Abstract
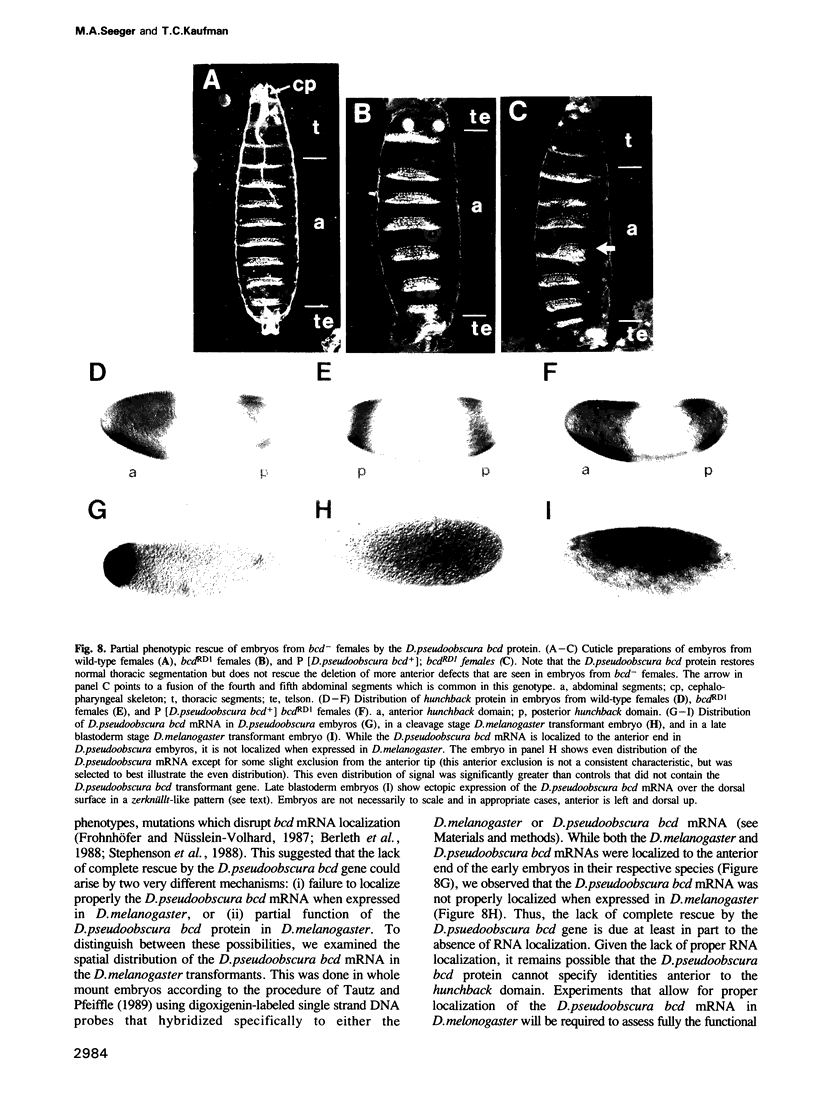
The specification of anterior positional information during Drosophila embryogenesis is largely dependent upon the function of the maternal-effect gene bicoid (bcd). Two aspects of bcd function are particularly striking. First, the bcd protein product forms a gradient during early embryogenesis, which regulates the transcription of at least one zygotic segmentation gene, hunchback, in a concentration dependent manner. Secondly, formation of the bcd protein gradient is dependent upon the specific localization of bcd mRNAs at the anterior end of the oocyte/embryo during oogenesis, a process which requires a cis-acting 625 nucleotide sequence within the 3' untranslated region of the bcd mRNA. We have cloned and sequenced the bcd gene from Drosophila pseudoobscura as a tool in identifying important functional domains within this transcription unit. DNA sequence comparisons reveal: (i) varying degrees of amino acid sequence conservation among the proposed functional domains of the bcd protein, (ii) the conservation of potential RNA secondary structures within the bcd mRNA localization element, and (iii) the maintenance of a short open reading frame within the 5' untranslated leader that may play a role in translational regulation. Finally, the D.pseudoobscura bcd gene partially rescues the phenotype of a bcd- mutation when placed into the D.melanogaster genome by germline transformation. The lack of full phenotypic rescue can be explained in part by the observed improper localization of the D.pseudoobscura bcd mRNA when expressed in D.melanogaster.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adam S. A., Nakagawa T., Swanson M. S., Woodruff T. K., Dreyfuss G. mRNA polyadenylate-binding protein: gene isolation and sequencing and identification of a ribonucleoprotein consensus sequence. Mol Cell Biol. 1986 Aug;6(8):2932–2943. doi: 10.1128/mcb.6.8.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguadé M. Nucleotide sequence comparison of the rp49 gene region between Drosophila subobscura and D. melanogaster. Mol Biol Evol. 1988 Jul;5(4):433–441. doi: 10.1093/oxfordjournals.molbev.a040502. [DOI] [PubMed] [Google Scholar]
- Bender M., Turner F. R., Kaufman T. C. A development genetic analysis of the gene regulator of postbithorax in Drosophila melanogaster. Dev Biol. 1987 Feb;119(2):418–432. doi: 10.1016/0012-1606(87)90046-7. [DOI] [PubMed] [Google Scholar]
- Berleth T., Burri M., Thoma G., Bopp D., Richstein S., Frigerio G., Noll M., Nüsslein-Volhard C. The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO J. 1988 Jun;7(6):1749–1756. doi: 10.1002/j.1460-2075.1988.tb03004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham J. R., Jr, Scott M. P. Developmentally regulated alternative splicing of transcripts from the Drosophila homeotic gene Antennapedia can produce four different proteins. EMBO J. 1988 Oct;7(10):3211–3222. doi: 10.1002/j.1460-2075.1988.tb03188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beverley S. M., Wilson A. C. Molecular evolution in Drosophila and the higher Diptera II. A time scale for fly evolution. J Mol Evol. 1984;21(1):1–13. doi: 10.1007/BF02100622. [DOI] [PubMed] [Google Scholar]
