Abstract
As a consequence of its difference in copy number between males and females, the X chromosome is subject to unique evolutionary forces and gene regulatory mechanisms. Previous studies of Drosophila melanogaster have shown that the expression of X-linked, testis-specific reporter genes is suppressed in the male germline. However, it is not known whether this phenomenon is restricted to testis-expressed genes or if it is a more general property of genes with tissue-specific expression, which are also underrepresented on the X chromosome. To test this, we compared the expression of three tissue-specific reporter genes (ovary, accessory gland and Malpighian tubule) inserted at various autosomal and X-chromosomal locations. In contrast to testis-specific reporter genes, we found no reduction of X-linked expression in any of the other tissues. In accessory gland and Malpighian tubule, we detected higher expression of the X-linked reporter genes, which suggests that they are at least partially dosage compensated. We found no difference in the tissue-specificity of X-linked and autosomal reporter genes. These findings indicate that, in general, the X chromosome is not a detrimental environment for tissue-specific gene expression and that the suppression of X-linked expression is limited to the male germline.
Introduction
In the well-studied XY sex determination system that is found in mammals and Drosophila, females are the homogametic sex with two copies of the X chromosome and males are heterogametic with one X and one Y chromosome. The difference in copy number between the sexes makes the X-chromosome subject to unique evolutionary forces and gene regulatory mechanisms, such as dosage compensation and meiotic sex chromosome inactivation (MSCI) (Vicoso and Charlesworth, 2006).
Several studies using Drosophila melanogaster found that, in the male germline, X-linked genes have reduced expression relative to autosomal genes (Hense et al., 2007; Meiklejohn et al., 2011). For example, the expression of a transgenic reporter gene driven by a testis-specific promoter was significantly lower when it was located on the X chromosome than on an autosome (Hense et al., 2007). A subsequent examination of over 100 unique reporter gene insertions driven by the same testis-specific promoter demonstrated that this pattern held for all regions of the X chromosome (Kemkemer et al., 2011). The effect was not limited to a single testis-specific promoter, as the same result was obtained for reporter genes under the control of four different testis-specific promoters, including three derived from X-linked genes (Kemkemer et al., 2014).
A recent study found that the suppression of X-linked expression in the male germline was not an artifact of using reporter genes in transposable element vectors. When small regions of the X chromosome were transposed to an autosome (without the use of a transposable element), the expression of the genes within the region increased in the male germline (Landeen et al., 2016). This was true for both testis-specific and housekeeping genes. The transposed housekeeping genes did not show increased expression in carcass or ovary (Landeen et al., 2016), indicating that the effect is either limited to testis-specific genes or is a more general property of tissue-specific genes.
It is important to note that the above results cannot be explained by differences in chromosome dose between the X and the autosomes or by the absence of dosage compensation in the male germline (Meiklejohn et al., 2011), as the copy number of the reporter (or transposed) genes was held constant at one copy in all comparisons. Furthermore, the patterns of expression seen in Drosophila are not entirely consistent with the specific mechanism of MSCI that has been described in mammals, in which X-linked gene expression is greatly suppressed at the prophase I stage of meiosis (da Cruz et al., 2016). Although a slight overrepresentation of X-linked genes that are downregulated during meiosis has been reported in D. melanogaster (Vibranovski et al., 2009), the reporter genes tested in Drosophila tend to have higher expression at the meiotic stage than at the premeiotic stage (Meiklejohn et al., 2011; Kemkemer et al., 2014). Since it remains unclear whether the phenomenon observed in Drosophila is analogous to mammalian MSCI (Meiklejohn et al., 2011; Mikhaylova and Nurminsky, 2011; Vibranovski et al., 2012; Vibranovski, 2014), we refer to it here as ‘X suppression’ (Landeen et al., 2016).
Although the suppression of X-linked gene expression has been demonstrated in testis, it is not known whether this phenomenon is unique to testis-specific genes, or if it occurs for genes expressed specifically in other tissues. Support for the latter possibility comes from genome-wide expression analyses, which indicate that tissue-specific genes are underrepresented on the X chromosome (Mikhaylova and Nurminsky, 2011; Meisel et al., 2012; but see Vibranovski et al., 2012). This suggests that the X chromosome may be an unfavorable environment for tissue-specific regulation. An exception is the ovary, where there is an excess of X-linked ovary-specific genes (Mikhaylova and Nurminsky, 2011; Meisel et al., 2012; Vibranovski et al., 2012).
Here we present an experimental test for X suppression of tissue-specific genes using the same reporter gene approach that has been used previously for testis-specific genes (Hense et al., 2007; Kemkemer et al., 2014), but with regulatory sequences that drive expression specifically in either ovary, accessory gland or Malpighian tubule. We chose the ovary to test if X suppression also occurs in the female germline. This tissue is also of interest because it is the only one to show an overrepresentation of X-linked tissue-specific genes (Mikhaylova and Nurminsky, 2011; Meisel et al., 2012), suggesting that the X chromosome might be a favorable environment for ovary-specific expression. We chose the accessory gland because it is a male-limited reproductive tissue, but not part of the germline. This allows us to test if X suppression occurs in a somatic male reproductive tissue. In addition, the accessory gland shows the greatest paucity of X-linked tissue-specific genes (Meisel et al., 2012), which is in agreement with the observed underrepresentation of accessory gland protein genes on the X chromosome (Swanson et al., 2001). Finally, we chose the Malpighian tubule, a somatic tissue present in both males and females, to test if X suppression is a common property of somatic tissue-specific genes in both sexes.
Materials and methods
Transformation vector construction
To identify tissue-specific regulatory sequences (here referred to as ‘promoters’), we used data from FlyAtlas (Chintapalli et al., 2007) to find genes with highly enriched expression in a single tissue. Our assumption was that the genomic regions directly upstream of such genes were likely to have tissue-specific regulatory function. When possible, we verified this by searching the literature for studies providing experimental evidence that the candidate sequence could drive gene expression in the tissue of interest. This approach was used to identify putative tissue-specific promoters of three genes: one ovary-specific (CG2175; dec-1), one accessory gland-specific (CG8982; Acp26Aa) and one Malpighian tubule-specific (CG15406). For the first two genes, the regulatory sequences had been tested previously (Park et al., 1994; Spangenberg and Waring, 2007) and corresponded to coordinates chrX:7 873 148–7 875 061 (1914 bp) and chr2L:5 896 212–5 893 873 (2340 bp) of release 6 of the D. melanogaster genome (Hoskins et al., 2015), respectively. Because no functional information was available for the CG15406 regulatory sequence, a putative promoter sequence corresponding to coordinates chr2L:3 307 394–3 309 228 (1835 bp) was used.
