FRI4 was identified and increased from VM-OB to TM-OB (Fig. 3b). These phenomena suggested that vernalization had a significant role in regulating the floral transition of GIG, whereas its roles in OB require further experiments to examine potential functional diversification in regulating floral transition.
Environmental and developmental cues eventually converge to a series of floral integrators and floral meristem identity genes. In this study, LFY and MADS-box genes, including AGL15/24, CAL-A, and FD were jointly up-regulated in VM-GIG vs. TM-GIG and SVM-GIG vs. TM-GIG (Fig. 3a,c). Additionally, MADS-box, AG, and AG26 increased from VM-GIG to TM-GIG. SOC1, AGL14, and MADS-box 25 were up-regulated in the SVM-GIG vs. TM-GIG comparison, but AP1, AP1 (FUL), PEBP, and SEP were down-regulated (Fig. 3a,c). Taken together, we concluded that SOC1, LFY, AGL14/15/24, CAL-A, MADS-box 25, KSN, and SPL6/7 function as vital activators controlling the floral transition in GIG. Moreover, a hierarchical clustering analysis showed that these genes exhibited high co-expression (Fig. 3c). In OB, LFY had high expression during the floral transition, and AGL11 increased from VM-OB to TM-OB, while DRM1 and MYB decreased (Fig. 3b). It is noteworthy that a hierarchical clustering analysis revealed high co-expression patterns among DRM1, REM14, PAN, VRN1, LFY, and MYB (Fig. 3b). Hence, these DEGs played a substantial role in the regulation of the floral transition in OB.
Verification of gene expression by RT-qPCR
To confirm the reliability of the RNA-seq data, we selected a few key candidate floral DEGs, involved in vernalization, photoperiod, and sugar metabolism pathways, for parallel RT-qPCR-based expression analysis in OB and GIG. The expression profiles of genes obtained by RT-qPCR and RNA-seq were highly similar, confirming the accuracy of our transcriptomic analysis (Fig. 4).
Figure 4.
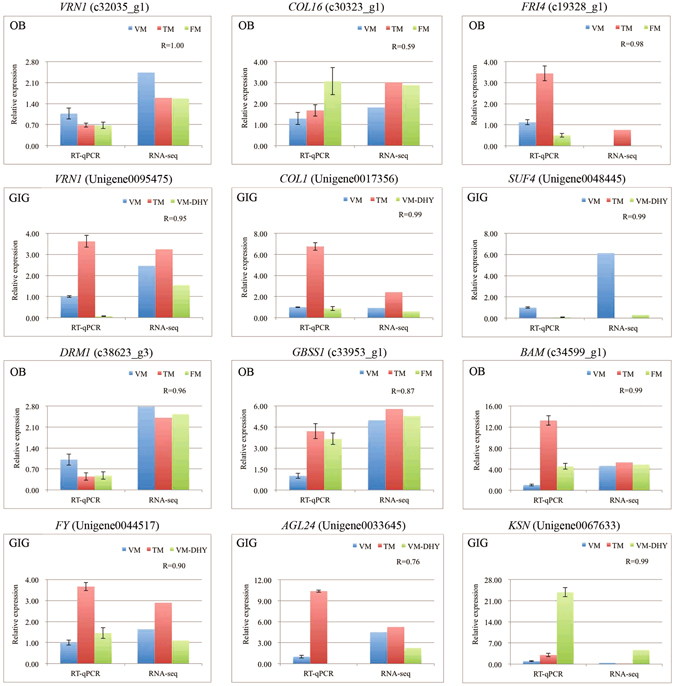
RT-qPCR validation of key candidate floral DEGs in different developmental stages of OB and GIG. The expression levels of genes revealed by RT-qPCR (left side) and RNA-seq (right side). Data from RT-qPCR were means of three replicates and bars represent SE. Data from RNA-seq were means of replicates and were normalized by Log2 transformed. The correlation coefficient (R) for each gene between RT-qPCR and RNA-seq data was shown and calculated by cor.test in R.
