Abstract
The opportunistic pathogen Candida albicans forms invasive filaments that grow into host tissues during disease. The glycosylated, integral plasma membrane protein Dfi1 is important for invasive filamentation in a laboratory model, and for lethality in murine disseminated candidiasis. However, Dfi1 topology and essential domains for Dfi1 biogenesis were undefined. Sequence analysis predicted that Dfi1 contains two transmembrane regions, located near the N- and C-termini. In this communication, we show that Dfi1 remains an integral membrane protein despite deletion of either predicted transmembrane region, whereas deletion of both regions results in a soluble protein. Additionally, Dfi1 that was properly oriented in the membrane, as indicated by N-linked glycosylation, was observed when either transmembrane region was deleted, but was absent when both transmembrane regions were deleted. Interestingly, deletion of the N-terminal transmembrane region resulted in production of two forms of Dfi1. Most of the protein molecules acquired normal N-linked glycosylation and a smaller population failed to become normally N-linked glycosylated. This defect was reversed by replacement of the N-terminal hydrophobic sequence with one synthetic transmembrane sequence but not another. Finally, microscopy studies revealed that Dfi1 lacking the N-terminal transmembrane region was observed at the cell periphery, where full-length Dfi1 normally localizes, whereas the double-truncation mutant was diffusely intracellular. Therefore, mature Dfi1 protein contains two transmembrane domains which contribute to its biogenesis.
Keywords: Candida albicans, invasive filamentation, Dfi1, topology, biogenesis, N-linked glycosylation
Introduction
Candida albicans is an opportunistic fungal pathogen that is a common commensal of the human gastrointestinal tract [1–3], but, in immunocompromised hosts, can cause disease [4]. Filamentation, growth as elongated filamentous cells, is a C. albicans virulence trait [5]. Many stimuli promote filamentation, including the embedding of cells within a semi-solid agar matrix [5]. Embedded filamentation may be related to tissue invasion during candidiasis, and the C. albicans protein Dfi1 is a key mediator of embedded filamentation [6]. Dfi1 is an integral plasma membrane protein that undergoes both O- and N-linked glycosylation [6]. The Dfi1 C-terminus contains motifs that are important for Dfi1 function [6,7], but the role of the Dfi1 N-terminus remains relatively unexplored.
Sequence analysis of the DFI1 gene reveals that the protein contains two predicted hydrophobic regions, each close to one of the protein termini, and Dfi1 was previously predicted to contain two transmembrane domains [8]. Furthermore, the five predicted canonical N-linked glycosylation motifs [9] of Dfi1 are all between the two hydrophobic regions. The central region of Dfi1 is thus predicted to be extracellular, and the N-terminal hydrophobic region is either the first transmembrane domain of a multispanning integral membrane protein or the cleaved signal sequence of a type I transmembrane protein.
In this communication, we analyzed the topology of Dfi1 and the role of the two Dfi1 hydrophobic regions, demonstrating that both regions contribute to membrane integration and are retained in the mature protein.
Materials and Methods
Strains and growth conditions
Strains are listed in Table S1. Escherichia coli strain DH5α was grown in LB broth plus antibiotic to propagate plasmids. C. albicans was grown in YPD (1% yeast extract, 2% peptone, 2% glucose) or CM-ura (complete medium minus uridine [10]) liquid media. Nourseothricin (Werner BioAgents) was used at 200 μg/mL.
Construction of strains
Mutant DFI1 alleles were constructed by overlap PCR with mutagenic primers using pSPLDFI1-TAP [6,7] as template. Primers are listed in Table S2.
For DFI1ΔC, the first round primers were pz290/pz63 and pz134/pz291, and the second round primers were pz134/pz63. This fragment was digested with XhoI and MluI and moved into pSPLDFI1-HA [6] (DFI1-HA in the SAT placer) or pSPLDFI1-GFP [6] (DFI1-GFP in the SAT placer) to generate pSPLDFI1ΔC-HA or pSPLDFI1ΔC-GFP plasmids.
For DFI1ΔN and DFI1ΔNΔC, the first round primers were SH01/SH20 and SH22/SH08, and the second round primers were SH01/SH08. For DFI1N-TM replacement C-HA, first-round primer pairs were SH37/SH08 and SH01/SH39, the SH37/SH08 product was amplified by SH38/SH08, then the SH38/SH08 and SH01/SH39 products were amplified by SH01/SH08. For DFI1N-TM replacement Z-HA, first-round primer pairs were SH40/SH08 and SH01/SH42, the SH40/SH08 product was amplified by SH41/SH08, then the SH41/SH08 and SH01/SH42 products were amplified by SH01/SH08. Fragments were digested with BglII and XhoI and ligated into pSPLDFI1-HA [6], pSPLDFI1ΔC-HA, pSPLDFI1-GFP [6], or pSPLDFI1ΔC-GFP. Inserts were confirmed by DNA sequencing. Plasmids were linearized with BglII and transformed into the dfi1Δ/Δ strain pcz5 [6].