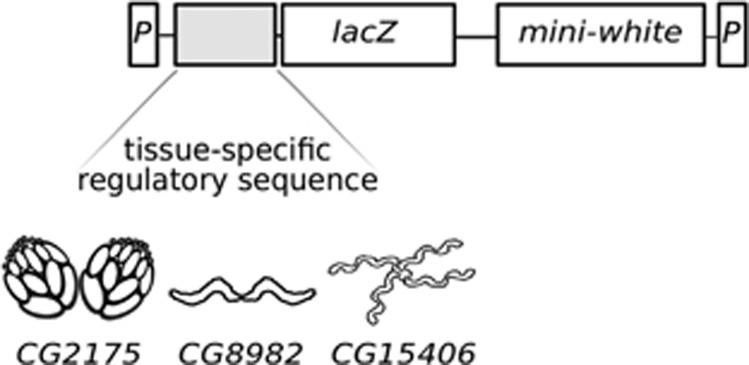
Each promoter sequence was PCR-amplified from genomic DNA of the Canton S strain of D. melanogaster and cloned into the pCR2.1 TOPO vector (Invitrogen, Carlsbad, CA, USA). The coding region of the lacZ gene was excised from the pCMV-SPORT-βgal vector (Invitrogen) with NotI and inserted into the unique NotI site located just downstream of the promoter sequence in the pCR2.1 TOPO vector. Subsequently, a BamHI–XbaI fragment containing both the promoter and the lacZ gene in the same transcriptional orientation was excised and ligated into the pP[wFl] transformation vector (Siegal and Hartl, 1996), which was previously linearized with BamHI and SpeI. This vector includes the D. melanogaster mini-white gene, which serves as a selectable eye color marker. The vector also contains the terminal inverted repeats of a P transposable element, which flank the inserted reporter gene and the mini-white gene (Figure 1).
Figure 1.
Schematic illustration of the reporter gene constructs. The upstream regulatory sequences of three genes with tissue-specific expression were cloned individually into a transposable element vector containing the lacZ reporter gene and the mini-white marker gene. The genes CG2175, CG8982 and CG15406 show highly enriched expression in ovary, accessory gland and Malpighian tubule, respectively. The terminal sequences of the P-element (P) represent the boundaries of the DNA fragment that is inserted into the genome.
Germline transformation
Transformation vectors were purified with the QIAfilter Plasmid Midi Kit (Qiagen, Hilden, Germany) and eluted in ultrapure water. They were then injected at a concentration of 200 ng μl−1 into D. melanogaster embryos of the strain yw;Δ2–3,Sb/TM6, which lacks functional P elements but contains a gene encoding the P-element transposase linked to the Stubble (Sb) phenotypic marker. Surviving adults were crossed to the white-eyed yw strain and their progeny was screened for transformants, which were identified by their red eye color. The red-eyed flies with wild-type bristles (i.e. those lacking the Sb mutation) were used to start a new stock that had a stable transgene insertion and lacked the transposase. Additional transgenic flies were obtained from an external injection service (Rainbow Transgenic Flies Inc., Camarillo, CA, USA). For these injections a helper plasmid containing the P-element transposase gene was coinjected with the reporter gene construct.
Generation of additional transgenic lines with insertions at different chromosomal locations was achieved by performing genetic crosses to mobilize transgenes from the X chromosome to an autosome or vice versa using the yw;Δ2–3,Sb/TM6 strain to provide a source of transposase as described by Hense et al. (2007).
Mapping insertion locations
The chromosomal location of the transgene (X-linked or autosomal) was determined through genetic crosses of transformed males with yw females. Males with X-linked insertions were expected to transmit the red eye color only to their female offspring, while those with autosomal insertions were expected to transmit the red eye color to 50% of their offspring of both sexes.
The precise genomic locations of the insertions were determined by inverse PCR (Bellen et al., 2004). Genomic DNA of each transgenic line was digested with either HinP1 or HpaII, which both cut at multiple locations within the D. melanogaster genome. The resulting fragments were then self-ligated with T4 DNA ligase (New England Biolabs, Ipswich, MA, USA). The fragment containing the inserted transgene construct was amplified by PCR with primers specific to the sequence of the pP[wFl] transformation vector. The following two primers pairs were used: Plac1-Plac4 (5′-CACCCAAGGCTCTGCTCCCACAAT-3′, 5′-ACTGTGCGTTAGGTCCTGTTCATTGTT-3′) and EY.3.F-EY.3.R (5′-CAATAAGTGCGAGTGAAAGG-3′, 5′-ACAATCATATCGCTGTCTCAC-3′). The resulting PCR product was sequenced with the primers Sp1 (5′-ACACAACCTTTCCTCTCAACAA-3′) and EY.3.F (above) using BigDye v.1.1 chemistry on an ABI 3730 Automated Sequencer (Applied Biosystems, Foster City, CA, USA). The insertion location was determined from the flanking genomic sequences, which were mapped to the D. melanogaster reference genome (release 6.09) using BLAST (Altschul et al., 1990).
Fly strains and maintenance
In addition to the newly generated transformed lines, we also used 17 transformed lines containing the β-galactosidase gene under the control of the testis-specific promoter of the CG7929 (ocnus) gene, which were originally designated as P[wFl-ocn-lacZ] lines by Hense et al. (2007). Of these lines, eight had the transgene inserted on an autosome and nine had the transgene inserted on the X chromosome.
All fly strains were maintained at 22 °C on cornmeal–agar–molasses medium with a 14 h light:10 h dark cycle. All flies used for tissue staining and β-galactosidase assays were 4–6 days old, mated and either heterozygous or hemizygous for the transgene insertion. Thus, all comparisons were of flies carrying a single copy of the reporter gene.