Discussion
Sugar metabolism is an important process in plant development; it not only provides an energy source, but also acts as a florigenic signal during the floral transition in A. thaliana 27. Starch is the key form of carbon reserve in plants, including storage starch and transitory starch, in which the dynamic polymer within transitory starch is regulated by the circadian clock and GBSS1, which directly promotes amylose synthesis28. Starch accumulation and the glycan composition change dramatically during the floral transition process and are controlled by CO 23. Moreover, increased sugar levels are concomitant with decreased starch levels during the floral transition process. The presence of CO in the promoter of GBSS1 was established by ChIP experiments, indicating that CO directly regulates the expression of GBSS1 23. In the current study, the soluble sugar contents increased slightly during the floral transition in GIG. The starch content decreased dramatically and subsequently increased at the SVM-GIG stage (Supplementary Fig. S3). At the same time, COL5 and GBSS1 synchronously increased in VM-GIG vs. TM-GIG and SVM-GIG vs. TM-GIG comparisons (Fig. 3c), suggesting that starch metabolism plays a significant role in regulating the floral transition in GIG. The trends in soluble sugar and starch contents in OB were similar to those in GIG during the floral transition (Supplementary Fig. S3), and COL16 was up-regulated from VM-OB to TM-OB (Fig. 3b). In addition to starch, trehalose-6-phosphate (T6P) is another floral stimulus that coordinates the floral transition in A. thaliana by itself or via the regulation of miR156 or SPL in the shoot apical meristem29. Interestingly, T6P also regulates the accumulation and turnover of transitory starch30. By contrast, the content of soluble sugar in OB was far more greater than that in GIG (Supplementary Fig. S3), and this property may be associated with continuous flowering, an economically valuable trait, in OB.
6PGDH activity is closely related to the floral transition in Brassica campestris and functions as a good enzyme marker for the floral transition31. In this study, the expression pattern of 6 PGDH coincided with the phase change from vegetative growth to flowering in the two roses (Fig. 3b). Another enzyme, FBA, is mediated by circadian clock and plays a dominant role in regulating flowering time via sugar metabolism32, 33. Hierarchical clustering analysis revealed that FBA was highly correlated with COP1, with up-regulated expression profiles (Fig. 3b). Indeed, COP1 mediates the control of light input to the circadian clock, involved in the floral transition of A. thaliana 34.
Sugar signals involve extensive interactions with hormone signals, affecting the transition from vegetative to reproductive growth35. Generally, T6P signaling is an indicator of sucrose transduction36. Various lines of evidence show that T6P represses the expression of SnRK when the content of sucrose exceeds a certain threshold concentration, and the T6P-SnRK module orchestrates changes in some floral genes, such as SPL and miR156, to regulate juvenility and floral signal transduction37. SnRK also participates in ABA signaling and functions as a positive regulator of ABA signaling. As shown in Supplementary Fig. S3, ABA levels and soluble sugar contents showed opposite trends during floral transition in the two roses. In GIG, SnRK decreased from VM-GIG to TM-GIG and had lower expression than that at SVM-GIG, but SnRK2/2.5 exhibited the opposite trend in expression. Likewise, SnRK2 was up-regulated from VM-OB to TM-OB. Further experiments will be needed to examine these patterns.
It has recently been demonstrated that GA operates positively and then negatively to mediate the floral transition in A. thaliana. GA initially activates the expression of LFY, whereas up-regulated LFY in turn reduces GA levels in a negative feedback loop, causing DELLA enrichment, which further promotes AP1 expression to induce the floral transition38. In OB, LFY exhibited the highest expression at the VM stage (Fig. 3b), indicating that LFY expression was activated quickly, before the floral transition commences, as observed in A. thaliana 39. Moreover, the GA levels decreased sharply from VM-OB to TM-OB (Supplementary Fig. S3). In that respect, GA might also influence LFY in OB. DELLA protein acts as a negative regulator of GA. Previous studies have shown that DELLA functions as an activator in the floral transition of Prunus avium and Citrus unshiu 40, 41. Furthermore, DELLA could interact with SPL via miR156, and further regulate SPL and MADS-box genes in A. thaliana 42. In addition, sugar is able to activate SPL via miR156 combined with the aging pathway43. Yuan and Wysocka-Diller also demonstrated that RGL2 participates in sugar metabolism and regulates the floral transition44. In this study, the expression the DELLA protein RGL1 and TPS1/9 corresponded with the phase-change of GIG (Fig. 3c), suggesting that RGL1 is probably involved in sugar metabolism. Furthermore, linkage between plant architecture and flowering behavior has been identified and is associated with some quantitative trait loci (QTLs). Moreover, some genes associated with GA biosynthesis and auxin signaling have been identified in the vicinity of these QTLs45.