- Blackman R. K., Meselson M. Interspecific nucleotide sequence comparisons used to identify regulatory and structural features of the Drosophila hsp82 gene. J Mol Biol. 1986 Apr 20;188(4):499–515. doi: 10.1016/s0022-2836(86)80001-8. [DOI] [PubMed] [Google Scholar]
- Cavener D. R. Comparison of the consensus sequence flanking translational start sites in Drosophila and vertebrates. Nucleic Acids Res. 1987 Feb 25;15(4):1353–1361. doi: 10.1093/nar/15.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot H. V., Hall J. C., Rosbash M. Interspecific comparison of the period gene of Drosophila reveals large blocks of non-conserved coding DNA. EMBO J. 1988 Dec 1;7(12):3929–3937. doi: 10.1002/j.1460-2075.1988.tb03279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle H. J., Kraut R., Levine M. Spatial regulation of zerknüllt: a dorsal-ventral patterning gene in Drosophila. Genes Dev. 1989 Oct;3(10):1518–1533. doi: 10.1101/gad.3.10.1518. [DOI] [PubMed] [Google Scholar]
- Driever W., Ma J., Nüsslein-Volhard C., Ptashne M. Rescue of bicoid mutant Drosophila embryos by bicoid fusion proteins containing heterologous activating sequences. Nature. 1989 Nov 9;342(6246):149–154. doi: 10.1038/342149a0. [DOI] [PubMed] [Google Scholar]
- Driever W., Nüsslein-Volhard C. A gradient of bicoid protein in Drosophila embryos. Cell. 1988 Jul 1;54(1):83–93. doi: 10.1016/0092-8674(88)90182-1. [DOI] [PubMed] [Google Scholar]
- Driever W., Nüsslein-Volhard C. The bicoid protein determines position in the Drosophila embryo in a concentration-dependent manner. Cell. 1988 Jul 1;54(1):95–104. doi: 10.1016/0092-8674(88)90183-3. [DOI] [PubMed] [Google Scholar]
- Driever W., Nüsslein-Volhard C. The bicoid protein is a positive regulator of hunchback transcription in the early Drosophila embryo. Nature. 1989 Jan 12;337(6203):138–143. doi: 10.1038/337138a0. [DOI] [PubMed] [Google Scholar]
- Driever W., Thoma G., Nüsslein-Volhard C. Determination of spatial domains of zygotic gene expression in the Drosophila embryo by the affinity of binding sites for the bicoid morphogen. Nature. 1989 Aug 3;340(6232):363–367. doi: 10.1038/340363a0. [DOI] [PubMed] [Google Scholar]
- Frigerio G., Burri M., Bopp D., Baumgartner S., Noll M. Structure of the segmentation gene paired and the Drosophila PRD gene set as part of a gene network. Cell. 1986 Dec 5;47(5):735–746. doi: 10.1016/0092-8674(86)90516-7. [DOI] [PubMed] [Google Scholar]
- Frohnhöfer H. G., Lehmann R., Nüsslein-Volhard C. Manipulating the anteroposterior pattern of the Drosophila embryo. J Embryol Exp Morphol. 1986 Oct;97 (Suppl):169–179. [PubMed] [Google Scholar]