Tissue staining
Flies from autosomal and X-linked fly lines of each construct, as well as flies of the yw strain (as a negative control) were dissected manually to extract ovaries, accessory glands or Malpighian tubules. Each tissue was incubated overnight at 37 °C in an Assay buffer (200 mM sodium phosphate (pH 7.3), 2 mM MgCl2, 100 mM 2-mercaptoethanol, 1.33 mg ml−1 o-nitrophenyl-b-d-galactopyranoside), with 1 mg ml−1 ferric ammonium citrate and 1.8 mg ml of S-GAL sodium salt (Sigma-Aldrich, St Louis, MO, USA).
β-Galactosidase assays
The expression of the lacZ reporter gene was quantified with a β-galactosidase activity assay. For each transformed line, soluble protein was extracted from eight whole flies (except for lines carrying the testis-specific construct, for which five whole flies were used) by homogenizing the flies in 200 μl of cold buffer (0.1 M Tris-HCl, 1 mM EDTA, 7 mm 2-mercaptoethanol; pH 7.5), incubating the homogenate on ice for 15 min, centrifuging at 12 000 r.p.m. for 15 min at 4 °C and collecting the supernatant. Subsequently, 50 μl of 2 × Assay buffer were added to 50 μl of protein extract, representing one technical replicate. Each sample was assayed in two technical replicates. Biological replication was carried out by extracting soluble protein from a new cohort of flies of the same genotype. The number of biological replicates varied among samples (see below). The enzymatic activity was measured spectrophotometrically by following absorbance at 420 nm at 37 °C. β-Galactosidase activity units were quantified as the change in absorbance per minute (maximum slope).
For the ovary-specific construct, 2–5 biological replicates were carried out using females. For the accessory gland-specific construct, 3–6 biological replicates were carried out using males. For the Malpighian tubule-specific construct, 3–6 biological replicates were performed separately for each sex. For the testis-specific construct, 2–4 biological replicates were performed for each line.
To test for reporter gene expression outside of the desired tissue, the above procedure was repeated using carcasses (here defined as the whole fly body, except for the target tissue). A total of 2–3 biological replicates were performed for each transformed line.
To test for differences in reporter gene activity between chromosomes or sexes, we used the mean activity of each transformed line as the input for the non-parametric Wilcoxon's (Mann–Whitney) test. For each line, the mean activity was calculated as the mean of the biological replicates, with each biological replicate representing the mean of its associated technical replicates.
Results
To determine the effects of X linkage on tissue-specific gene expression, we generated transgenic flies containing the lacZ reporter gene under the control of an ovary-specific, accessory gland-specific or Malpighian tubule-specific promoter (Figure 1). For the ovary and accessory gland expression constructs, we obtained 12 independent autosomal insertions and seven independent X-linked insertions each (Supplementary Table S1). For the Malpighian tubule expression construct, we obtained 12 independent autosomal insertions and six independent X-linked insertions (Supplementary Table S1). This allowed us to compare the expression of the same transgene when located on an autosome or on the X chromosome. To avoid the potentially confounding effects of gene dose, we only compared flies carrying a single copy of the reporter gene (i.e. flies heterozygous for autosomal insertions or hemizygous for X-linked insertions). For each reporter gene, we also compared levels of expression outside the tissue of interest (here designated as the carcass) between X-linked and autosomal insertions of each construct. This allowed us to test whether the ability to regulate tissue-specific expression differed between the autosomes and the X chromosome.
Expression in ovary
The gene CG2175 encodes a structural constituent of the chorion and shows very high and specific expression in the ovary (Table 1). Using a 1.9-kb sequence immediately upstream of the CG2175 coding region, we were able to drive high expression of the reporter gene in ovary (Figure 2). Reporter gene expression was very low outside the ovary, with expression in the carcass (whole body excluding the ovaries) being 25-fold lower than the expression in whole flies (Figures 3a and c).
Table 1. Expression of the genes from which promoter sequences were derived.
| Gene | Tissue | Tissue/fly a | T1/T2b | Tau c | M/F d |
|---|---|---|---|---|---|
| GC2175 | Ovary | 32 | 108 | 0.83 | 0.07 |
| CG8982 | Accessory gland | 193 | 170 | 0.74 | 25.79 |
| CG15406 | Malpighian tubule | 274 | 190 | 0.69 | 1.96 |
Ratio of the expression level in the tissue of interest to whole body of adult fly (Chintapalli et al., 2007).
Ratio of the expression level in the tissue of interest (T1) to the tissue with the next highest expression (T2) in adult fly (Chintapalli et al., 2007).
Tissue-specificity index calculated using FlyAtlas data (Chintapalli et al., 2007). Values range from 0 (broad expression) to 1 (highly tissue-specific).
Male/female expression ratio from SEBIDA database release 3.2 (Gnad and Parsch, 2006).
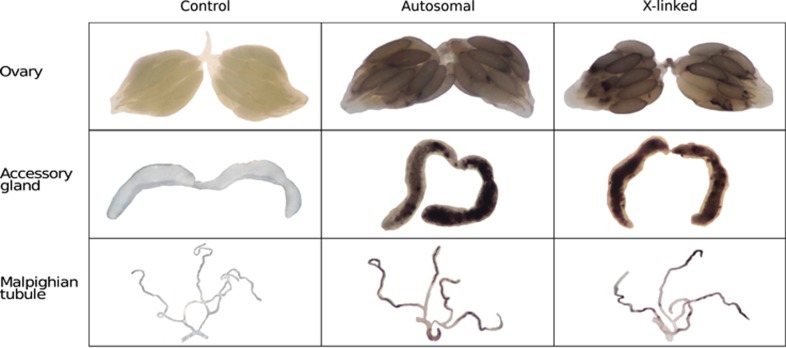
Figure 2.
Expression of the reporter genes in their target tissues. Ovaries, accessory glands and Malpighian tubules were dissected and incubated with the β-galactosidase substrate, S-gal. Dark areas indicate regions of reporter gene expression (β-galactosidase activity). Tissues from yw flies, which lack the reporter gene, are shown as negative controls. The remaining panels show tissues from flies carrying an autosomal or an X-linked insertion of the transgene.
