Abstract
Introduction
Many screening platforms are prone to assay interferences that can be avoided by directly measuring the target or enzymatic product. Capillary electrophoresis (CE) and microchip electrophoresis (MCE) have been applied in a variety of formats to drug discovery. CE provides direct detection of the product allowing for the identification of some forms of assay interference. The high efficiency, rapid separations, and low volume requirements make CE amenable to drug discovery.
Areas Covered
This article describes advances in capillary electrophoresis throughput, sample introduction, and target assays as they pertain to drug discovery and screening. Instrumental advances discussed include integrated droplet microfluidics platforms and multiplexed arrays. Applications of CE to assays of diverse drug discovery targets, including enzymes and affinity interactions are also described.
Expert opinion
Current screening with CE does not fully take advantage of the throughputs or low sample volumes possible with CE and is most suitable as a secondary screening method or for screens that are inaccessible with more common platforms. With further development, droplet microfluidics coupled to MCE could take advantage of the low sample requirements by performing assays on the nanoliter scale at high throughput.
Keywords: capillary electrophoresis, microchip electrophoresis, screening, microfluidics
1. Introduction
Capillary electrophoresis (CE), and its microfluidic counterpart microchip electrophoresis (MCE), have emerged as promising techniques with growing use in the pharmaceutical industry for characterizing biopharmaceuticals, quality control, and for drug discovery. In this review, we focus on the application of CE to drug discovery and screening. CE separates molecules based on their differential migration in an electric field. In free solution, the migration of a molecule in an applied field is dependent upon its charge and size, enabling separation by either property. Modification of the separation media can alter the separation selectivity, e.g. electrophoresis within gels enable separation on size only. Separated molecules are detected by a variety of methods including UV absorbance, capacitively coupled contactless conductivity, mass spectrometry (MS), and laser-induced fluorescence (LIF). The combination of separation and direct, on-line detection enable CE and MCE to be used for many kinds of assays useful for screening. Of relevance to drug screening, optical detection is well-suited for detecting enzyme activity and affinity interactions.
Compared to other drug screening platforms, such as fluorescent plate readers, CE offers a number of potential advantages including low sample volume requirements (nanoliter or less), rapid separations, and sensitive detection of analytes. Combining fractionation with direct detection of product can simplify assay development because it is not necessary to develop coupled reactions or engineer optical changes to generate a selective signal. CE also allows for resolution of confounding components in the assay such as interference from optically active test compounds [1–3], non-specific protein aggregation [3], and compound precipitation [3]. A comparison of methods for a kinase screen against fluorescent test compounds determined electrophoresis to be preferred over fluorescence polarization, amplified luminescent proximity homogenous assay, and enzyme fragment complementation for quantifying fluorescent inhibitors. The preference for CE was due to the tolerance of the CE assay to fluorescent compounds, the assay’s sensitivity, and comparatively low substrate and enzyme requirements. In the CE assay the fluorescent compound was tolerated because test compound was resolved from the substrate and product.[2] An example of similar benefits for a protein-protein interaction assay is illustrated in Figure 1.[3]
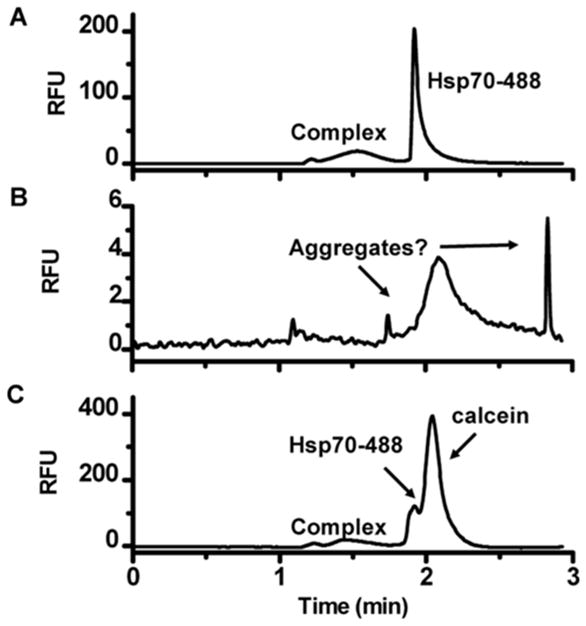
Figure 1.
