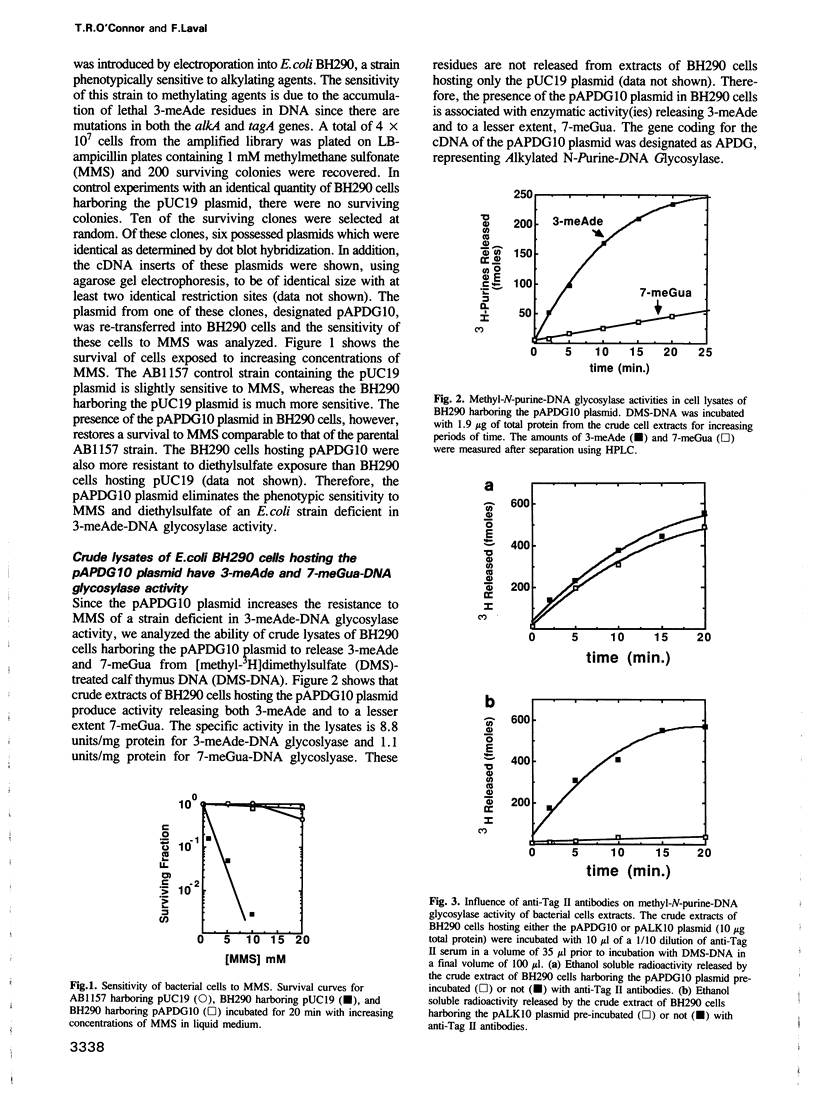
Abstract
A cDNA plasmid expression library was constructed from the poly(A)+ mRNA of H4 cells, a rat hepatoma cell line. The library was introduced into Escherichia coli strain BH290 deficient in the repair of 3-methyladenine (3-meAde) residues in DNA. This DNA repair deficiency renders the stain phenotypically sensitive to treatment with alkylating agents. The cDNA library was screened for survivors to methylmethane sulfonate. BH290 cells hosting one of the plasmids, pAPDG10 (Alkylated N-Purine-DNA Glycosylase), from surviving cells had a sensitivity to MMS equivalent to that of the wild type strain. Crude extracts of BH290 cells harboring the pAPDG10 plasmid released 3-meAde and 7-methylguanine residues from DNA methylated with [methyl-3H]dimethylsulfate. The cDNA sequence of 993 bp inserted in pAPDG10 has a single open reading frame greater than 85 amino acids in length. The derived APDG protein sequence of 253 amino acids and the 3-meAde-DNA glycosylase II of E. coli coded for by the alkA gene have regions of conserved sequences. Analysis of the genomic DNA using Southern hybridization suggests that the APDG gene has a minimal size of 6.5-12 kb. Northern blot analysis shows that the transcript produced in H4 cells is also present in normal rat liver cells.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beranek D. T., Weis C. C., Evans F. E., Chetsanga C. J., Kadlubar F. F. Identification of N5-methyl-N5-formyl-2,5,6-triamino-4-hydroxypyrimidine as a major adduct in rat liver DNA after treatment with the carcinogens, N,N-dimethylnitrosamine or 1,2-dimethylhydrazine. Biochem Biophys Res Commun. 1983 Jan 27;110(2):625–631. doi: 10.1016/0006-291x(83)91195-6. [DOI] [PubMed] [Google Scholar]
- Boiteux S., Huisman O., Laval J. 3-Methyladenine residues in DNA induce the SOS function sfiA in Escherichia coli. EMBO J. 1984 Nov;3(11):2569–2573. doi: 10.1002/j.1460-2075.1984.tb02175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux S., O'Connor T. R., Laval J. Formamidopyrimidine-DNA glycosylase of Escherichia coli: cloning and sequencing of the fpg structural gene and overproduction of the protein. EMBO J. 1987 Oct;6(10):3177–3183. doi: 10.1002/j.1460-2075.1987.tb02629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Derfler B., Maskati A., Samson L. Cloning a eukaryotic DNA glycosylase repair gene by the suppression of a DNA repair defect in Escherichia coli. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7961–7965. doi: 10.1073/pnas.86.20.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. D., Dixon J. E., Zalkin H. Cloning of a chicken liver cDNA encoding 5-aminoimidazole ribonucleotide carboxylase and 5-aminoimidazole-4-N-succinocarboxamide ribonucleotide synthetase by functional complementation of Escherichia coli pur mutants. Proc Natl Acad Sci U S A. 1990 Apr;87(8):3097–3101. doi: 10.1073/pnas.87.8.