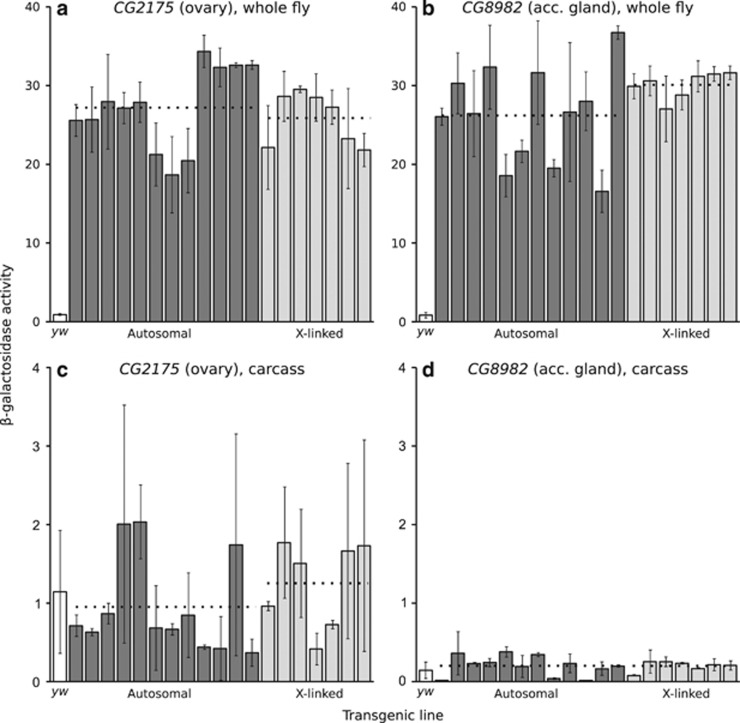
Figure 3.
Reporter gene expression of the ovary-specific (a, c) and accessory gland-specific (b, d) constructs in whole fly and in carcass. Each bar represents a transformed line with the reporter gene inserted at a unique autosomal (dark) or X-linked (light) location. The white bar indicates the activity of the control yw (non-transgenic) strain. Expression was measured spectrophotometrically as β-galactosidase activity in units of mOD min−1. Error bars indicate the standard deviation across biological replicates. Dotted lines indicate the average activities of all autosomal or X-linked lines. There was not a significant difference between autosomal and X-linked expression for either construct in whole flies (Wilcoxon's test; P=0.84 for CG2175, P=0.17 for CG8982) or in carcasses (P=0.34 for CG2175, P=0.90 for CG8982).
In whole females, the mean (median) β-galactosidase activities of autosomal and X-linked lines were 27.2 (27.5) and 25.9 (27.2) mOD min−1, respectively, and did not differ significantly (Wilcoxon's test, P=0.84) (Figure 3a). Thus, there was no evidence for X suppression in the female germline.
In carcass, the mean (median) β-galactosidase activities of autosomal and X-linked lines were 0.95 (0.70) and 1.25 (1.50) mOD min−1, respectively, and did not differ significantly (Wilcoxon's test, P=0.34) (Figure 3c). Thus, there was no evidence of a difference in tissue-specificity between the autosomal and X-linked reporter genes.
Expression in accessory gland
The gene CG8982 encodes an accessory gland protein (Acp26Aa), which shows very high and specific expression in the accessory gland (Table 1). Using a 2.3-kb sequence immediately upstream of the CG8982 coding region, we were able to drive high expression of the reporter gene in accessory gland (Figure 2). Reporter gene expression was very low outside the accessory gland, with expression in the carcass (whole body excluding the accessory gland, ejaculatory duct and bulb) being 148-fold lower than the expression in whole flies (Figures 3b and d).
In whole males, the mean (median) β-galactosidase activities of autosomal and X-linked lines were 26.2 (26.5) and 30.1 (30.6) mOD min−1, respectively. There was not a significant difference between autosomal and X-linked expression (Wilcoxon's test, P=0.17) (Figure 3b). Thus, there was no evidence for X suppression in the accessory gland. The higher expression observed on the X chromosome may be a consequence of dosage compensation of the X-linked transgenes in this somatic tissue.
In carcass, the mean (median) β-galactosidase activities of autosomal and X-linked lines were 0.18 (0.20) and 0.20 (0.20) mOD min−1, respectively, and did not differ significantly (Wilcoxon's test, P=0.90) (Figure 3d). Thus, there was no evidence of a difference in tissue-specificity between the autosomal and X-linked reporter genes.
Expression in Malpighian tubule
The gene CG15406 encodes a fructose transmembrane transporter that shows very high and specific expression in the Malpighian tubule (Table 1). Using a 1.8-kb sequence immediately upstream of the CG15406 coding region, we were able to drive high expression of the reporter gene in Malpighian tubule (Figure 2). Reporter gene expression was very low outside the Malpighian tubule, with expression in the carcass (whole body excluding the tubule and the directly adjacent segment of midgut) being at least 34-fold lower than the expression in whole flies of both sexes (Figure 4).
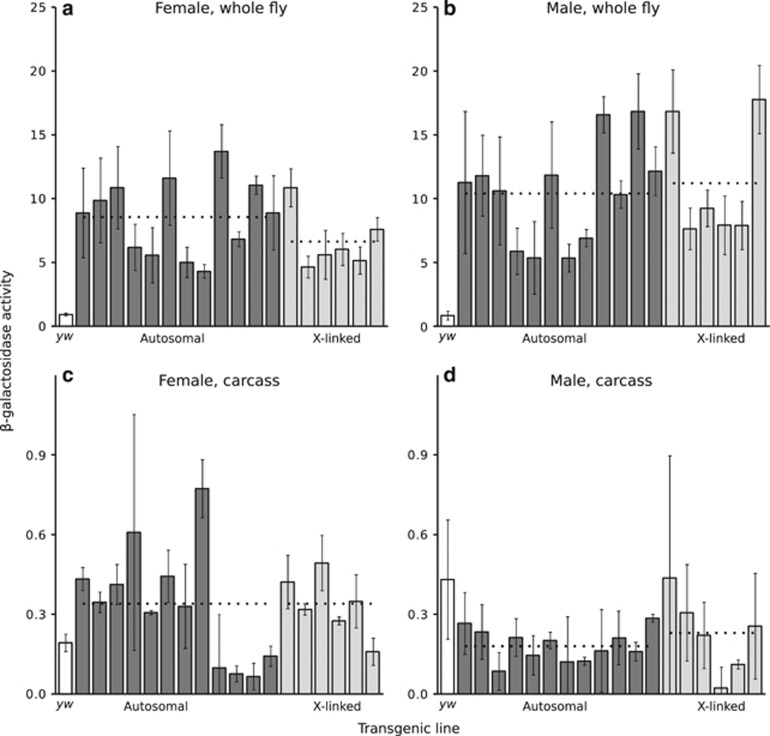
Figure 4.