Identification of assay interference in CE based on electropherograms. Electropherograms of Hsp70-488 and Bag3 interaction in the presence of (A) complex inhibitor epigallocatechin gallate, (B) hematoxylin which caused aggregation identified by loss of signal and sharp, unexpected peaks, (C) fluorescent test compound calcein causing optical interference.[3] Reprinted with permission from reference [3]. Copyright 2013 American Chemical Society.
MCE systems have been commercialized for screening. Perhaps the most popular system is the LabChip instrument (PerkinElmer), which uses vacuum to pull sample from a multiwell plate (MWP) into the microfluidic separation channel with fluorescence detection. This system has been used to screen 10 × 384 well plates in 10 h.[4] This platform has been applied to diverse targets including kinases, phosphatases, proteases, phosphodiesterases, epigenetic targets, and nucleic acid binding proteins.[1,2,4–9] Previous reviews on the use of CE in drug discovery have focused on the use of this system and specific enzyme target assays.[1,10]
Aside from this impressive technology, a variety of strategies for improving throughput and sample requirements have emerged, expanding the possibilities for using CE. In this review, we discuss new instruments and methods that have the potential to improve throughput and reduce sample consumption of CE or MCE for screening. Because one of the biggest limitations to throughput is sample injection onto the chip or capillary, many of these studies involve improvements in injection method. We also review assays, such as affinity assays for protein-protein interaction, where CE may be useful.
2. Improvements to Throughput
High throughput screening (HTS) using MWP with optical readouts can perform > 104 assays per day. The throughput of CE and MCE assays is limited by the separation time required to resolve the molecules of interest. As commonly practiced, CE separation times are a few minutes per sample; however, it is possible to achieve separations in < 1 s. Throughput may also be improved by running separations in parallel or by multiplexing the assay.
2.1. Rapid separations
Strategies for increasing the speed of a separation can be appreciated with a brief review of electrophoresis principles. The migration time of an analyte (tmig) in an electrophoresis separation channel with length (L) is dependent upon its electrophoretic mobility (μep) under an applied voltage (V):[11]
| (1) |
Therefore, to decrease the separation time the applied voltage can be increased or the separation length can be decreased. For example, in an enzyme inhibition assay of metalloproteinase performed on a commercial CE instrument the separation time was decreased from about 250 s to 70 s by simply reversing the polarity of the applied electric field and injecting from the ‘outlet’ side of the capillary, near the detection window, resulting in a shorter separation length.[12] In general however, commercial CE instruments have limited accessible separation lengths so that it is difficult to reduce separations to shorter than this. Some custom-built CE systems have allowed sub-second separations;[13,14] however, with one exception [15,16], they have not been investigated for screening.
MCE allows for use of shorter separation channel lengths and smaller internal diameters than commercial CE systems. Increasing the applied voltage over a given separation length can increase the separation speed; however, this approach is limited because eventually Joule heating becomes significant enough to create mixing effects that destroy the separation. Effective heat dissipation can be achieved by using liquid cooling or by lowering separation channel diameters. These strategies enable higher voltages and faster separations. MCE with micrometer dimension channels enable separations in microseconds to seconds.[17] Decreasing the separation channel internal diameter places greater demands on the detector to achieve adequate sensitivity. LIF detection is often used when low internal diameters are employed because of the inherent sensitivity of fluorescence detection. Despite inherent challenges in coupling electrophoresis to MS recent advances in MS sensitivity[18] may eventually allow detection by this label-free, highly selective detector.
Another approach to improving throughput is to perform multiple injections rapidly so that multiple separations are overlapping in the separation channel at one time. This technique, which requires proper spacing of the injections relative to the separation times of peaks of interest, increases throughput by eliminating time between runs and taking advantage of the time between resolved peaks in a single separation. In one study, the total analysis time per sample was reduced by about half with sequential injection.[19] This approach is best used with CE where the separation time tends to be longer than MCE.
2.2. Sample Introduction
Although very fast separations are possible by CE and MCE, sample introduction and injection are critical parameters to achieving high throughput. Rapid separations in short, low volume separation channels require special considerations for sample injection to avoid overloading the separation channel with sample. Typical injection volumes are less than 1% of separation channel volume, corresponding to 120 pL volume on a 2 cm long x 20 μm tall x 30 μm wide channel. Rapid, small volume injection techniques include gated injection[20], optically gated injection[13,14,21,22], flow-gating interfaces[23], and spontaneous injection[24–26]. These methods allow adequate control to inject small volumes onto the channel and therefore achieve high quality separations in < 10 s; however, commonly used designs require that much larger samples (microliters) be loaded into a sample reservoir mounted on the chip. This approach therefore requires more sample than necessary for the actual electrophoresis separation and is not compatible with rapidly changing from one sample to the next. Recent advances in sample introduction methods are showing potential for improved throughput of screening.