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison A. J., Scott J. E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986 Sep;67(Pt 9):1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- Demple B., Sedgwick B., Robins P., Totty N., Waterfield M. D., Lindahl T. Active site and complete sequence of the suicidal methyltransferase that counters alkylation mutagenesis. Proc Natl Acad Sci U S A. 1985 May;82(9):2688–2692. doi: 10.1073/pnas.82.9.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower W. J., Miller J. F., Ragsdale C. W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988 Jul 11;16(13):6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaboriaud C., Bissery V., Benchetrit T., Mornon J. P. Hydrophobic cluster analysis: an efficient new way to compare and analyse amino acid sequences. FEBS Lett. 1987 Nov 16;224(1):149–155. doi: 10.1016/0014-5793(87)80439-8. [DOI] [PubMed] [Google Scholar]
- Gallagher P. E., Brent T. P. Partial purification and characterization of 3-methyladenine-DNA glycosylase from human placenta. Biochemistry. 1982 Dec 7;21(25):6404–6409. doi: 10.1021/bi00268a013. [DOI] [PubMed] [Google Scholar]
- Karran P., Hjelmgren T., Lindahl T. Induction of a DNA glycosylase for N-methylated purines is part of the adaptive response to alkylating agents. Nature. 1982 Apr 22;296(5859):770–773. doi: 10.1038/296770a0. [DOI] [PubMed] [Google Scholar]
- Larson K., Sahm J., Shenkar R., Strauss B. Methylation-induced blocks to in vitro DNA replication. Mutat Res. 1985 Jun-Jul;150(1-2):77–84. doi: 10.1016/0027-5107(85)90103-4. [DOI] [PubMed] [Google Scholar]
- Laval F., Laval J. Adaptive response in mammalian cells: crossreactivity of different pretreatments on cytotoxicity as contrasted to mutagenicity. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1062–1066. doi: 10.1073/pnas.81.4.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laval F., Little J. B. Enhancement of survival of X-irradiated mammalian cells by the uncoupler of oxidative phosphorylation, m-chloro carbonyl cyanide phenylhydrazone. Radiat Res. 1977 Sep;71(3):571–578. [PubMed] [Google Scholar]
- Laval J. Two enzymes are required from strand incision in repair of alkylated DNA. Nature. 1977 Oct 27;269(5631):829–832. doi: 10.1038/269829a0. [DOI] [PubMed] [Google Scholar]
- Lawley P. D., Warren W. Removal of minor methylation products 7-methyladenine and 3-methylguanine from DNA of Escherichia coli treated with dimethyl sulphate. Chem Biol Interact. 1976 Feb;12(2):211–220. doi: 10.1016/0009-2797(76)90100-9. [DOI] [PubMed] [Google Scholar]
- Lefebvre P., Laval F. Enhancement of O6-methylguanine-DNA-methyltransferase activity induced by various treatments in mammalian cells. Cancer Res. 1986 Nov;46(11):5701–5705. [PubMed] [Google Scholar]