Analyses of membrane association
Experiments were performed as previously described [6], with the following modifications: Extraction buffer (EB) was supplemented with 1 mM PMSF and 1% Fungal Specific Protease Inhibitor (v/v; FSPI; Sigma). After lysis by beadbeating, extracts were centrifuged twice at 16,100g for 2 min at 4°C, then centrifuged at 300,000g for 20 min at 4°C. The resulting pellet was homogenized in the original volume of EB with 1% SDS, or underwent extraction with equal volumes of either EB alone, EB with 4M urea (final concentration), or EB with 5% Triton X-100 (final concentration; v/v). Samples were incubated on ice for 30 min, homogenized, and centrifuged at 300,000g for 20 min. The resulting pellet was homogenized in the same volume of EB supplemented with 1% SDS. An SDS polyacrylamide gel was loaded with 50 μL of each sample (except the DF1ΔC pellet extraction samples, which used 12.5 μL to avoid excessive signal).
Western blotting
Samples were run on an 8% SDS-PAGE gel, and transferred to PVDF (BioRad). Blots were blocked, incubated overnight with 1:1000 primary antibody, washed and incubated with 1:4000 secondary antibody. HA was detected with Purified anti-HA.11 Epitope Tag antibody (16B12, BioLegend) and Goat Anti-Mouse HRP Conjugate antibody (cat# 170-5047, BioRad). Tubulin was detected with Rat Anti-Tubulin Alpha antibody (MCA78G, BioRad) and HRP-Goat Anti-Rat IgG(H+L) (cat# 62-9520, Invitrogen). Blots were developed using Pierce ECL Western Blotting Substrate on a G:BOX developer (Syngene). Each experiment was performed at least three times.
PNGase F treatment
Crude cell lysates were made as described above, except cell pellets were resuspended in 880 μL EB supplemented with 1 mM PMSF and 1% FSPI (v/v). Samples were incubated as described by the manufacturer, with or without 750U PNGase F (NEB), for 4 hr at 30°C. Samples were assessed by western blotting for HA.
GFP localization
Exponential phase cells grown in CM-ura at 30°C were pelleted at 6000g for 30 seconds and resuspended in PBS. 15 μL of cell suspension was placed onto an agarose pad. Images were taken on a Leica SP8 confocal microscope with a 63x objective, controlled by the LAS X program. Z-stacks were reconstructed into 3D and analyzed using Volocity software (Perkin Elmer).
Results
Mature Dfi1 contains two transmembrane domains
To analyze Dfi1 topology, strains encoding HA tagged full length Dfi1 or Dfi1 lacking the N-terminal hydrophobic region, lacking the C-terminal hydrophobic region, or lacking both regions were employed (Figure 1A). Cells of these four strains were extracted as described in the Materials and Methods. Extracts were centrifuged at 300,000g for 20 min to pellet membrane fractions and other large subcellular material. The resulting supernatants and pellets, and the original crude lysates, were examined via Western blotting for the presence of HA-tagged Dfi1.
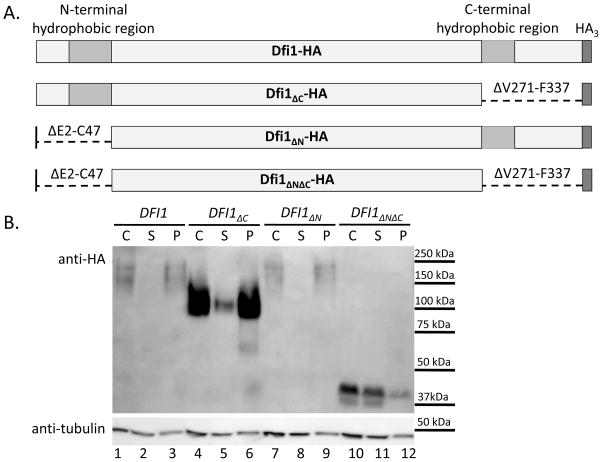
Figure 1. Either hydrophobic region is sufficient for membrane association of Dfi1.
Panel A. Diagrams of Dfi1 protein variants. Diagrams show full-length Dfi1 (Dfi1-HA), Dfi1 lacking the C-terminal hydrophobic region (Dfi1ΔC-HA), Dfi1 lacking the N-terminal hydrophobic region (Dfi1ΔN-HA), or Dfi1 in which the both hydrophobic regions were deleted (Dfi1ΔNΔC-HA). Diagrams are not to scale.