2.2.1. Direct Injection from Multi-well Plate
Most screens are performed from MWP. Transferring samples rapidly and in series from the MWP to MCE requires specific fluidic handling components. The LabChip system uses “sippers” (capillaries attached to the chip) to dip into the MWP and pull as little as 10 nL of sample into the chip through a vacuum system. Interestingly, this system does not use the high efficiency injection methods described above. Instead, vacuum is used to pull samples into the separation channel. Therefore, a limitation of this commercial system is that the vacuum-induced flow within the separation channel causes band broadening, limiting the separation efficiencies that can be achieved. Nevertheless, this is the only commercial system that allows robust sampling for thousands of assays with MCE separations. Furthermore, high Z′-scores (>0.8), a measure of a screen’s data quality, have been reported using this platform.[1,7]
When assay reactions are performed in a MWP, the system does not reduce sample requirements even though only small volumes are removed from the MWP. The use of microfluidics, however, does allow enzymatic reactions to be performed on-chip. In on-chip mode, compounds are sipped from MWP and enzyme reagents are added in channels so that reactions occur on-chip. In one kinase screen, on-chip reactions reduced enzyme consumption 7-fold.[4] Due to short on-chip incubation times, however, the on-chip format requires high concentrations of enzymes and is limited to enzymes with rapid turnover (> 30% in 1 min).[6]
2.2.2. Injection from Small Sample Volumes
To reduce sample volumes required for injection, new designs for picoliter injections from low sample volumes have been developed.[24,27] One platform utilizes a “Slipchip.”[28] This design consists of two plates; one contains sample well and a discontinuous separation channel and the other contains a small volume sample well aligned with the bottom of the sample well on the other plate. The plates are aligned such that by moving one plate relative to the other discrete sub-nanoliter volume samples can be formed in the small volume sample well and alignment of this well with the separation channel allows for subsequent injection. The design is also parallelized for analysis from 10 discrete samples with 30 parallel separations.[27] Changing samples beyond the parallelized number, however, currently requires manual manipulations. In another platform, an array of nanoliter sample droplets was covered with immiscible oil and picoliter injections were achieved spontaneously by surface tension when the capillary tip was removed from the sample droplet. An array of 25 samples were injected with RSDs for peak height and migration time < 5%.[24] Potential exists to scale both of these systems to higher sample numbers to achieve higher throughput.
2.2.3. Integration of Segmented Flow
Another strategy for rapidly introducing new samples and miniaturizing sample requirements is use of droplet microfluidics. In droplet microfluidics, discrete aqueous samples are compartmentalized by an immiscible, often fluorinated, carrier fluid. Flow focusing and t-junctions can be used to make many droplets from one sample. [29–32] However, to make a few droplets from many samples, as is necessary in screening, different methods are required. In one approach, samples in MWPs are reformatted into segmented droplets of nanoliter volume inside tubing by using aspiration to sequentially draw up plugs of sample and carrier fluid.[15,16,33–36] Microfluidic droplets can then undergo further manipulations such as mixing,[37–41] merging,[42–46] splitting,[43,47–50] addition,[35,37,38,51,52] incubation and extraction,[15,53–59] which can enable entire assays to be performed at small scale. Indeed, this approach has been used with fluorescence detection to screen 704 compounds against protein tyrosine phosphatase and high-resolution dose response curves were obtained. Sample consumption per data point was reduced 25,000-fold.[60] Droplet microfluidics therefore is emerging as an exciting way to perform screens at much reduced volume and instrument overhead relative to MWP. Use of droplet microfluidics offers the potential to take better advantage of the throughput and miniaturization possible with MCE.