After a period of vernalization, several flowering inhibitors, such as FLC and FRI, are silenced epigenetically via histone modifications26. FLC is also inhibited by the autonomous pathway, including FY, FCA, and FLD 46. In addition, VRN1/REM5, activated by vernalization, act as promoters in inducing floral transition46. In GIG, key genes involved in vernalization and autonomous pathways play activating roles in inducing flowering, as observed in A. thaliana 26, suggesting that vernalization and autonomous pathways have significant influences on the floral transition in parallel. Unexpectedly, genes associated with vernalization, including FRI4, VRN1, and REM14, were also identified in OB, which exhibits continuous flowering throughout the year. However, their expression trends were different from those of GIG during the floral transition (Fig. 3b,c), suggesting that vernalization is not a dominant pathway affecting the floral transition of OB or that these genes underwent functional diversification. DRM1 is an important locus that promotes flowering; it mediates the expression of SOC1 and LFY and is involved in the autonomous pathway47. In addition, DRM1 is also related with apical dominance or paradormancy, and is repressed by increasing auxin and soluble sugar contents, but is activated by ABA48, 49. DRM1 expression was associated with physiological changes, such as auxin, soluble sugar, and ABA (Figs 3b and S3), indicating a role in the floral transition in OB.
It has been demonstrated that the bZIP transcription factor PAN interacts with various developmental pathways, such as light, hormones, and meristem control systems50. Moreover, PAN can activate the expression of AG 51. As shown in Fig. 3b, PAN, DRM1, and REM14 were highly correlated, suggesting that methylation and epigenetic regulation caused by the autonomous or vernalization pathways mediated the expression of PAN, and further regulated the floral transition from vegetative to reproductive growth. Additionally, a plethora of MADS-box transcription factors were identified in this study. It was noteworthy that AGL24 was identified in GIG but not in OB, and it originated from a branch of DAM involved in perennial dormancy that is controlled by seasonal factors, including vernalization and photoperiod52. In this way, the floral transition in GIG was influenced by vernalization.
Methods
Plant material and experimental procedures
Samples of OB and GIG were collected from Kunming Yang Chinese Rose Gardening Co., Ltd in the Yunnan Province of China (24°45′N, 102°53′E). The vegetative meristem (VM), pre-floral meristem (TM), floral meristem (FM), and secondary vegetative meristem (SVM) stages of OB and GIG were identified by a histological analysis53 (Supplementary Fig. S1). Samples of each stage of OB are abbreviated VM-OB, TM-OB, FM-OB, and samples of different developmental stages in GIG are abbreviated VM-GIG, TM-GIG, FM-GIG, and SVM-GIG. The samples obtained for the VM-OB, TM-OB, and FM-OB stages were collected on December 29, 2014, January 2, 2015, and January 8, 2015, respectively, while samples of VM-OB, TM-OB, FM-OB, and SVM-GIG were obtained on December 27, 2014, January 2, 2015, January 8, 2015, and March 8, 2015, respectively. The scales and young leaves outside of buds were peeled off as soon as possible, so that the remaining tissue was closer to the shoot apical meristem. Each biological replicate contained mixed buds (~0.30 g), and each stage had three biological replicates. All samples were collected from 12:00 to 17:00, and quickly placed in liquid nitrogen and stored at −80 °C until further use. Samples for paraffin sections, RNA extraction, and hormone, soluble sugar, and starch measurements were collected at the same time.
Measurements of soluble sugar, starch, and hormone contents
Almost 0.1 g fresh samples at different developmental stages of OB and GIG were respectively collected for the measurement of soluble sugar and starch, and at least 0.05 g for the hormone measurement. The former was measured according to the sulfuric acid-anthrone colorimetric method54. High-performance liquid chromatography-mass spectrometry (HPLC - MS) (AB 5500, Beijing, China) was used to identify and quantify hormones as previously described55.