- Lindahl T., Sedgwick B., Sekiguchi M., Nakabeppu Y. Regulation and expression of the adaptive response to alkylating agents. Annu Rev Biochem. 1988;57:133–157. doi: 10.1146/annurev.bi.57.070188.001025. [DOI] [PubMed] [Google Scholar]
- Loechler E. L., Green C. L., Essigmann J. M. In vivo mutagenesis by O6-methylguanine built into a unique site in a viral genome. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6271–6275. doi: 10.1073/pnas.81.20.6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Male R., Helland D. E., Kleppe K. Purification and characterization of 3-methyladenine-DNA glycosylase from calf thymus. J Biol Chem. 1985 Feb 10;260(3):1623–1629. [PubMed] [Google Scholar]
- Margison G. P., Pegg A. E. Enzymatic release of 7-methylguanine from methylated DNA by rodent liver extracts. Proc Natl Acad Sci U S A. 1981 Feb;78(2):861–865. doi: 10.1073/pnas.78.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch D. J., Dalrymple M. A., Davison A. J., Dolan A., Frame M. C., McNab D., Perry L. J., Scott J. E., Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988 Jul;69(Pt 7):1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- Nakabeppu Y., Miyata T., Kondo H., Iwanaga S., Sekiguchi M. Structure and expression of the alkA gene of Escherichia coli involved in adaptive response to alkylating agents. J Biol Chem. 1984 Nov 25;259(22):13730–13736. [PubMed] [Google Scholar]
- O'Connor T. R., Boiteux S., Laval J. Ring-opened 7-methylguanine residues in DNA are a block to in vitro DNA synthesis. Nucleic Acids Res. 1988 Jul 11;16(13):5879–5894. doi: 10.1093/nar/16.13.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor T. R., Laval J. Physical association of the 2,6-diamino-4-hydroxy-5N-formamidopyrimidine-DNA glycosylase of Escherichia coli and an activity nicking DNA at apurinic/apyrimidinic sites. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5222–5226. doi: 10.1073/pnas.86.14.5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen L. C., Aasland R., Wittwer C. U., Krokan H. E., Helland D. E. Molecular cloning of human uracil-DNA glycosylase, a highly conserved DNA repair enzyme. EMBO J. 1989 Oct;8(10):3121–3125. doi: 10.1002/j.1460-2075.1989.tb08464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival K. J., Klein M. B., Burgers P. M. Molecular cloning and primary structure of the uracil-DNA-glycosylase gene from Saccharomyces cerevisiae. J Biol Chem. 1989 Feb 15;264(5):2593–2598. [PubMed] [Google Scholar]
- Pierre J., Laval J. Cloning of Micrococcus luteus 3-methyladenine-DNA glycosylase genes in Escherichia coli. Gene. 1986;43(1-2):139–146. doi: 10.1016/0378-1119(86)90017-x. [DOI] [PubMed] [Google Scholar]