Reporter gene expression of the Malpighian tubule-specific construct in females (a, c) and males (b, d) in whole fly and in carcass. Within each sex, each bar represents a transformed line with the reporter gene inserted at a unique autosomal (dark) or X-linked (light) location. The white bar indicates the activity of the control yw (non-transgenic) strain. The transformed lines are presented in the same order in all panels. Expression was measured spectrophotometrically as β-galactosidase activity in units of mOD min−1. Error bars indicate the standard deviation across biological replicates. Dotted lines indicate the average activities of all autosomal or X-linked lines within each sex. There was not a significant difference between autosomal and X-linked expression for either sex in whole flies (Wilcoxon's test; P=0.21 for females, P=0.82 for males) or in carcasses (P=0.89 for females, P=0.49 for males). However, in whole flies male expression was significantly higher than female expression on both the autosomes (Wilcoxon's signed-rank test, P=0.012) and the X chromosome (P=0.031).
In whole females, the mean (median) β-galactosidase activities of autosomal and X-linked lines were 8.50 (8.90) and 6.60 (5.80) mOD min−1, respectively, and did not differ significantly (Wilcoxon's test, P=0.21) (Figure 4a). In whole males, the mean (median) β-galactosidase activities of autosomal and X-linked lines were 10.40 (10.90) and 11.20 (8.60) mOD min−1, respectively, and also did not differ significantly (Wilcoxon's test, P=0.82) (Figure 4b). Thus, there was no evidence for X suppression in the Malpighian tubules of either sex.
Because the CG15406 reporter gene is expressed in both sexes, we could compare the expression of the same transgene (at the same genomic location) between males and females. This revealed that the expression was significantly higher in males than in females for both autosomal (Wilcoxon's signed-rank test, P=0.012) and X-linked (P=0.031) insertions. This difference between the sexes is likely the result of sex-specific regulation, as the native CG15406 gene is known to show male-biased expression (Table 1). Partial dosage compensation of X-linked transgenes in males may also contribute to this pattern, as the ratio of male-to-female expression is significantly greater for X-linked insertions (1.67) than for autosomal insertions (1.22) (Wilcoxon's test, P=0.003).
In female carcass, the mean (median) β-galactosidase activities of autosomal and X-linked lines were 0.33 (0.34) and 0.33 (0.33) mOD min−1, respectively, and did not differ significantly (Wilcoxon's test, P=0.89) (Figure 4c). In male carcass, the mean (median) β-galactosidase activities of autosomal and X-linked lines were 0.18 (0.18) and 0.23 (0.24) mOD min−1, respectively, and did not differ significantly (P=0.49) (Figure 4d). Thus, there was no evidence of a difference in tissue-specificity between the autosomal and X-linked reporter genes in either sex.
Comparison with testis-specific reporter genes
To compare our results for the ovary, accessory gland and Malpighian tubule with those previously obtained for testis, we measured reporter gene expression in 17 transgenic lines (8 autosomal and 9 X-linked) carrying the lacZ reporter gene under the control of the testis-specific CG7929 (ocnus) promoter. The creation of these lines and the initial analysis of their reporter gene expression were described by Hense et al. (2007).
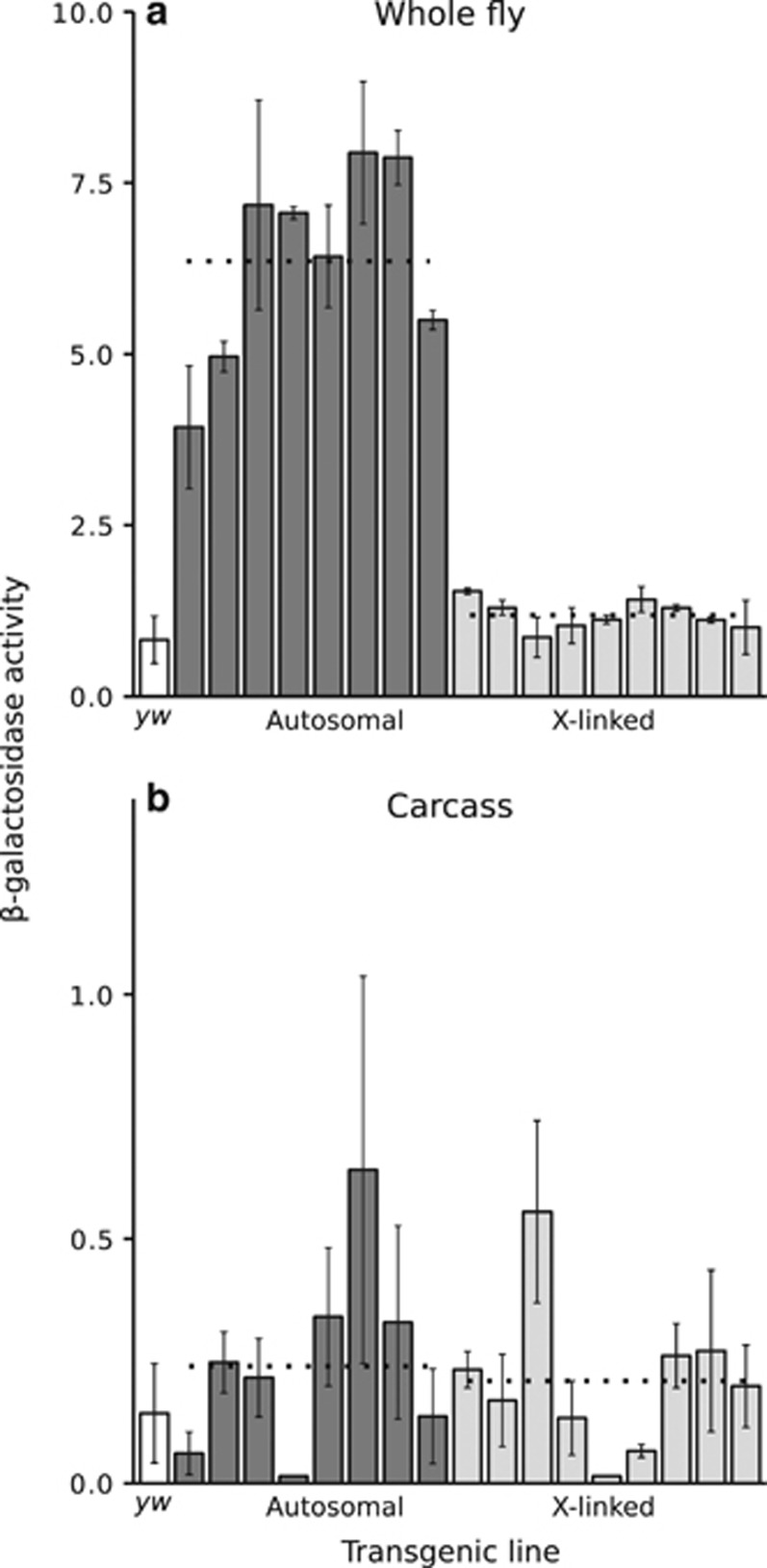
In whole males, the mean (median) β-galactosidase activities of autosomal and X-linked lines were 6.36 (6.74) and 1.19 (1.12) mOD min−1, respectively. The difference between autosomal and X-linked expression was highly significant (Wilcoxon's test, P=8.3 × 10−5) (Figure 5a). This result is in line with previous findings (Hense et al., 2007) and demonstrates that X-linkage has a much greater effect on expression in testis than in any other tissue.