Panel B. Membrane association. Cell lysates of C. albicans strains encoding HA-tagged full-length Dfi1, Dfi1ΔC, Dfi1ΔN, or Dfi1ΔNΔC were made as described in the Materials and Methods. Lysates underwent ultracentrifugation at 300,000g for 20 min to pellet cell membranes. Equal volumes of the crude lysate before ultracentrifugation (C), and the supernatant (S) and pellet (P) resulting from ultracentrifugation, were loaded into each lane, and the resulting membrane was probed for HA (top image) and tubulin (bottom image). This blot is representative of three independent experiments.
Full-length HA-tagged Dfi1 (DFI1) is predicted to encode a 36.5 kDa, glycosylated protein [6], but, as seen in previous studies [6], migrates at a wide range of apparent molecular weights centered around approximately 200 kDa (Figure 1B). As observed previously [6], full-length Dfi1 was observed in the crude lysate and the pellet (Figure 1B, lanes 1 and 3). The vast majority of Dfi1 mutant protein lacking the C-terminal hydrophobic region (Dfi1ΔC) fractionated into the membrane-containing pellet (Figure 1B, lane 6), with a small amount in the supernatant (Figure 1B, lane 5). The Dfi1ΔC protein migrated at a lower apparent molecular weight, approximately 100 kDa, and formed a narrower band (Figure 2, lanes 13–18). Dfi1ΔC protein (Figure 1B, lane 4) was present at higher levels than wild-type protein (Figure 1B, lane 1), indicated by the tubulin loading control.
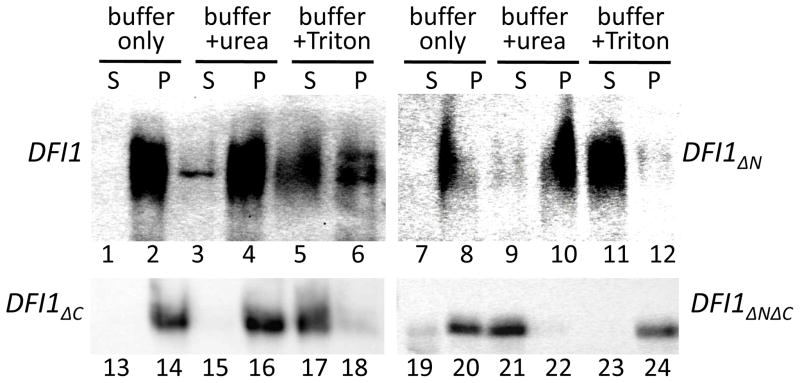
Figure 2. Dfi1 is an integral membrane protein when at least one hydrophobic region is present.
Cell lysates of C. albicans strains encoding HA-tagged full-length Dfi1, Dfi1ΔN, Dfi1ΔC, or Dfi1ΔNΔC underwent ultracentrifugation to pellet cell membranes at 300,000g for 20 min, as described in the Materials and Methods. The pellets were then homogenized with extraction buffer alone, buffer plus 4M urea (final concentration), or buffer plus 5% Triton X-100 (final concentration; v/v). These homogenates then underwent a second round of ultracentrifugation and the resulting supernatants (S) and pellets (P) were probed for HA by Western blotting. The lanes from the DF1ΔC strain were each loaded with 12.5 μL of sample, to avoid excessive signal, and all other lanes were each loaded with 50 μL of sample. Each blot is representative of at least three experiments.
Dfi1 mutant protein lacking the N-terminal hydrophobic region (Dfi1ΔN) was detected in similar quantities to full-length Dfi1, and migrated within the same size range. Dfi1ΔN protein was detected in the membrane-containing pellet (Figure 1B, lane 9). Therefore, Dfi1 protein containing either one of the two hydrophobic regions localized almost exclusively to the membrane-containing pellet fraction. Finally, Dfi1 mutant protein lacking both the N-terminal and C-terminal hydrophobic regions (Dfi1ΔNΔC) migrated as a narrow band around 40 kDa, and fractionated to the supernatant (Figure 1B, lane 11), not the membrane-containing pellet (Figure 1B, lane 12). Thus, the Dfi1ΔNΔC mutant lost its membrane association. Therefore, the membrane association characteristic of Dfi1 requires at least one of its two hydrophobic regions.
To determine whether these truncated Dfi1 proteins were integral membrane proteins, membrane-containing pellets were extracted with buffer alone, buffer containing urea, or buffer containing Triton X-100. After extraction, the samples were centrifuged at 300,000g for 20 min and the supernatants and pellets were probed for Dfi1 protein. Consistent with a previous report [6], the majority of full-length Dfi1 (DFI1) was extracted by Triton X-100 (Figure 2, lanes 5 and 6), but not by urea (Figure 2, lanes 3 and 4) or buffer alone (Figure 2, lanes 1 and 2).