One hurdle associated with using droplet microfluidics in CE screening is automated injection of droplet samples into the CE separation channel. Injection of immiscible, non-conductive, segmenting liquid is not compatible with CE separation and has been observed to cause electroosmotic flow instability and plug formation in the separation channel leading to shorting and dielectric breakdown of the channel and device.[53,54,61] Therefore, extraction of aqueous sample from the segmented flow is necessary. Passive, active, whole, and partial droplet extraction and injection strategies have been reported.[15,53–59]
Active extraction uses an electric field to destabilize the fluorinated liquid-aqueous interface and merge the aqueous sample with a parallel aqueous stream providing robust and selective extraction.[55] Active extraction coupled to electrophoretic separation has yet to be reported.
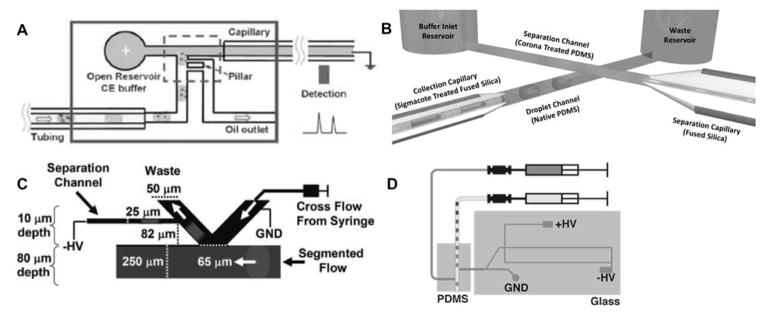
Passive extraction is somewhat simpler to integrate, as it does not require external input. As seen in Figure 2, several passive droplet extraction strategies have been coupled to downstream CE separations. Pillar arrays have been applied for complete extraction of carrier phase[56] and injection of the whole droplet into an electrophoresis channel (Figure 2A).[57] Passive droplet extraction strategies often rely on surface modifications to extract the hydrophilic aqueous sample droplet from the hydrophobic carrier phase. In one strategy, whole droplets are injected into an electrophoresis channel by a multilayer device where a portion of the separation channel is open to the segmented flow channel. The droplet is simultaneously extracted and injected when the aqueous sample passes the junction and coalesces with the separation buffer (Figure 2B).[62] Interestingly, passive whole droplet extraction and injection assisted by a hydrophobic and oleophilic foam was also effectively coupled to capillary gel electrophoretic and free solution separations.[59] The limitation of whole droplet injection strategies is that large injection volumes limit the separation quality.
Figure 2.
Passive extraction and injection strategies for coupling segmented flow sample droplets to electrophoresis separations. (A) Pillar array extraction of oil.[57] (B) Intersecting segmented flow and separation channel geometry for simultaneous extraction and injection.[62] (C) Virtual wall used for extraction.[53] (D) Hybrid PDMS-glass device used for decoupling extraction and injection processes (http://pubs.acs.org/doi/full/10.1021/ac502758h).[15] Reprinted with permission from references listed in sub captions. Copyright Royal Society of Chemistry and American Chemical Society.
In another strategy, a microchip device with a ‘virtual wall’ between hydrophobic and hydrophilic channels was fabricated by derivatizing a channel surface to make it hydrophobic, allowing for aqueous sample to be discreetly injected (Figure 2C).[53,54] A hybrid polydimethylsiloxane (PDMS)-glass device passively extracted droplets from a segmented stream within the hydrophobic PDMS device into a hydrophilic capillary connected to a glass microchip for gated injection could be used to introduce small volume injections for electrophoretic separation (Figure 2D). For screening, droplets were catalogued by the presence of a positively charged, rapidly migrating analyte in every other droplet which was also used to visualize rinsing between droplets. High efficiency, rapid separations were reported with this platform (Figure 3).[15,16]
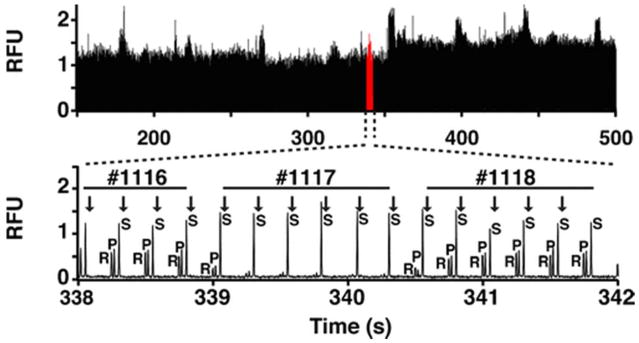
Figure 3.