Figure 5.
Reporter gene expression of the testis-specific CG7929 construct in whole fly (a) and carcass (b). Each bar represents a transformed line with the reporter gene inserted at a unique autosomal (dark) or X-linked (light) location. The white bar indicates the activity of the control yw (non-transgenic) strain. Expression was measured spectrophotometrically as β-galactosidase activity in units of mOD min−1. Error bars indicate the standard deviation across biological replicates. Dotted lines indicate the average activities of all autosomal or X-linked lines. There was a highly significant difference between autosomal and X-linked expression in whole flies (Wilcoxon's test; P=8.3 × 10−5), but not in carcasses (P=0.74).
In carcass, the mean (median) β-galactosidase activities of autosomal and X-linked lines were 0.24 (0.23) and 0.21 (0.20) mOD min−1, respectively, and did not differ significantly (Wilcoxon's test, P=0.74) (Figure 5b). Thus, there was no evidence of a difference in tissue-specificity between the autosomal and X-linked reporter genes.
Discussion
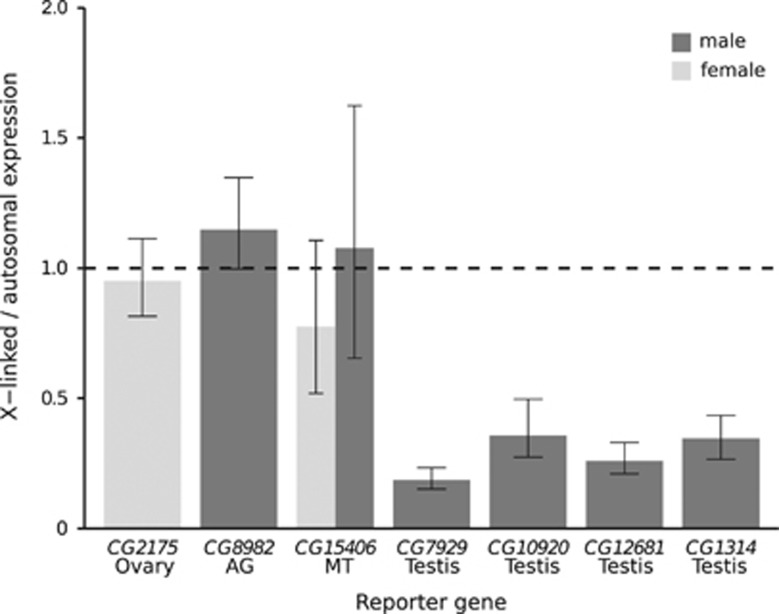
The present study was designed to determine the extent of tissue-specific gene suppression on the X chromosome. Overall, we found no evidence for X-chromosome suppression in any tissue aside from the testis, as the X/autosome ratio of reporter gene expression was very close to one in all of the other tissues that we tested (Figure 6). In contrast, the X/autosome ratio of reporter gene expression was well below one for the testis-specific reporter gene tested in the current study, as well as for three other testis-specific reporter genes reported previously (Figure 6). The average reduction of X-linked expression relative to autosomal expression for all of the testis-specific reporter genes was greater than threefold, with a range of 2.8-fold to 5.4-fold. Furthermore, for each of the testis-specific reporter genes, there was no overlap among the autosomal and the X-linked expression values. That is, the X-linked transgene with the highest expression still had lower expression than the autosomal transgene with the lowest expression (Figure 5; Kemkemer et al., 2014). In such a situation, a significant difference between X-linked and autosomal expression could be detected with a sample size as small as eight (four of each category) using a Wilcoxon's test and a two-tailed α of 0.05. Since our samples sizes were larger than this for all of the tested transgenes, we can conclude that the marked suppression of X-linked expression that is seen for testis-specific genes is unique to the male germline and is not a general property of tissue-specific genes. However, we cannot rule out the possibility that some tissue-specific genes may show more subtle expression differences between the X chromosome and the autosomes that are beyond the detection limits of our experiments.
Figure 6.
Mean X-to-autosomal expression ratio of tissue-specific reporter genes. For the reporter genes expressed in ovary, accessory gland (AG), Malpighian tubule (MT) and testis (CG7929), the bars represent the mean X/autosome expression as measured in this study. For comparison, the mean X/autosome expression of three additional testis-specific reporter genes (CG10920, CG12681 and CG1314) is shown. These data were taken from Kemkemer et al. (2014). Error bars indicate the 95% confidence interval of the ratio. The dashed line reflects the ratio expected if there is no difference between autosomal and X-linked expression.
