Abstract
The EcoRI restriction endonuclease cleaves DNA molecules at the sequence GAATTC. We devised a genetic screen to isolate EcoRI mutants with altered or broadened substrate specificity. In vitro, the purified mutant enzymes cleave both the wild-type substrate and sites which differ from this by one nucleotide (EcoRI star sites). These mutations identify four residues involved in substrate recognition and catalysis that are different from the amino acids proposed to recognize the substrate based on the EcoRI-DNA co-crystal structure. In fact, these mutations suppress EcoRI mutants altered at some of the proposed substrate binding residues (R145, R200). We argue that these mutations permit cleavage of additional DNA sequences either by perturbing or removing direct DNA-protein interactions or by facilitating conformational changes that allosterically couple substrate binding to DNA scission.
Full text
PDF

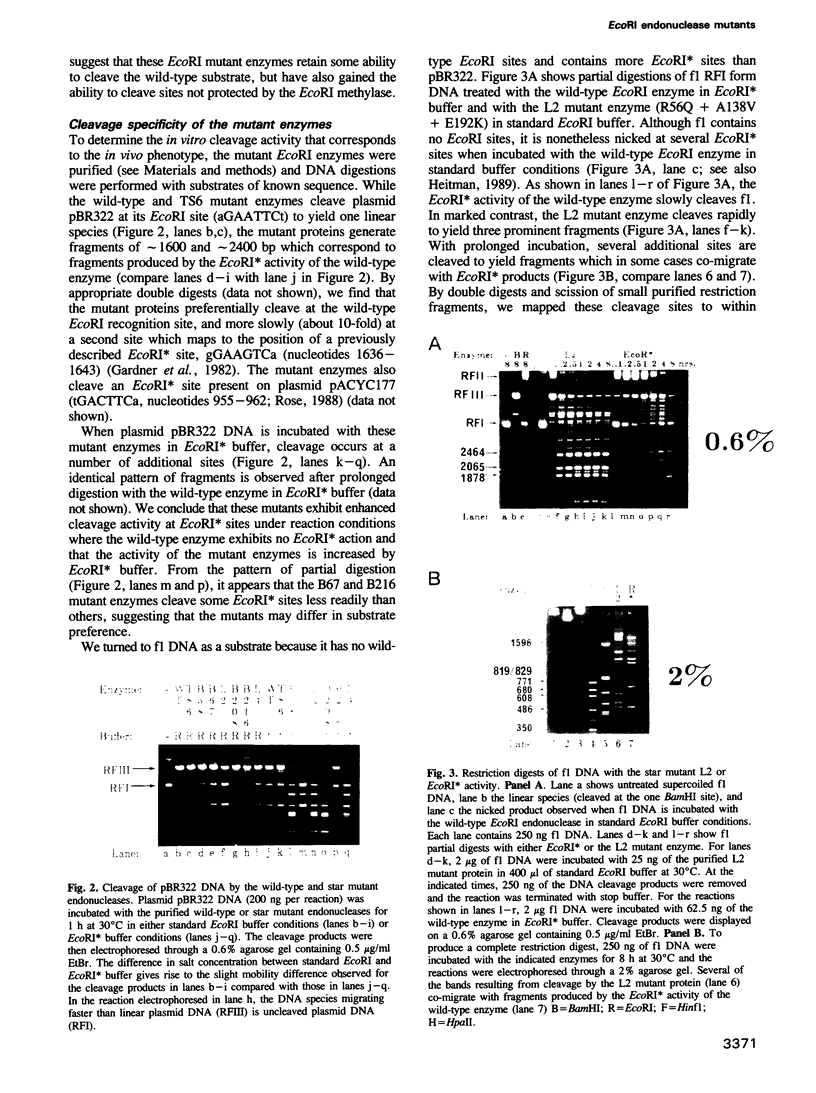


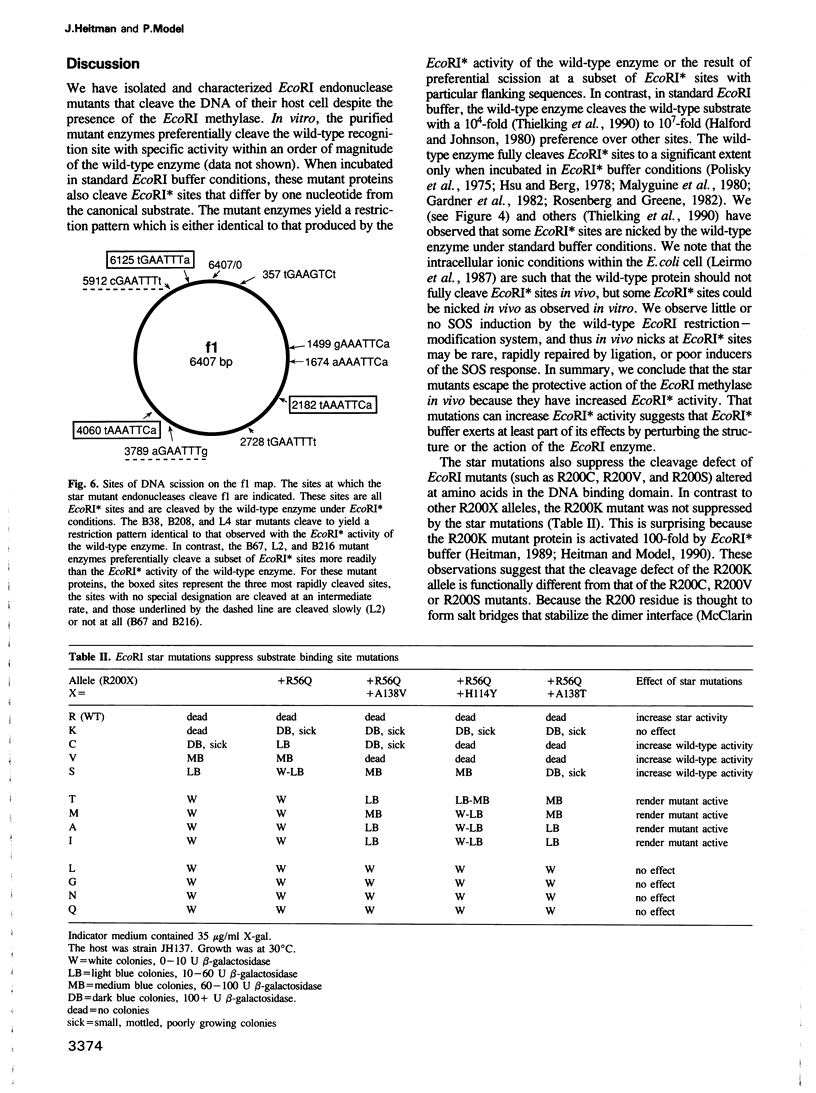




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiken C., Gumport R. I. Restriction endonuclease RsrI from Rhodobacter sphaeroides, an isoschizomer of EcoRI: purification and properties. Nucleic Acids Res. 1988 Aug 25;16(16):7901–7916. doi: 10.1093/nar/16.16.7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves J., Rüter T., Geiger R., Fliess A., Maass G., Pingoud A. Changing the hydrogen-bonding potential in the DNA binding site of EcoRI by site-directed mutagenesis drastically reduces the enzymatic activity, not, however, the preference of this restriction endonuclease for cleavage within the site-GAATTC-. Biochemistry. 1989 Mar 21;28(6):2678–2684. doi: 10.1021/bi00432a047. [DOI] [PubMed] [Google Scholar]
- Barany F. The TaqI 'star' reaction: strand preferences reveal hydrogen-bond donor and acceptor sites in canonical sequence recognition. Gene. 1988 May 30;65(2):149–165. doi: 10.1016/0378-1119(88)90452-0. [DOI] [PubMed] [Google Scholar]
- Becker M. M., Lesser D., Kurpiewski M., Baranger A., Jen-Jacobson L. "Ultraviolet footprinting" accurately maps sequence-specific contacts and DNA kinking in the EcoRI endonuclease-DNA complex. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6247–6251. doi: 10.1073/pnas.85.17.6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkner K. L., Folk W. R. Overmethylation of DNAs by the EcoRI methylase. Nucleic Acids Res. 1978 Feb;5(2):435–450. doi: 10.1093/nar/5.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C. A., Van Cleve M. D., Gumport R. I. The effects of base analogue substitutions on the cleavage by the EcoRI restriction endonuclease of octadeoxyribonucleotides containing modified EcoRI recognition sequences. J Biol Chem. 1986 Jun 5;261(16):7270–7278. [PubMed] [Google Scholar]