The Dfi1 mutant proteins lacking either the N-terminal or C-terminal hydrophobic region (DFI1ΔN or DFI1ΔC, respectively) were also extracted by Triton X-100 (Figure 2, lanes 11, 12, 17, and 18), but not by urea (Figure 2, lanes 9, 10, 15, and 16) or buffer alone (Figure 2, lanes 7, 8, 13, and 14). Therefore, the single-truncation Dfi1 proteins represented integral membrane proteins. Finally, doubly-truncated Dfi1 protein (DFI1ΔNΔC) yielded a small amount of protein that pelleted (Figure 1B, lane 12), and this protein was extracted with urea (Figure 2, lanes 21 and 22), but not with Triton X-100 (Figure 2, lanes 23 and 24). This protein likely represented aggregated protein.
To summarize, at least one of the two hydrophobic regions was required, and either was sufficient, for Dfi1 to behave as an integral membrane protein. Therefore, mature Dfi1 protein contains two transmembrane domains, each represented by one of these two hydrophobic regions.
Normal Dfi1 membrane orientation but abnormal glycosylation patterns resulted from deletion of either transmembrane domain
The roles of the two Dfi1 transmembrane domains in Dfi1 biogenesis were examined, first focusing on Dfi1 membrane orientation and glycan maturation. The five canonical N-linked glycosylation motifs [9] of Dfi1 are located between the two transmembrane domains and preserved in each Dfi1 mutant. The N-linked glycosylation patterns of full-length Dfi1 and the three truncation mutants (Dfi1ΔC, Dfi1ΔN, and Dfi1ΔNΔC) were analyzed to determine whether this central region of the protein was exposed to the lumen of the endoplasmic reticulum, where N-linked glycosylation initiates during translation [11,12]. N-linked glycans are further processed through the endoplasmic reticulum (ER) and Golgi, but glycoproteins must be correctly folded to exit the ER [12]. As such, the presence and pattern of N-linked glycosylation of the Dfi1 mutants will indicate the proteins’ orientation in the membrane and glycan maturation, respectively.
N-linked glycosylation of Dfi1 can be detected as a shift in apparent molecular weight following treatment with peptide: N-glycosidase F (PNGase F), an endoglycosidase that removes N-linked oligosaccharides by cleaving at the sugar-asparagine junction [6,13]. Therefore, crude cellular lysates from strains encoding one of the four versions of Dfi1 (HA-tagged full-length DFI1, DFI1ΔC, DFI1ΔN and DFI1ΔNΔC) underwent treatment or sham-treatment with PNGase F and were examined via Western blotting for HA-tagged Dfi1.
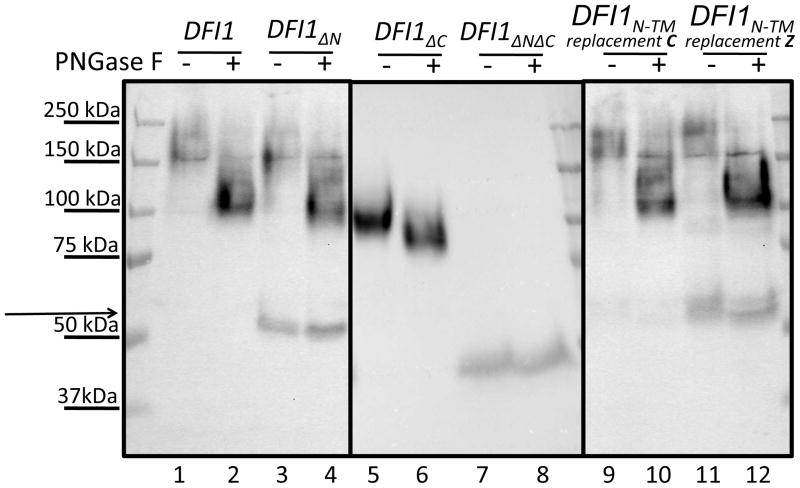
Full-length Dfi1 (DFI1) showed a large shift in mobility upon PNGase F treatment (Figure 3, lanes 1 and 2), as previously reported [6]. Dfi1ΔN yielded a large, wide band migrating in the 200 kDa region that shifted mobility following PNGase F treatment (Figure 3, lanes 3 and 4). A second, narrow band migrating at 55 kDa (arrow in Figure 3, lane 3) was detected when longer exposures were used. This species of Dfi1ΔN did not undergo a large change in apparent molecular weight upon PNGase F treatment (Figure 3, lanes 3 and 4). Therefore, the major species of Dfi1ΔN was N-linked glycosylated and had achieved the native membrane orientation, while the minor species was aberrant. The smaller Dfi1ΔN species may represent protein with non-native membrane orientation causing mislocalization of the glycosylation motifs, improperly-folded glycoprotein that failed to mature, or proteolytically-cleaved protein.