Our results complement those of Landeen et al. (2016), who found that housekeeping genes transposed from the X chromosome to an autosome showed an increase in expression in the testis, but not in ovaries or carcass. This suggests that all expression from the X chromosome is suppressed in the male germline regardless of whether or not the gene has tissue-specific expression. Similarly, we can conclude that the expression of X-linked genes, whether tissue-specific or not, is not suppressed in tissues outside the male germline. Interestingly, X-linked, testis-expressed genes are enriched for an upstream regulatory motif that drives high expression in testis (Landeen et al., 2016), suggesting that gene-specific regulatory mechanisms have evolved to at least partially compensate for X suppression in the male germline. In this context, it is noteworthy that the ovary-specific promoter used in our experiments was derived from an X-linked gene. It is possible that this promoter (or the promoters of other X-linked, ovary-specific genes) may contain regulatory sequences that increase expression in ovary. However, we see no evidence for a promoter-by-chromosome interaction, as expression of the ovary-specific reporter gene is not higher when it is X-linked (Figure 3a). Similarly, the regulatory motif studied by Landeen et al. (2016) appears to enhance testis expression equally well whether it is autosomal or X-linked.
Despite the significant excess of ovary-specific genes on the X chromosome (Mikhaylova and Nurminsky, 2011; Meisel et al., 2012; Vibranovski et al., 2012), we found no evidence that X-linked transgenes have higher expression than autosomal transgenes in the ovary (Figures 3a and 6). Hence, the overabundance of ovary-expressed genes on the X chromosome cannot be explained by a chromosome-wide regulatory mechanism. Instead, it may be that sexual antagonism is involved. Dominant mutations that are beneficial to females, but deleterious to males, are expected to accumulate on the X chromosome, and the accumulation of such mutations could drive the fixation of expression modifiers that suppress expression in males (Rice, 1984; Charlesworth et al., 1987; Ellegren and Parsch, 2007). In line with this interpretation, a general ‘feminization’ of the Drosophila X chromosome has been observed, in which genes with female-biased expression are enriched on the X chromosome in whole flies and in multiple tissues (Parisi et al., 2003; Meisel et al., 2012; Huylmans and Parsch, 2015).
The X chromosome exhibits a significant paucity of accessory gland-specific genes (Mikhaylova and Nurminsky, 2011; Meisel et al., 2012). However, we found no evidence for X suppression in this tissue (Figures 3b and 6). This suggests that the underrepresentation of X-linked accessory gland genes is not a consequence of tissue-specific regulation. It may be that sexual antagonism also has a role in shaping the genomic distribution of accessory gland-specific genes. It has been observed that the expression of some accessory gland proteins is beneficial to male reproduction and/or sperm competition, but deleterious to female survival (Wolfner, 1997). Thus, genes expressed in accessory gland may be subject to sexually antagonistic evolution, which is expected to be more prevalent among X-linked genes (Rice, 1984; Charlesworth et al., 1987). If the male-beneficial/female-detrimental effect of mutations in these genes is, on average, dominant, then one would expect such mutations to be removed more efficiently from the X chromosome (Rice, 1984; Charlesworth et al., 1987; Ellegren and Parsch, 2007). Over evolutionary time, this process could make it less likely for genes encoding accessory gland proteins to arise or be maintained on the X chromosome.
We found no difference between the expression of X-linked and autosomal reporter genes expression in the Malpighian tubules of either males or females (Figures 4a and b), indicating that X suppression does not occur in this somatic tissue that is common to both sexes. However, the reporter genes did show a general pattern of male-biased expression, which is consistent with the expression of the native CG15406 gene from which the promoter sequence was derived (Table 1). This indicates that the regulatory elements needed to drive male-biased expression are contained within the 1.8- kb promoter sequence included in our expression construct. At present, it is not known whether this promoter sequence confers male-biased expression by enhancing expression in males or by repressing expression in females, or through a combination of both mechanisms. In a genome-wide analysis of sex-biased expression, Gallach and Betrán (2016) found that strong male-biased expression was accompanied by downregulation of expression in females.
In males, there was evidence for at least partial dosage compensation of X-linked transgenes in somatic tissues (accessory gland and Malpighian tubule), where the ratio of X/autosomal expression was greater in males than in females (Figure 6). In Malpighian tubule, the ratio of male/female expression was significantly greater for X-linked reporter genes than for autosomal ones, which also suggests a global upregulation of the X chromosome in males. In contrast, there was no evidence for dosage compensation of the X-linked, testis-specific transgenes tested here or in previous studies, as all of these transgenes had ratios of X/autosomal expression much less than one (Figure 6). Although an early microarray study of gonadal gene expression suggested that the X chromosome was dosage compensated in the male germline (Gupta et al., 2006), a more recent RNA-seq study found that X-linked genes exhibit lower expression than autosomal genes in the male germline, which is consistent with the absence of dosage compensation in this tissue (Meiklejohn et al., 2011). Our results agree with the latter study. Since transgene copy number was held at one for all lines used in our experiments, global X-chromosome dosage compensation should result in the higher expression of transgenes inserted on the X chromosome than on the autosomes. For the testis-expressed transgenes, the opposite pattern is observed (Figure 6), suggesting that not only does the male germline lack dosage compensation, but that there is a mechanism to suppress expression from the X chromosome in this tissue.
It has been proposed that the paucity of genes with tissue-specific expression on the X chromosome may be a result of the X chromosome being less efficient at activating or repressing expression in a tissue-specific manner (Mikhaylova and Nurminsky, 2011). We found no evidence that the ability to regulate tissue-specific expression was compromised on the X chromosome. For all reporter genes, expression in the carcass was very low and there was no significant difference in carcass expression between transgenes on the X chromosome and the autosomes. Thus, we conclude that even relatively short regulatory sequences (in the range of 1.8–2.3 kb) are sufficient to drive highly tissue-specific expression on both the autosomes and the X chromosome.