- Carter P., Wells J. A. Dissecting the catalytic triad of a serine protease. Nature. 1988 Apr 7;332(6164):564–568. doi: 10.1038/332564a0. [DOI] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Cheng S. C., Kim R., King K., Kim S. H., Modrich P. Isolation of gram quantities of EcoRI restriction and modification enzymes from an overproducing strain. J Biol Chem. 1984 Sep 25;259(18):11571–11575. [PubMed] [Google Scholar]
- Cheng S. C., Modrich P. Positive-selection cloning vehicle useful for overproduction of hybrid proteins. J Bacteriol. 1983 May;154(2):1005–1008. doi: 10.1128/jb.154.2.1005-1008.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox E. C., Horner D. L. Dominant mutators in Escherichia coli. Genetics. 1982 Jan;100(1):7–18. doi: 10.1093/genetics/100.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson R. E. Base sequence and helix structure variation in B and A DNA. J Mol Biol. 1983 May 25;166(3):419–441. doi: 10.1016/s0022-2836(83)80093-x. [DOI] [PubMed] [Google Scholar]
- Diekmann S., McLaughlin L. W. DNA curvature in native and modified EcoRI recognition sites and possible influence upon the endonuclease cleavage reaction. J Mol Biol. 1988 Aug 20;202(4):823–834. doi: 10.1016/0022-2836(88)90561-x. [DOI] [PubMed] [Google Scholar]
- Estell D. A., Graycar T. P., Miller J. V., Powers D. B., Wells J. A., Burnier J. P., Ng P. G. Probing steric and hydrophobic effects on enzyme-substrate interactions by protein engineering. Science. 1986 Aug 8;233(4764):659–663. doi: 10.1126/science.233.4764.659. [DOI] [PubMed] [Google Scholar]
- Evans R. M., Hollenberg S. M. Zinc fingers: gilt by association. Cell. 1988 Jan 15;52(1):1–3. doi: 10.1016/0092-8674(88)90522-3. [DOI] [PubMed] [Google Scholar]
- Frederick C. A., Grable J., Melia M., Samudzi C., Jen-Jacobson L., Wang B. C., Greene P., Boyer H. W., Rosenberg J. M. Kinked DNA in crystalline complex with EcoRI endonuclease. Nature. 1984 May 24;309(5966):327–331. doi: 10.1038/309327a0. [DOI] [PubMed] [Google Scholar]
- Gardner R. C., Howarth A. J., Messing J., Shepherd R. J. Cloning and sequencing of restriction fragments generated by Eco RI*. DNA. 1982;1(2):109–115. doi: 10.1089/dna.1.1982.1.109. [DOI] [PubMed] [Google Scholar]
- Garges S., Adhya S. Sites of allosteric shift in the structure of the cyclic AMP receptor protein. Cell. 1985 Jul;41(3):745–751. doi: 10.1016/s0092-8674(85)80055-6. [DOI] [PubMed] [Google Scholar]
- Geiger R., Rüter T., Alves J., Fliess A., Wolfes H., Pingoud V., Urbanke C., Maass G., Pingoud A., Düsterhöft A. Genetic engineering of EcoRI mutants with altered amino acid residues in the DNA binding site: physicochemical investigations give evidence for an altered monomer/dimer equilibrium for the Gln144Lys145 and Gln144Lys145Lys200 mutants. Biochemistry. 1989 Mar 21;28(6):2667–2677. doi: 10.1021/bi00432a046. [DOI] [PubMed] [Google Scholar]