- Hanes S. D., Brent R. DNA specificity of the bicoid activator protein is determined by homeodomain recognition helix residue 9. Cell. 1989 Jun 30;57(7):1275–1283. doi: 10.1016/0092-8674(89)90063-9. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Eghtedarzadeh M. K. Conserved arrangement of nested genes at the Drosophila Gart locus. Genetics. 1987 Dec;117(4):711–725. doi: 10.1093/genetics/117.4.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassis J. A., Poole S. J., Wright D. K., O'Farrell P. H. Sequence conservation in the protein coding and intron regions of the engrailed transcription unit. EMBO J. 1986 Dec 20;5(13):3583–3589. doi: 10.1002/j.1460-2075.1986.tb04686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann R., Nüsslein-Volhard C. hunchback, a gene required for segmentation of an anterior and posterior region of the Drosophila embryo. Dev Biol. 1987 Feb;119(2):402–417. doi: 10.1016/0012-1606(87)90045-5. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Macdonald P. M., Struhl G. A molecular gradient in early Drosophila embryos and its role in specifying the body pattern. Nature. 1986 Dec 11;324(6097):537–545. doi: 10.1038/324537a0. [DOI] [PubMed] [Google Scholar]
- Macdonald P. M., Struhl G. cis-acting sequences responsible for anterior localization of bicoid mRNA in Drosophila embryos. Nature. 1988 Dec 8;336(6199):595–598. doi: 10.1038/336595a0. [DOI] [PubMed] [Google Scholar]
- Mlodzik M., Fjose A., Gehring W. J. Molecular structure and spatial expression of a homeobox gene from the labial region of the Antennapedia-complex. EMBO J. 1988 Aug;7(8):2569–2578. doi: 10.1002/j.1460-2075.1988.tb03106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller P. P., Hinnebusch A. G. Multiple upstream AUG codons mediate translational control of GCN4. Cell. 1986 Apr 25;45(2):201–207. doi: 10.1016/0092-8674(86)90384-3. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., Frohnhöfer H. G., Lehmann R. Determination of anteroposterior polarity in Drosophila. Science. 1987 Dec 18;238(4834):1675–1681. doi: 10.1126/science.3686007. [DOI] [PubMed] [Google Scholar]
- O'Connor M. B., Binari R., Perkins L. A., Bender W. Alternative RNA products from the Ultrabithorax domain of the bithorax complex. EMBO J. 1988 Feb;7(2):435–445. doi: 10.1002/j.1460-2075.1988.tb02831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace N. R., Smith D. K., Olsen G. J., James B. D. Phylogenetic comparative analysis and the secondary structure of ribonuclease P RNA--a review. Gene. 1989 Oct 15;82(1):65–75. doi: 10.1016/0378-1119(89)90031-0. [DOI] [PubMed] [Google Scholar]
- Query C. C., Bentley R. C., Keene J. D. A common RNA recognition motif identified within a defined U1 RNA binding domain of the 70K U1 snRNP protein. Cell. 1989 Apr 7;57(1):89–101. doi: 10.1016/0092-8674(89)90175-x. [DOI] [PubMed] [Google Scholar]
- Rebagliati M. An RNA recognition motif in the bicoid protein. Cell. 1989 Jul 28;58(2):231–232. doi: 10.1016/0092-8674(89)90834-9. [DOI] [PubMed] [Google Scholar]
- Rechsteiner M. Do myc, fos and E1A function as protein phosphatase inhibitors? Biochem Biophys Res Commun. 1987 Feb 27;143(1):194–198. doi: 10.1016/0006-291x(87)90649-8. [DOI] [PubMed] [Google Scholar]