In summary, we found no evidence that the expression of X-linked, tissue-specific genes is suppressed in the female germline, in a somatic male-limited reproductive tissue, or in a somatic tissue present in both sexes. These results strongly suggest that the phenomenon is restricted to the male germline. In this sense, the process is comparable to the MSCI that occurs in mammals. Nevertheless, there appear to be differences in the specific molecular and cellular mechanisms used to achieve these processes in the two taxa, which is not surprising, given that male heterogametic sex determination evolved independently on these two lineages. Thus, MSCI and X suppression may not be completely analogous. Our results further suggest that the general paucity of tissue-specific genes (or the overrepresentation of ovary-specific genes) on the X chromosome is not the result of a chromosome-wide regulatory mechanism or an inherent inability to limit the breadth of expression of X-linked genes. Therefore, these patterns must have other causes, such as sexually antagonistic selection.
Data archiving
Data available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.02f6r.
Acknowledgments
We thank Hilde Lainer and Gaby Kumpfmüller for excellent technical assistance. We also thank the Munich evolutionary biology group for helpful comments and discussions. This work was supported by Deutsche Forschungsgemeinschaft grant PA 903/6.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
The authors declare no conflict of interest.
Supplementary Material
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990). Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Bellen HJ, Levis RW, Liao G, He Y, Carlson JW, Tsang G et al. (2004). The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167: 761–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Coyne JA, Barton NH. (1987). The relative rates of evolution of sex chromosomes and autosomes. Am Nat 130: 113–146. [Google Scholar]
- Chintapalli VR, Wang J, Dow JA. (2007). Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet 39: 715–720. [DOI] [PubMed] [Google Scholar]
- da Cruz I, Rodriguez-Casuriaga R, Santinaque FF, Farias J, Curti G, Capoano CA et al. (2016). Transcriptome analysis of highly purified mouse spermatogenic cell populations: gene expression signatures switch from meiotic-to postmeiotic-related processes at pachytene stage. BMC Genomics 17: 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H, Parsch J. (2007). The evolution of sex-biased genes and sex-biased gene expression. Nat Rev Genet 8: 689–698. [DOI] [PubMed] [Google Scholar]
- Gallach M, Betrán E. (2016). Dosage compensation and the distribution of sex-biased gene expression in Drosophila: considerations and genomic constraints. J Mol Evol 82: 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnad F, Parsch J. (2006). Sebida: a database for the functional and evolutionary analysis of genes with sex-biased expression. Bioinformatics 22: 2577–2579. [DOI] [PubMed] [Google Scholar]
- Gupta V, Parisi M, Sturgill D, Nuttall R, Doctolero M, Dudko OK et al. (2006). Global analysis of X-chromosome dosage compensation. J Biol 5: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hense W, Baines JF, Parsch J. (2007). X chromosome inactivation during Drosophila spermatogenesis. PLoS Biol 5: e273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins RA, Carlson JW, Wan KH, Park S, Mendez I, Galle SE et al. (2015). The Release 6 reference sequence of the Drosophila melanogaster genome. Genome Res 25: 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huylmans AK, Parsch J. (2015). Variation in the X:autosome distribution of male-biased genes among Drosophila melanogaster tissues and its relationship with dosage compensation. Genome Biol Evol 7: 1960–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemkemer C, Catalan A, Parsch J. (2014). 'Escaping' the X chromosome leads to increased gene expression in the male germline of Drosophila melanogaster. Heredity (Edinb) 112: 149–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemkemer C, Hense W, Parsch J. (2011). Fine-scale analysis of X chromosome inactivation in the male germ line of Drosophila melanogaster. Mol Biol Evol 28: 1561–1563. [DOI] [PubMed] [Google Scholar]
- Landeen EL, Muirhead CA, Wright L, Meiklejohn CD, Presgraves DC. (2016). Sex chromosome-wide transcriptional suppression and compensatory cis-regulatory evolution mediate gene expression in the Drosophila male germline. PLoS Biol 14: e1002499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn CD, Landeen EL, Cook JM, Kingan SB, Presgraves DC. (2011). Sex chromosome-specific regulation in the Drosophila male germline but little evidence for chromosomal dosage compensation or meiotic inactivation. PLoS Biol 9: e1001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel RP, Malone JH, Clark AG. (2012). Disentangling the relationship between sex-biased gene expression and X-linkage. Genome Res 22: 1255–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhaylova LM, Nurminsky DI. (2011). Lack of global meiotic sex chromosome inactivation, and paucity of tissue-specific gene expression on the Drosophila X chromosome. BMC Biol 9: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M, Nuttall R, Naiman D, Bouffard G, Malley J, Andrews J et al. (2003). Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science 299: 697–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M, Monsma SA, Wolfner MF. (1994). Two tightly-linked Drosophila male accessory gland transcripts with the same developmental expression derive from independent transcription units. Mech Dev 48: 51–57. [DOI] [PubMed] [Google Scholar]
- Rice WR. (1984). Sex chromosomes and the evolution of sexual dimorphism. Evolution 38: 735–742. [DOI] [PubMed] [Google Scholar]
- Siegal ML, Hartl DL. (1996). Transgene coplacement and high efficiency site-specific recombination with the Cre/loxP system in Drosophila. Genetics 144: 715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangenberg DK, Waring GL. (2007). A mutant dec-1 transgene induces dominant female sterility in Drosophila melanogaster. Genetics 177: 1595–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson WJ, Clark AG, Waldrip-Dail HM, Wolfner MF, Aquadro CF. (2001). Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. Proc Natl Acad Sci USA 98: 7375–7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibranovski MD. (2014). Meiotic sex chromosome inactivation in Drosophila. J Genomics 2: 104–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibranovski MD, Lopes HF, Karr TL, Long M. (2009). Stage-specific expression profiling of Drosophila spermatogenesis suggests that meiotic sex chromosome inactivation drives genomic relocation of testis-expressed genes. PLoS Genet 5: e1000731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibranovski MD, Zhang YE, Kemkemer C, Lopes HF, Karr TL, Long M. (2012). Re-analysis of the larval testis data on meiotic sex chromosome inactivation revealed evidence for tissue-specific gene expression related to the Drosophila X chromosome. BMC Biol 10: 49; author reply 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Charlesworth B. (2006). Evolution on the X chromosome: unusual patterns and processes. Nat Rev Genet 7: 645–653. [DOI] [PubMed] [Google Scholar]
- Wolfner MF. (1997). Tokens of love: functions and regulation of Drosophila male accessory gland products. Insect Biochem Mol Biol 27: 179–192. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.