- George J., Chirikjian J. G. Sequence-specific endonuclease BamHI: relaxation of sequence recognition. Proc Natl Acad Sci U S A. 1982 Apr;79(8):2432–2436. doi: 10.1073/pnas.79.8.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene P. J., Gupta M., Boyer H. W., Brown W. E., Rosenberg J. M. Sequence analysis of the DNA encoding the Eco RI endonuclease and methylase. J Biol Chem. 1981 Mar 10;256(5):2143–2153. [PubMed] [Google Scholar]
- Ha J. H., Spolar R. S., Record M. T., Jr Role of the hydrophobic effect in stability of site-specific protein-DNA complexes. J Mol Biol. 1989 Oct 20;209(4):801–816. doi: 10.1016/0022-2836(89)90608-6. [DOI] [PubMed] [Google Scholar]
- Halford S. E., Johnson N. P. The EcoRI restriction endonuclease with bacteriophage lambda DNA. Equilibrium binding studies. Biochem J. 1980 Nov 1;191(2):593–604. doi: 10.1042/bj1910593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgpeth J., Goodman H. M., Boyer H. W. DNA nucleotide sequence restricted by the RI endonuclease. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3448–3452. doi: 10.1073/pnas.69.11.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J., Fulford W., Model P. Phage Trojan horses: a conditional expression system for lethal genes. Gene. 1989 Dec 21;85(1):193–197. doi: 10.1016/0378-1119(89)90480-0. [DOI] [PubMed] [Google Scholar]
- Heitman J., Model P. Site-specific methylases induce the SOS DNA repair response in Escherichia coli. J Bacteriol. 1987 Jul;169(7):3243–3250. doi: 10.1128/jb.169.7.3243-3250.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J., Model P. Substrate recognition by the EcoRI endonuclease. Proteins. 1990;7(2):185–197. doi: 10.1002/prot.340070207. [DOI] [PubMed] [Google Scholar]
- Heitman J., Treisman J., Davis N. G., Russel M. Cassettes of the f1 intergenic region. Nucleic Acids Res. 1989 Jun 12;17(11):4413–4413. doi: 10.1093/nar/17.11.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J., Zinder N. D., Model P. Repair of the Escherichia coli chromosome after in vivo scission by the EcoRI endonuclease. Proc Natl Acad Sci U S A. 1989 Apr;86(7):2281–2285. doi: 10.1073/pnas.86.7.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill D. F., Petersen G. B. Nucleotide sequence of bacteriophage f1 DNA. J Virol. 1982 Oct;44(1):32–46. doi: 10.1128/jvi.44.1.32-46.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu M., Berg P. Altering the specificity of restriction endonuclease: effect of replacing Mg2+ with Mn2+. Biochemistry. 1978 Jan 10;17(1):131–138. doi: 10.1021/bi00594a019. [DOI] [PubMed] [Google Scholar]