Transcriptome analysis
The quality and quantity of RNA was determined using the NanoPhotometer@ spectrophotometer (IMPLEN, Westlake Village, CA, USA). Approximately, 3 μg of RNA per sample was used for the construction of libraries using the NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, Ipswich, MA, USA) following manufacturer’s instructions. Library quality was assessed using the Agilent Bioanalyzer 2100 system. Three biological replicates were sequenced, paired-end sequencing was conducted using an Illumina HiSeqTM 4000. After filtering raw reads, clean reads were de novo assembled using the Trinity short reads assembling program56, producing two transcriptome reference databases for OB and GIG, respectively. All sequence data of OB and GIG were deposited in the Genome Sequence Archive with project ID PRJCA000258 and project ID PRJCA000286, respectively. RPKM (Reads Per kb per Million reads) was used to calculate the relative expression levels57. Differential expression analysis of each comparison was performed using the DESeq R package (1.10.1), and the p-values correction (false discovery rate, FDR) was adjusted using the Benjamini and Hochberg method58. The identification of differentially expressed genes (DEGs) between two groups was evaluated with |fold change|>1.5 and FDR < 0.05.
The web-based ReviGO software (http://revigo.irb.hr) was used to identify gene ontology (GO) functional categories (p < 0.05) associated with each set of compared samples59. The DEGs were annotated using the Mercator web tool and then loaded into MapMan software for function enrichment analysis60, 61.
Data validation by real-time quantitative PCR (RT-qPCR)
Total RNA treated with DNase (Takara, Dalian, China) was reverse transcribed to produce cDNA templates for reactions according to the manufacturer’s instructions (Takara, Dalian, China). For RT-qPCR, the reaction was carried out with a qTOWER 2.2 PCR System (Jena, Germany) and SYBR Green PCR Master Mix (Takara, Dalian, China) according to the manufacturer’s protocol. The primer sequences for RT-qPCR were designed using the PrimerQuest Tool (Supplementary Table S4), and synthesized by Sangon Biotech Co., Ltd. (Beijing, China). The RcActin gene was used as a reference gene62. Each 20 μL reaction mixture contained 2 μL cDNA template. The amplification program was as follows: 3 min at 95 °C and 40 cycles of 10 s at 95 °C and 30 s at 60 °C. Relative expression levels of candidate genes were calculated using the 2−ΔΔCt method63. Each reaction was performed with three biological replicates. RNA-seq data were Log-transformed prior to the Pearson’s correlation evaluation. Pearson’s correlation values between RNA-seq and RT-qPCR data of selected genes were calculated using cor. test in R version 3.3.
Electronic supplementary material
Acknowledgements
We thank Yuyong Yang (Kunming Yang Chinese Rose Gardening Co., Ltd) and Haiquan Huang (Southwest Forestry University) for the help of collecting samples. This work was supported by the Fundamental Research Funds for the Central Universities(NO.2016ZCQ02), the 12th Five Years Key Programs for Science and Technology Development of China (2013BAD01B07), and Special Fund for Beijing Common Construction Project.
Author Contributions
Q.-X.Z., H.-T.P., X.-L.G., and C.Y., conceived the research. X.-L.G. and C.Y. collected plant materials, carried out data analysis and wrote the paper. L.L. and H.-H.W. contributed to draw graphs, Y.-S.L., J.W., and T.-R.C. helped to modify the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Xuelian Guo and Chao Yu contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-05850-8
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ortizmarchena MI, et al. Photoperiodic Control of Carbon Distribution during the Floral Transition in Arabidopsis. The Plant cell. 2014;26:565–584. doi: 10.1105/tpc.114.122721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blumel M, Dally N, Jung C. Flowering time regulation in crops-what did we learn from Arabidopsis? Current opinion in biotechnology. 2015;32:121–129. doi: 10.1016/j.copbio.2014.11.023. [DOI] [PubMed] [Google Scholar]
- 3.Bouché F, et al. Integrating roots into a whole plant network of flowering time genes inArabidopsis thaliana. Scientific reports. 2016;6:203–214. doi: 10.1038/srep29042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foster T, Johnston R, Seleznyova A. A Morphological and Quantitative Characterization of Early Floral Development in Apple (Malus × domestica Borkh.) Annals of Botany. 2003;92:199–206. doi: 10.1093/aob/mcg120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xing LB, et al. Transcription Profiles Reveal Sugar and Hormone Signaling Pathways Mediating Flower Induction in Apple (Malus domestica Borkh) Plant and Cell Physiology. 2015;56:364–369. doi: 10.1093/pcp/pcv124. [DOI] [PubMed] [Google Scholar]