Figure 3. N-linked glycosylation is abnormal in some truncated Dfi1 proteins.
Crude lysates of C. albicans encoding HA-tagged full-length Dfi1, Dfi1ΔC, Dfi1ΔN, or Dfi1ΔNΔC were either mock-treated (−) or treated (+) with PNGase F, and these samples were probed for HA by Western blotting, as described in the Materials and Methods. Arrow indicates a faster-migrating species. C. albicans strains encoding DFI1N-TM replacement C or DFI1N-TM replacement Z underwent identical treatment. The lanes from the DF11ΔC strain were each loaded with 11uL of sample, and all other lanes were each loaded with 45 μL of sample. Each blot is representative of at least three experiments.
Dfi1 mutant protein lacking the C-terminal transmembrane domain (Dfi1ΔC) showed an appreciable decrease in apparent molecular weight upon PNGase F treatment (Figure 3, lanes 5 and 6), indicating normal membrane orientation. Notably, the decrease in apparent molecular weight upon PNGase treatment was less substantial for Dfi1ΔC than for full-length Dfi1. This indicates that the N-linked glycan of Dfi1ΔC was present but abnormal, with the Dfi1ΔC glycoprotein achieving normal membrane orientation but not maturing normally thereafter. Finally, the Dfi1 mutant protein lacking both the N- and C-terminal transmembrane domains (Dfi1ΔNΔC) showed no appreciable shift in molecular weight upon PNGase F treatment, indicating that this polypeptide is not N-linked glycosylated. This lack of N-linked glycosylation may occur because the doubly-truncated protein is cytosolic, and not accessible to the glycosylation machinery.
To summarize, at least one of the two Dfi1 transmembrane domains was required, and either was sufficient, to produce Dfi1 protein with normal membrane orientation. Furthermore, apparently-normal glycoprotein maturation was achieved in the absence of the N-terminal transmembrane domain, but not the C-terminal transmembrane domain.
The Dfi1 N-terminal transmembrane domain is not required for localization to the cell periphery
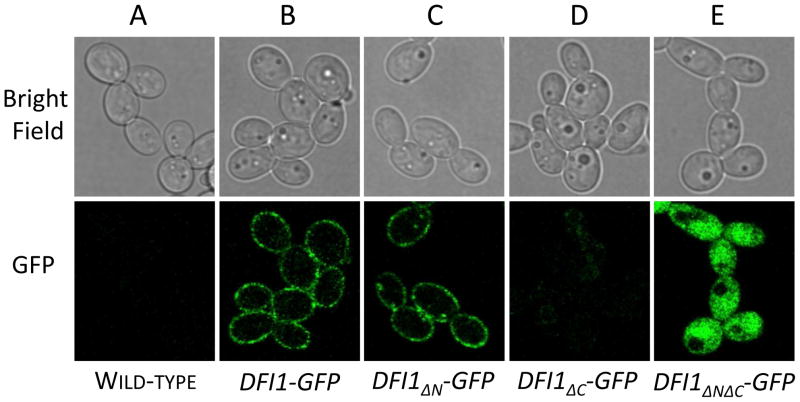
Deletion of either Dfi1 transmembrane domain could affect Dfi1 subcellular localization. As Dfi1 characteristically localizes to the cell periphery [6], the localization of the truncated Dfi1 mutant proteins was determined. Strains encoding GFP-tagged versions of full-length Dfi1 or a truncation mutant (Dfi1ΔN, Dfi1ΔC, and Dfi1ΔNΔC) were observed by confocal bright-field and GFP fluorescence microscopy. An untagged strain showed no fluorescence (Figure 4, panel A). The strain encoding full-length Dfi1-GFP showed fluorescence at the cell periphery (Figure 4, panel B), as previously described [6,7]. The strain encoding Dfi1ΔN-GFP showed fluorescent signal at the cell periphery (Figure 4, panel C). The strain encoding Dfi1ΔC-GFP showed faint or no GFP signal (Figure 4, panel D). Dfi1ΔC-GFP is the only examined version of Dfi1 in which the GFP tag is predicted to be on the extracellular side of the plasma membrane; This localization may prevent GFP activity. Finally, the strain encoding Dfi1ΔNΔC-GFP showed diffuse intracellular signal. Therefore, the Dfi1 N-terminal transmembrane domain was dispensable for localization to the cell periphery, but removal of both transmembrane domains resulted in a cytoplasmic protein.