- Huo L., Martin K. J., Schleif R. Alternative DNA loops regulate the arabinose operon in Escherichia coli. Proc Natl Acad Sci U S A. 1988 Aug;85(15):5444–5448. doi: 10.1073/pnas.85.15.5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack W. E., Terry B. J., Modrich P. Involvement of outside DNA sequences in the major kinetic path by which EcoRI endonuclease locates and leaves its recognition sequence. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4010–4014. doi: 10.1073/pnas.79.13.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jen-Jacobson L., Kurpiewski M., Lesser D., Grable J., Boyer H. W., Rosenberg J. M., Greene P. J. Coordinate ion pair formation between EcoRI endonuclease and DNA. J Biol Chem. 1983 Dec 10;258(23):14638–14646. [PubMed] [Google Scholar]
- Jen-Jacobson L., Lesser D., Kurpiewski M. The enfolding arms of EcoRI endonuclease: role in DNA binding and cleavage. Cell. 1986 May 23;45(4):619–629. doi: 10.1016/0092-8674(86)90294-1. [DOI] [PubMed] [Google Scholar]
- Kenyon C. J., Walker G. C. DNA-damaging agents stimulate gene expression at specific loci in Escherichia coli. Proc Natl Acad Sci U S A. 1980 May;77(5):2819–2823. doi: 10.1073/pnas.77.5.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King K., Benkovic S. J., Modrich P. Glu-111 is required for activation of the DNA cleavage center of EcoRI endonuclease. J Biol Chem. 1989 Jul 15;264(20):11807–11815. [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988 Jun 24;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Leirmo S., Harrison C., Cayley D. S., Burgess R. R., Record M. T., Jr Replacement of potassium chloride by potassium glutamate dramatically enhances protein-DNA interactions in vitro. Biochemistry. 1987 Apr 21;26(8):2095–2101. doi: 10.1021/bi00382a006. [DOI] [PubMed] [Google Scholar]
- Malyguine E., Vannier P., Yot P. Alteration of the specificity of restriction endonucleases in the presence of organic solvents. Gene. 1980 Jan;8(2):163–177. doi: 10.1016/0378-1119(80)90035-9. [DOI] [PubMed] [Google Scholar]
- Martin K., Huo L., Schleif R. F. The DNA loop model for ara repression: AraC protein occupies the proposed loop sites in vivo and repression-negative mutations lie in these same sites. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3654–3658. doi: 10.1073/pnas.83.11.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClarin J. A., Frederick C. A., Wang B. C., Greene P., Boyer H. W., Grable J., Rosenberg J. M. Structure of the DNA-Eco RI endonuclease recognition complex at 3 A resolution. Science. 1986 Dec 19;234(4783):1526–1541. doi: 10.1126/science.3024321. [DOI] [PubMed] [Google Scholar]