- 6.Fu J, et al. Photoperiodic control of FT- like gene ClFT initiates flowering in Chrysanthemum lavandulifolium. Plant Physiology & Biochemistry Ppb. 2014;74:230–238. doi: 10.1016/j.plaphy.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 7.Böhlenius H, et al. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science. 2006;312:1040–1043. doi: 10.1126/science.1126038. [DOI] [PubMed] [Google Scholar]
- 8.Fan, Z. et al. Genome-wide transcriptome profiling provides insights into floral bud development of summer-flowering Camellia azalea. Scientific reports5 (2015). [DOI] [PMC free article] [PubMed]
- 9.Jameson, P. E. & Clemens, J. Phase change and flowering in woody plants of the New Zealand flora. Journal of experimental botany (2015). [DOI] [PMC free article] [PubMed]
- 10.Henriod RE, Jameson PE, Clemens J. Effect of irradiance during floral induction on floral initiation and subsequent development in buds of different size in Metrosideros excelsa (Myrtaceae) Journal of Horticultural Science & Biotechnology. 2003;78:204–212. doi: 10.1080/14620316.2003.11511607. [DOI] [Google Scholar]
- 11.Song J, Clemens J, Jameson PE. Quantitative expression analysis of the ABC genes in Sophora tetraptera, a woody legume with an unusual sequence of floral organ development. Journal of experimental botany. 2008;59:247–259. doi: 10.1093/jxb/erm305. [DOI] [PubMed] [Google Scholar]
- 12.Song J, Clemens J, Jameson PE. Expression of floral identity genes in Clianthus maximus during mass inflorescence abortion and floral development. Annals of Botany. 2011;107:1501–1509. doi: 10.1093/aob/mcr035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang R, et al. PEP1 regulates perennial flowering in Arabis alpina. Nature. 2009;459:423–427. doi: 10.1038/nature07988. [DOI] [PubMed] [Google Scholar]
- 14.Angel A, et al. Vernalizing cold is registered digitally at FLC. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:4146–4151. doi: 10.1073/pnas.1503100112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iwata H, et al. The TFL1 homologue KSN is a regulator of continuous flowering in rose and strawberry. The Plant journal: for cell and molecular biology. 2012;69:116–125. doi: 10.1111/j.1365-313X.2011.04776.x. [DOI] [PubMed] [Google Scholar]
- 16.Roberts AV, Blake PS, Lewis R, Taylor JM, Dunstan DI. The effect of gibberellins on flowering in roses. Journal of plant growth regulation. 1999;18:113–119. doi: 10.1007/PL00007058. [DOI] [PubMed] [Google Scholar]
- 17.Randoux M, et al. RoKSN, a floral repressor, forms protein complexes with RoFD and RoFT to regulate vegetative and reproductive development in rose. New Phytologist. 2014;202:161–173. doi: 10.1111/nph.12625. [DOI] [PubMed] [Google Scholar]
- 18.Horibe, T., Yamada, K., Otagaki, S., Matsumoto, S. & Kawamura, K. Molecular genetic studies on continuous-flowering roses that do not originate from Rosa chinensis. Acta Horticulturae1064 (2013).
- 19.Wang F, Lian LJ, Liu QL. Low expression of RTFL1c releases expression of RLFY, which induces recurrent flowering in rose. Acta Horticulturae. 2015;1064:131–134. doi: 10.17660/ActaHortic.2015.1064.16. [DOI] [Google Scholar]
- 20.Albani, M. C. & Coupland, G. Comparative Analysis of Flowering in Annual and Perennial Plants. 91, 323–348, doi:10.1016/s0070-2153(10)91011-9 (2010). [DOI] [PubMed]
- 21.Khan MRG, Ai XY, Zhang JZ. Genetic regulation of flowering time in annual and perennial plants. Wiley Interdisciplinary Reviews Rna. 2014;5:347–359. doi: 10.1002/wrna.1215. [DOI] [PubMed] [Google Scholar]
- 22.Colasanti J, Sundaresan V. ‘Florigen’ enters the molecular age: long-distance signals that cause plants to flower. Trends in Biochemical Sciences. 2000;25:236–240. doi: 10.1016/S0968-0004(00)01542-5. [DOI] [PubMed] [Google Scholar]
- 23.Ortiz-Marchena MI, et al. Photoperiodic control of carbon distribution during the floral transition in Arabidopsis. The Plant cell. 2014;26:565–584. doi: 10.1105/tpc.114.122721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Claeys H, Bodt SD, Inzé D. Gibberellins and DELLAs: central nodes in growth regulatory networks. Trends in plant science. 2014;19:231–239. doi: 10.1016/j.tplants.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 25.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic Acid: Emergence of a Core Signaling Network. Annual review of plant biology. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 26.Kim DH, Doyle MR, Sung SB, Amasino RM. Vernalization: winter and the timing of flowering in plants. Annual Review of Cell & Developmental Biology. 2009;25:277–299. doi: 10.1146/annurev.cellbio.042308.113411. [DOI] [PubMed] [Google Scholar]