Figure 4. The Dfi1 N-terminal TM domain is not required for localization to the cell periphery.
C. albicans strains encoding no GFP-tagged Dfi1, or encoding GFP-tagged full-length Dfi1, Dfi1ΔN, Dfi1ΔC, or Dfi1ΔNΔC were grown in liquid culture and imaged by confocal microscopy, as described in Materials and Methods. Images shown are xy-slices through 3D reconstructed images. Scale bar 5 μm.
Production of the abnormal, low molecular-weight species of Dfi1ΔN can be abrogated by the substitution of a generic transmembrane region
The main defect in Dfi1 biogenesis that was detected upon deletion of the Dfi1 N-terminal transmembrane domain was the appearance of an aberrant, poorly-glycosylated Dfi1ΔN species. We proposed that this abnormal Dfi1ΔN species occurs because the Dfi1 N-terminal transmembrane domain is required for the efficient biogenesis of Dfi1. To test this, we constructed DFI1 alleles in which a region of twenty amino acids in the native N-terminal transmembrane region was substituted with a sequence of twenty amino acids from an artificial, generic transmembrane helix [14,15]. One such sequence was LLIAVLLLIAVVLILLIAVA, based on Choma et al. [14] (denoted DFI1N-TM replacement C). A second sequence was LLLLLLVLLLLLLVLLLLLL, based on Zhou et al. [15] (denoted DFI1N-TM replacement Z). The effect of these artificial transmembrane sequences on Dfi1 N-linked glycosylation was analyzed using western blotting of cell lysates that had been PNGase F- or mock-treated.
Both Dfi1 substitution mutant proteins migrated as wide bands of large apparent molecular weights (Figure 3, lanes 9 and 11). These species underwent a dramatic shift upon PNGase F treatment (Figure 3, lanes 10 and 12). The Dfi1N-TM replacement C protein did not yield a lower molecular-weight, aberrant species (Figure 3, lanes 9 and 10). In contrast, the Dfi1N-TM replacement Z version showed a distinct smaller species (Figure 3, lanes 11 and 12) that was similar to the abnormal species of Dfi1ΔN in its apparent molecular weight, relative abundance, and lack of appreciable N-linked glycosylation. Therefore, substitution of a generic transmembrane sequence could promote efficient Dfi1 biogenesis but not all sequences had the same effect.
Finally, to examine whether the abnormal Dfi1ΔN species was inserted into the membrane, we examined blots of DFI1ΔN membrane-containing pellets after extraction. The small species was not extracted from the pellet using buffer alone or urea (Figure S1, lanes 1, 2, 3, and 4). Some protein was extracted by treatment with Triton X-100 (Figure S1, lanes 5 and 6). This suggests that the lack of N-linked glycosylation was not due to complete failure of membrane insertion. As such, some of this population likely represents mis-oriented membrane-inserted protein.
Discussion
The C. albicans protein Dfi1 is required for normal invasive filamentation and virulence in murine disseminated candidiasis [6]. Here we experimentally determined that mature Dfi1 contains two transmembrane domains. This and other data [7] suggest a model of Dfi1 topology and membrane orientation where the N- and C-termini are cytoplasmic, and the inter-transmembrane region is extracellular. Because the N-terminal transmembrane domain is retained in mature Dfi1 protein, this domain and/or the intracellular N-terminal region may mediate or regulate Dfi1 signaling. The topology of Dfi1, containing two transmembrane domains with an extracellular inter-transmembrane region, is similar to that of Sln1 [16], an osmosensor of S. cerevisiae [17]. It has been proposed that cell wall or periplasmic molecules can contact the Sln1 extracellular domain and activate Sln1 [17]. Contact sensing by Dfi1, which itself covalently binds to the cell wall [6], may share some mechanistic features with sensing mediated by Sln1.
We also examined the role of the two Dfi1 transmembrane domains in Dfi1 biogenesis, including membrane orientation, glycoprotein maturation, and subcellular localization. Removal of both transmembrane domains disrupted multiple features of Dfi1 biogenesis, yielding a soluble protein without N-linked glycosylation that localized diffusely within the cell. In contrast, removal of only one of the two Dfi1 transmembrane domains produced more subtle effects.
Dfi1 protein lacking the C-terminal transmembrane domain (Dfi1ΔC) was present at dramatically higher levels than wild-type Dfi1 protein in equivalent amounts of cell extract. Interestingly, mutation of other functionally-important motifs within the Dfi1 C-terminus [6,7] did not cause this marked increase in Dfi1 quantity. Therefore, a yet-uncharacterized feature of the Dfi1 C-terminus may regulate Dfi1 protein levels.