- McLaughlin L. W., Benseler F., Graeser E., Piel N., Scholtissek S. Effects of functional group changes in the EcoRI recognition site on the cleavage reaction catalyzed by the endonuclease. Biochemistry. 1987 Nov 17;26(23):7238–7245. doi: 10.1021/bi00397a007. [DOI] [PubMed] [Google Scholar]
- Miller J., McLachlan A. D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989 Mar 10;56(5):777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Nasri M., Thomas D. Relaxation of recognition sequence of specific endonuclease HindIII. Nucleic Acids Res. 1986 Jan 24;14(2):811–821. doi: 10.1093/nar/14.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needels M. C., Fried S. R., Love R., Rosenberg J. M., Boyer H. W., Greene P. J. Determinants of EcoRI endonuclease sequence discrimination. Proc Natl Acad Sci U S A. 1989 May;86(10):3579–3583. doi: 10.1073/pnas.86.10.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A. K., Rubin R. A., Kim S. H., Modrich P. DNA sequences of structural genes for Eco RI DNA restriction and modification enzymes. J Biol Chem. 1981 Mar 10;256(5):2131–2139. [PubMed] [Google Scholar]
- Otwinowski Z., Schevitz R. W., Zhang R. G., Lawson C. L., Joachimiak A., Marmorstein R. Q., Luisi B. F., Sigler P. B. Crystal structure of trp repressor/operator complex at atomic resolution. Nature. 1988 Sep 22;335(6188):321–329. doi: 10.1038/335321a0. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Polisky B., Greene P., Garfin D. E., McCarthy B. J., Goodman H. M., Boyer H. W. Specificity of substrate recognition by the EcoRI restriction endonuclease. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3310–3314. doi: 10.1073/pnas.72.9.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J. Restriction endonucleases. CRC Crit Rev Biochem. 1976 Nov;4(2):123–164. doi: 10.3109/10409237609105456. [DOI] [PubMed] [Google Scholar]
- Rose R. E. The nucleotide sequence of pACYC177. Nucleic Acids Res. 1988 Jan 11;16(1):356–356. doi: 10.1093/nar/16.1.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg J. M., Greene P. Eco RI* specificity and hydrogen bonding. DNA. 1982;1(2):117–124. doi: 10.1089/dna.1.1982.1.117. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortle D. A genetic system for analysis of staphylococcal nuclease. Gene. 1983 May-Jun;22(2-3):181–189. doi: 10.1016/0378-1119(83)90102-6. [DOI] [PubMed] [Google Scholar]
- Stephenson F. H., Ballard B. T., Boyer H. W., Rosenberg J. M., Greene P. J. Comparison of the nucleotide and amino acid sequences of the RsrI and EcoRI restriction endonucleases. Gene. 1989 Dec 21;85(1):1–13. doi: 10.1016/0378-1119(89)90458-7. [DOI] [PubMed] [Google Scholar]