- 27.Ortiz-Marchena MI, Romero JM, Valverde F. Photoperiodic control of sugar release during the floral transition: What is the role of sugars in the florigenic signal? Plant signaling & behavior. 2015;10:e1017168. doi: 10.1080/15592324.2015.1017168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeeman SC, Kossmann J, Smith AM. Starch: its metabolism, evolution, and biotechnological modification in plants. Plant Biology. 2010;61:209–234. doi: 10.1146/annurev-arplant-042809-112301. [DOI] [PubMed] [Google Scholar]
- 29.Wahl V, et al. Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science. 2013;339:704–707. doi: 10.1126/science.1230406. [DOI] [PubMed] [Google Scholar]
- 30.Martins MC, et al. Feedback inhibition of starch degradation in Arabidopsis leaves mediated by trehalose 6-phosphate. Plant physiology. 2013;163:1142–1163. doi: 10.1104/pp.113.226787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orr AR. Histochemical Study of Enzyme Activity in the Shoot Apical Meristem of Brassica campestris L. During Transition to Flowering. III. Glucose6- Phosphate Dehydrogenase and 6Phosphogluconate Dehydrogenase. International Journal of Plant Sciences. 1985;7:347–349. [Google Scholar]
- 32.Harmer SL, et al. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 33.Haake V, Zrenner R, Sonnewald U, Stitt M. A moderate decrease of plastid aldolase activity inhibits photosynthesis, alters the levels of sugars and starch, and inhibits growth of potato plants. Plant Journal. 1998;14:147–157. doi: 10.1046/j.1365-313X.1998.00089.x. [DOI] [PubMed] [Google Scholar]
- 34.Yu JW, et al. COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Molecular Cell. 2009;32:617–630. doi: 10.1016/j.molcel.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsoukas IG. Interplay between sugar and hormone signaling pathways modulate floral signal transduction. Frontiers in Genetics. 2014;5:218–218. doi: 10.3389/fgene.2014.00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.John EL, et al. Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. Biochemical Journal. 2006;397:139–148. doi: 10.1042/BJ20060083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gómez LD, Gilday A, Feil R, Lunn JE, Graham IA. AtTPS1-mediated trehalose 6-phosphate synthesis is essential for embryogenic and vegetative growth and responsiveness to ABA in germinating seeds and stomatal guard cells. Plant Journal. 2010;64:1–13. doi: 10.1111/j.1365-313X.2010.04312.x. [DOI] [PubMed] [Google Scholar]
- 38.Yamaguchi N, et al. Gibberellin acts positively then negatively to control onset of flower formation in Arabidopsis. Science. 2014;344:638–641. doi: 10.1126/science.1250498. [DOI] [PubMed] [Google Scholar]
- 39.Blázquez MA, Soowal LN, Lee I, Weigel D. LEAFY expression and flower initiation in Arabidopsis. Development. 1997;124:3835–3844. doi: 10.1242/dev.124.19.3835. [DOI] [PubMed] [Google Scholar]
- 40.Boss PK, Thomas MR. Association of dwarfism and floral induction with a grape ‘green revolution’ mutation. Nature. 2002;416:847–850. doi: 10.1038/416847a. [DOI] [PubMed] [Google Scholar]
- 41.Nishikawa F, et al. Differences in seasonal expression of flowering genes between deciduous trifoliate orange and evergreen Satsuma mandarin. Tree Physiology. 2009;29:921–926. doi: 10.1093/treephys/tpp021. [DOI] [PubMed] [Google Scholar]
- 42.Yu S, et al. Gibberellin Regulates the Arabidopsis Floral Transition through miR156-Targeted squamosa Promoter binding–like Transcription Factors. The Plant cell. 2012;24:3320–3332. doi: 10.1105/tpc.112.101014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang L, Conway SR, Poethig RS. Vegetative phase change is mediated by a leaf-derived signal that represses the transcription of miR156. Development. 2011;138:245–249. doi: 10.1242/dev.058578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan K, Wysocka-Diller J. Phytohormone signalling pathways interact with sugars during seed germination and seedling development. Journal of experimental botany. 2006;57:3359–3367. doi: 10.1093/jxb/erl096. [DOI] [PubMed] [Google Scholar]
- 45.Kawamura K, Oyant HS, Thouroude T, Jeauffre J, Foucher F. Inheritance of garden rose architecture and its association with flowering behaviour. Tree Genetics & Genomes. 2015;11:1–12. doi: 10.1007/s11295-015-0844-3. [DOI] [Google Scholar]
- 46.Levy, Y. Y., Mesnage, S., Mylne, J. S., Gendall, A. R. & Dean, C. Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science297, págs, 243–246 (2002). [DOI] [PubMed]
- 47.Yong Z, et al. Characterization of a novel developmentally retarded mutant (drm1) associated with the autonomous flowering pathway in Arabidopsis. Cell Research. 2005;15:133–140. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- 48.Rae, G. M., David, K. & Wood, M. The Dormancy Marker DRM1/ARP Associated with Dormancy but a Broader Role In Planta. Developmental Biology Journal2013 (2013).