Removal of the N-terminal transmembrane domain of Dfi1 resulted in an extensively glycosylated protein subpopulation, consistent with proper membrane orientation and glycoprotein maturation, and a second, aberrant subpopulation with defective glycosylation. Dfi1 lacking the N-terminal transmembrane domain localized predominantly to the cell periphery. These results show that a substantial amount of apparently-normal Dfi1 biogenesis can occur in the absence of the N-terminal transmembrane domain. The presence of the second, aberrant species of Dfi1ΔN is not unexpected, because the membrane orientation of multispanning integral membrane proteins like Dfi1 is strongly affected by the first transmembrane domain [18], which is missing in Dfi1ΔN. Substitution of one generic transmembrane sequence, LLIAVLLLIAVVLILLIAVA, but not another, similar generic transmembrane sequence, LLLLLLVLLLLLLVLLLLLL, at the Dfi1 N-terminus was sufficient to promote efficient normal Dfi1 biogenesis. The native Dfi1 N-terminal transmembrane domain thus has characteristics, beyond mere hydrophobicity, which are required for normal Dfi1 biogenesis.
Since an N-terminal transmembrane sequence is important for Dfi1 biogenesis, conservation of an N-terminal transmembrane domain would be expected in Dfi1 orthologs of non-albicans Candida species. A BLAST search using the Dfi1 amino acid sequence yielded closely-related proteins in C. dubliniensis, C. orthopsilosis, C. parapsilosis, and C. tropicalis. Dfi1 and the four Dfi1-like proteins were individually analyzed for transmembrane tendency, based on a published model [19], using the ProtScale website (http://web.expasy.org/protscale/pscale/Transmembranetendency.html, Accessed Jan. 2017). C. albicans Dfi1 and the Dfi1-like proteins of C. dubliniensis, C. orthopsilosis, C. parapsilosis were each predicted to contain two transmembrane domains, suggesting that the resulting protein topology contributes to Dfi1 function. In contrast, the Dfi1-like protein of C. tropicalis was predicted to contain only one transmembrane domain, near the C-terminus. This suggests that the Dfi1-like protein of C. tropicalis may differ from C. albicans Dfi1 in biogenesis and function.
The topology and biogenesis of any protein underlies its phenotypic function and mechanism of action. In the case of Dfi1, the demonstration of the presence of two transmembrane domains and elucidation of their respective roles in biogenesis promotes our understanding of this important mediator of invasive filamentation in the opportunistic pathogen C. albicans.
Supplementary Material
The C. albicans strain encoding HA-tagged Dfi1ΔN was grown in liquid culture and lysed by bead beating. Lysates underwent ultracentrifugation to pellet cell membranes, which were extracted with extraction buffer alone, buffer plus 4M urea (final concentration), or buffer plus 5% Triton X-100 (final concentration; v/v), as described in Materials and Methods. These homogenates then underwent a second round of ultracentrifugation, and the resulting supernatants (S) and pellets (P) were probed for HA by western blotting. Each lane was loaded with 50 μL of sample. This blot is representative of three experiments.
C. albicans strains used in this study.
Primers used in this study.
Highlights.
Two hydrophobic regions of Dfi1 contribute to its integration into cellular membranes [85 char]
The two Dfi1 hydrophobic sequences contribute differently to N-linked glycosylation [83 char]
An artificial hydrophobic sequence at the Dfi1 N-terminus rescues glycosylation [81 char]
Dfi1 lacking the N-terminal hydrophobic sequence localizes to the cell periphery [81 char]
Acknowledgments
We gratefully acknowledge Drs. Claire Moore, Gavin Schnitzler, and Honorine Ward for helpful discussion. We are grateful to Dr. Kim Davis for cloning assistance, and Dr. Susan Lee for assistance with ultracentrifugation. This research was supported by grants R01AI081794 and R01AI118898 from the National Institutes of Health (NIH) to C.A.K.. S.E.H. was partially supported by T32GM008448 from the NIH. S.T. was partially supported by grant R00AI114952 from the NIH. We also acknowledge the Tufts Center for Neuroscience Research, supported by P30 NS047243 to F.R. Jackson. The funders had no role in study design, data analysis or the decision to publish.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pierce CG, López-Ribot JL. Candidiasis drug discovery and development: new approaches targeting virulence for discovering and identifying new drugs. Expert Opin Drug Discov. 2013 doi: 10.1517/17460441.2013.807245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schulze J, Sonnenborn U. Yeasts in the gut: from commensals to infectious agents. Dtsch Arztebl Int. 2009;106:837–842. doi: 10.3238/arztebl.2009.0837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akpan A, Morgan R. Oral candidiasis. Postgraduate Medical Journal. 2002;78:455–459. doi: 10.1136/pmj.78.922.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cassone A, Cauda R. Candida and candidiasis in HIV-infected patients: where commensalism, opportunistic behavior and frank pathogenicity lose their borders. Aids. 2012;26:1457–1472. doi: 10.1097/QAD.0b013e3283536ba8. [DOI] [PubMed] [Google Scholar]