- Tasseron-de Jong J. G., Aker J., Giphart-Gassler M. The ability of the restriction endonuclease EcoRI to digest hemi-methylated versus fully cytosine-methylated DNA of the herpes tk promoter region. Gene. 1988 Dec 25;74(1):147–149. doi: 10.1016/0378-1119(88)90272-7. [DOI] [PubMed] [Google Scholar]
- Terry B. J., Jack W. E., Modrich P. Facilitated diffusion during catalysis by EcoRI endonuclease. Nonspecific interactions in EcoRI catalysis. J Biol Chem. 1985 Oct 25;260(24):13130–13137. [PubMed] [Google Scholar]
- Thielking V., Alves J., Fliess A., Maass G., Pingoud A. Accuracy of the EcoRI restriction endonuclease: binding and cleavage studies with oligodeoxynucleotide substrates containing degenerate recognition sequences. Biochemistry. 1990 May 15;29(19):4682–4691. doi: 10.1021/bi00471a024. [DOI] [PubMed] [Google Scholar]
- Thomas M., Davis R. W. Studies on the cleavage of bacteriophage lambda DNA with EcoRI Restriction endonuclease. J Mol Biol. 1975 Jan 25;91(3):315–328. doi: 10.1016/0022-2836(75)90383-6. [DOI] [PubMed] [Google Scholar]
- Vershon A. K., Bowie J. U., Karplus T. M., Sauer R. T. Isolation and analysis of arc repressor mutants: evidence for an unusual mechanism of DNA binding. Proteins. 1986 Dec;1(4):302–311. doi: 10.1002/prot.340010404. [DOI] [PubMed] [Google Scholar]
- Vinson C. R., Sigler P. B., McKnight S. L. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989 Nov 17;246(4932):911–916. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]
- Wharton R. P., Ptashne M. A new-specificity mutant of 434 repressor that defines an amino acid-base pair contact. 1987 Apr 30-May 6Nature. 326(6116):888–891. doi: 10.1038/326888a0. [DOI] [PubMed] [Google Scholar]
- Wolfes H., Alves J., Fliess A., Geiger R., Pingoud A. Site directed mutagenesis experiments suggest that Glu 111, Glu 144 and Arg 145 are essential for endonucleolytic activity of EcoRI. Nucleic Acids Res. 1986 Nov 25;14(22):9063–9080. doi: 10.1093/nar/14.22.9063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodbury C. P., Jr, Downey R. L., von Hippel P. H. DNA site recognition and overmethylation by the Eco RI methylase. J Biol Chem. 1980 Dec 10;255(23):11526–11533. [PubMed] [Google Scholar]
- Woodbury C. P., Jr, Hagenbüchle O., von Hippel P. H. DNA site recognition and reduced specificity of the Eco RI endonuclease. J Biol Chem. 1980 Dec 10;255(23):11534–11548. [PubMed] [Google Scholar]
- Wright D. J., King K., Modrich P. The negative charge of Glu-111 is required to activate the cleavage center of EcoRI endonuclease. J Biol Chem. 1989 Jul 15;264(20):11816–11821. [PubMed] [Google Scholar]
- Yanofsky S. D., Love R., McClarin J. A., Rosenberg J. M., Boyer H. W., Greene P. J. Clustering of null mutations in the EcoRI endonuclease. Proteins. 1987;2(4):273–282. doi: 10.1002/prot.340020403. [DOI] [PubMed] [Google Scholar]