- Robertson H. M., Preston C. R., Phillis R. W., Johnson-Schlitz D. M., Benz W. K., Engels W. R. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics. 1988 Mar;118(3):461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S., Wells R., Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986 Oct 17;234(4774):364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M. A., Haffley L., Kaufman T. C. Characterization of amalgam: a member of the immunoglobulin superfamily from Drosophila. Cell. 1988 Nov 18;55(4):589–600. doi: 10.1016/0092-8674(88)90217-6. [DOI] [PubMed] [Google Scholar]
- Spradling A. C., Rubin G. M. Transposition of cloned P elements into Drosophila germ line chromosomes. Science. 1982 Oct 22;218(4570):341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- Stephenson E. C., Chao Y. C., Fackenthal J. D. Molecular analysis of the swallow gene of Drosophila melanogaster. Genes Dev. 1988 Dec;2(12A):1655–1665. doi: 10.1101/gad.2.12a.1655. [DOI] [PubMed] [Google Scholar]
- Stroeher V. L., Gaiser J. C., Garber R. L. Alternative RNA splicing that is spatially regulated: generation of transcripts from the Antennapedia gene of Drosophila melanogaster with different protein-coding regions. Mol Cell Biol. 1988 Oct;8(10):4143–4154. doi: 10.1128/mcb.8.10.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G., Struhl K., Macdonald P. M. The gradient morphogen bicoid is a concentration-dependent transcriptional activator. Cell. 1989 Jun 30;57(7):1259–1273. doi: 10.1016/0092-8674(89)90062-7. [DOI] [PubMed] [Google Scholar]
- Tautz D., Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989 Aug;98(2):81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Tautz D. Regulation of the Drosophila segmentation gene hunchback by two maternal morphogenetic centres. Nature. 1988 Mar 17;332(6161):281–284. doi: 10.1038/332281a0. [DOI] [PubMed] [Google Scholar]
- Treier M., Pfeifle C., Tautz D. Comparison of the gap segmentation gene hunchback between Drosophila melanogaster and Drosophila virilis reveals novel modes of evolutionary change. EMBO J. 1989 May;8(5):1517–1525. doi: 10.1002/j.1460-2075.1989.tb03536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton K. A., Johansen K. M., Xu T., Artavanis-Tsakonas S. Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell. 1985 Dec;43(3 Pt 2):567–581. doi: 10.1016/0092-8674(85)90229-6. [DOI] [PubMed] [Google Scholar]
- Wilde C. D., Akam M. Conserved sequence elements in the 5' region of the Ultrabithorax transcription unit. EMBO J. 1987 May;6(5):1393–1401. doi: 10.1002/j.1460-2075.1987.tb02380.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAJIMA H. Studies on embryonic determination of the harlequin-fly, Chironomus dorsalis. I. Effects of centrifugation and of its combination with constriction and puncturing. J Embryol Exp Morphol. 1960 Jun;8:198–215. [PubMed] [Google Scholar]
- Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989 Apr 7;244(4900):48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]