- 5.Sudbery PE. Growth of Candida albicans hyphae. Nat Rev Microbiol. 2011;9:737–748. doi: 10.1038/nrmicro2636. [DOI] [PubMed] [Google Scholar]
- 6.Zucchi PC, Davis TR, Kumamoto CA. A Candida albicans cell wall-linked protein promotes invasive filamentation into semi-solid medium. Mol Microbiol. 2010;76:733–748. doi: 10.1111/j.1365-2958.2010.07137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis TR, Zucchi PC, Kumamoto CA. Calmodulin binding to Dfi1p promotes invasiveness of Candida albicans. PLoS ONE. 2013;8:e76239. doi: 10.1371/journal.pone.0076239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabezón V, Llama-Palacios A, Nombela C, Monteoliva L, Gil C. Analysis of Candida albicans plasma membrane proteome. Proteomics. 2009;9:4770–4786. doi: 10.1002/pmic.200800988. [DOI] [PubMed] [Google Scholar]
- 9.Mellquist JL, Kasturi L, Spitalnik SL, Shakin-Eshleman SH. The amino acid following an asn-X-Ser/Thr sequon is an important determinant of N-linked core glycosylation efficiency. Biochemistry. 1998;37:6833–6837. doi: 10.1021/bi972217k. [DOI] [PubMed] [Google Scholar]
- 10.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, et al. Current protocols in molecular biology. New York: John Wiley, New York: J. Wiley & Sons; 2003. [Google Scholar]
- 11.van Geest M, Lolkema JS. Membrane topology and insertion of membrane proteins: search for topogenic signals. Microbiol Mol Biol Rev. 2000;64:13–33. doi: 10.1128/mmbr.64.1.13-33.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martínez-Duncker I, Díaz-Jiménez DF, Mora-Montes HM. Comparative Analysis of Protein Glycosylation Pathways in Humans and the Fungal Pathogen Candida albicans. Int J Microbiol. 2014;2014:267497. doi: 10.1155/2014/267497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tretter V, Altmann F, März L. Peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase F cannot release glycans with fucose attached alpha 1----3 to the asparagine-linked N-acetylglucosamine residue. Eur J Biochem. 1991;199:647–652. doi: 10.1111/j.1432-1033.1991.tb16166.x. [DOI] [PubMed] [Google Scholar]
- 14.Choma C, Gratkowski H, Lear JD, DeGrado WF. Asparagine-mediated self-association of a model transmembrane helix. Nat Struct Biol. 2000;7:161–166. doi: 10.1038/72440. [DOI] [PubMed] [Google Scholar]
- 15.Zhou FX, Cocco MJ, Russ WP, Brunger AT, Engelman DM. Interhelical hydrogen bonding drives strong interactions in membrane proteins. Nat Struct Biol. 2000;7:154–160. doi: 10.1038/72430. [DOI] [PubMed] [Google Scholar]
- 16.Maeda T, Wurgler-Murphy SM, Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- 17.Narang SS, Malone CL, Deschenes RJ, Fassler JS. Modulation of yeast Sln1 kinase activity by the CCW12 cell wall protein. J Biol Chem. 2008;283:1962–1973. doi: 10.1074/jbc.M706877200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shao S, Hegde RS. Membrane Protein Insertion at the Endoplasmic Reticulum. Annu Rev Cell Dev Biol. 2011;27:25–56. doi: 10.1146/annurev-cellbio-092910-154125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao G, London E. An amino acid “transmembrane tendency” scale that approaches the theoretical limit to accuracy for prediction of transmembrane helices: Relationship to biological hydrophobicity. Protein Science. 2006;15:1987–2001. doi: 10.1110/ps.062286306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The C. albicans strain encoding HA-tagged Dfi1ΔN was grown in liquid culture and lysed by bead beating. Lysates underwent ultracentrifugation to pellet cell membranes, which were extracted with extraction buffer alone, buffer plus 4M urea (final concentration), or buffer plus 5% Triton X-100 (final concentration; v/v), as described in Materials and Methods. These homogenates then underwent a second round of ultracentrifugation, and the resulting supernatants (S) and pellets (P) were probed for HA by western blotting. Each lane was loaded with 50 μL of sample. This blot is representative of three experiments.
C. albicans strains used in this study.
Primers used in this study.