- 49.Gonzali S, et al. Identification of sugar-modulated genes and evidence for in vivo sugar sensing in Arabidopsis. Journal of Plant Research. 2006;119:115–123. doi: 10.1007/s10265-005-0251-1. [DOI] [PubMed] [Google Scholar]
- 50.Maier AT, Stehlingsun S, Offenburger SL, Lohmann JU. The bZIP Transcription Factor PERIANTHIA: A Multifunctional Hub for Meristem Control. Frontiers in plant science. 2011;2:1085–1091. doi: 10.3389/fpls.2011.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maier AT, et al. Dual roles of the bZIP transcription factor PERIANTHIA in the control of floral architecture and homeotic gene expression. Development. 2009;136:1613–1620. doi: 10.1242/dev.033647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Z, Reighard GL, Abbott AG, Bielenberg DG. Dormancy-associated MADS genes from the EVG locus of peach [Prunus persica (L.) Batsch] have distinct seasonal and photoperiodic expression patterns. Journal of experimental botany. 2009;60:3521–3530. doi: 10.1093/jxb/erp195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kurokura T, Inaba Y, Neri D, Sugiyama N. A morphological study of the development of the second inflorescences in strawberry (Fragaria x ananassa Duch.) Annals of Applied Biology. 2005;146:511–515. doi: 10.1111/j.1744-7348.2005.040087.x. [DOI] [Google Scholar]
- 54.He, Z. F. The fast determination method for content of glucose, frutose, sucrose and starch by anthrone colorimetry. In: He, Z. F., editor. Grain Quality and it’s Analysis Technology. Beijing: Agricultural Press p. 144–150 (1985).
- 55.Pan X, Welti R, Wang X. Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography–mass spectrometry. Nature Protocols. 2010;5:986–992. doi: 10.1038/nprot.2010.37. [DOI] [PubMed] [Google Scholar]
- 56.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature biotechnology. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 58.Thissen D, Kuang D. Quick and Easy Implementation of the Benjamini-Hochberg Procedure for Controlling the False Positive Rate in Multiple Comparisons. Journal of Educational & Behavioral Statistics. 2010;27:77–83. doi: 10.3102/10769986027001077. [DOI] [Google Scholar]
- 59.Supek F, Bošnjak M, Škunca N. Šmuc & Tomislav. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PloS one. 2011;6:e21800–e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lohse M, et al. Mercator: a fast and simple web server for genome scale functional annotation of plant sequence data. Plant, cell & environment. 2014;37:1250–1258. doi: 10.1111/pce.12231. [DOI] [PubMed] [Google Scholar]
- 61.Usadel B, et al. A guide to using MapMan to visualize and compare Omics data in plants: a case study in the crop species, Maize. Plant, cell & environment. 2009;32:1211–1229. doi: 10.1111/j.1365-3040.2009.01978.x. [DOI] [PubMed] [Google Scholar]
- 62.Dubois A, et al. Genomic approach to study floral development genes in Rosa sp. PloS one. 2011;6:e28455. doi: 10.1371/journal.pone.0028455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nature Protocols. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