- Potter P. M., Wilkinson M. C., Fitton J., Carr F. J., Brennand J., Cooper D. P., Margison G. P. Characterisation and nucleotide sequence of ogt, the O6-alkylguanine-DNA-alkyltransferase gene of E. coli. Nucleic Acids Res. 1987 Nov 25;15(22):9177–9193. doi: 10.1093/nar/15.22.9177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riazuddin S., Lindahl T. Properties of 3-methyladenine-DNA glycosylase from Escherichia coli. Biochemistry. 1978 May 30;17(11):2110–2118. doi: 10.1021/bi00604a014. [DOI] [PubMed] [Google Scholar]
- Sakumi K., Nakabeppu Y., Yamamoto Y., Kawabata S., Iwanaga S., Sekiguchi M. Purification and structure of 3-methyladenine-DNA glycosylase I of Escherichia coli. J Biol Chem. 1986 Nov 25;261(33):15761–15766. [PubMed] [Google Scholar]
- Steinum A. L., Seeberg E. Nucleotide sequence of the tag gene from Escherichia coli. Nucleic Acids Res. 1986 May 12;14(9):3763–3772. doi: 10.1093/nar/14.9.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tano K., Shiota S., Collier J., Foote R. S., Mitra S. Isolation and structural characterization of a cDNA clone encoding the human DNA repair protein for O6-alkylguanine. Proc Natl Acad Sci U S A. 1990 Jan;87(2):686–690. doi: 10.1073/pnas.87.2.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney U., Hutcheon T., van de Sande J. H. Sequence analysis, expression, and conservation of Escherichia coli uracil DNA glycosylase and its gene (ung). J Biol Chem. 1988 Jun 5;263(16):7776–7784. [PubMed] [Google Scholar]
- Vollberg T. M., Siegler K. M., Cool B. L., Sirover M. A. Isolation and characterization of the human uracil DNA glycosylase gene. Proc Natl Acad Sci U S A. 1989 Nov;86(22):8693–8697. doi: 10.1073/pnas.86.22.8693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrad D. M., Caradonna S. Identification of the coding sequence for herpes simplex virus uracil-DNA glycosylase. J Virol. 1988 Dec;62(12):4774–4777. doi: 10.1128/jvi.62.12.4774-4777.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y., Katsuki M., Sekiguchi M., Otsuji N. Escherichia coli gene that controls sensitivity to alkylating agents. J Bacteriol. 1978 Jul;135(1):144–152. doi: 10.1128/jb.135.1.144-152.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y., Sekiguchi M. Pathways for repair of DNA damaged by alkylating agent in Escherichia coli. Mol Gen Genet. 1979 Mar 27;171(3):251–256. doi: 10.1007/BF00267579. [DOI] [PubMed] [Google Scholar]
- Yeung C. Y., Ingolia D. E., Roth D. B., Shoemaker C., Al-Ubaidi M. R., Yen J. Y., Ching C., Bobonis C., Kaufman R. J., Kellems R. E. Identification of functional murine adenosine deaminase cDNA clones by complementation in Escherichia coli. J Biol Chem. 1985 Aug 25;260(18):10299–10307. [PubMed] [Google Scholar]
- van Duin M., van den Tol J., Warmerdam P., Odijk H., Meijer D., Westerveld A., Bootsma D., Hoeijmakers J. H. Evolution and mutagenesis of the mammalian excision repair gene ERCC-1. Nucleic Acids Res. 1988 Jun 24;16(12):5305–5322. doi: 10.1093/nar/16.12.5305. [DOI] [PMC free article] [PubMed] [Google Scholar]