Example of rapid enzyme assay separations achievable by MCE for screen of SIRT5 1,280 compounds. Separations of internal standard (R), product (P), and substrate (S) were achieved in 250 ms, #1117 is an enzyme inhibitor. Reprinted from reference [16], copyright 2016, with permission from Springer.
Several screens have been reported using this technology. Z′-scores of 0.8 were reported for protein kinase A and Sirtuin 5 screens using the hybrid PDMS-glass extraction approach. Throughputs of 0.2 to 0.5 samples/s have been reported with this platform.[15,16] To date, these approaches have only been used for up to 1,408 compounds. If these rates could be scaled to a large number of samples, then throughput could be 14,400 samples/8 h day on a single channel system.
2.3. Parallelization and Multiplexing Strategies
Operating CE or MCE in parallel can also improve the throughput. Challenges in developing platforms for running parallel separations include achieving multichannel detection, connections to peripheral power supplies, and attaining reliable simultaneous separations across parallel separation channels.[63] Many of these challenges were met when CE was being developed for DNA analysis. The necessity of high throughput sequencing led to using arrays of capillaries to run many parallel assays.[64] This concept has been adapted to drug discovery. A commercial instrumentation with a 96-capillary array was successfully applied to enzyme screening with UV absorption detection and throughputs of about 30 min per 96 samples without overlapping injections, high fields, or short capillaries.[65]
With MCE, parallel separation channel arrays can be made with small footprints at no additional fabrication cost. Up to 384 parallel separations have been reported for genotyping on a single microfluidic device.[66] Such highly parallel chips for screening have not yet been reported; however, the LabChip system can be used with 12 channels. Higher parallelization may be possible. A 36 channel microfluidic device with gated injection has been demonstrated for a model enzyme inhibitor screen with 36 parallel assays completed in 30 s.[67] Parallelization on chip with fast optically-gated injection has been used to perform for a high throughput enzyme assay with 4 parallel separations and 30 s separation time.[21] The throughput of a droplet extraction ‘virtual walls’ device was improved by parallelization with three extraction channels on one device and was demonstrated for an enzyme assay achieving throughputs of 120 samples in 10 min.[58] It seems likely that higher throughput could be achieved by parallelizing newer droplet extraction techniques.[15,16]
Another way to improve throughput is by test compound sample pooling. With sample pooling, versus assaying each test compound individually to find hits, a mixture of test compounds is assayed. If a hit is identified then each compound in the original test compound mixture is assayed individually. The low hit rates typical with screening allow far fewer assays to be done if sample pooling is used. Sample pooling for CE screening has been demonstrated with 10 test compounds assayed at a time.[68]
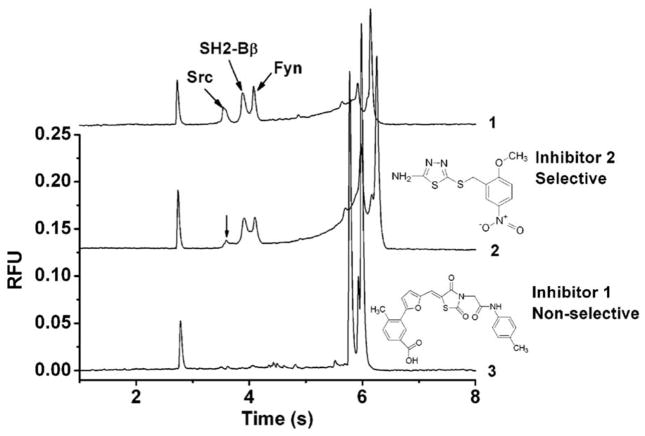
Multiplexing takes advantage of the separation power of CE to assay multiple targets simultaneously.[69,70] A four-plexed assay of protein kinases demonstrated simultaneous resolution of distinct substrates and products.[69] A study of protein-peptide interaction targets with src homology 2 (SH2) domains simultaneously assayed three proteins (Figure 4).[70] Such multiplexed assays also provide additional information about hit specificity by readily identifying selective and non-selective inhibitors.[69,70]
Figure 4.
Multiplexed assay of affinity interaction between SH2 domain proteins and phosphopeptides. Electropherograms identifying selective (middle) and non-selective inhibitors (bottom) of these interactions. Reprinted with permission from [70], copyright 2007, American Chemical Society.
2.4. Data Processing
A challenge of using CE for screening is that large numbers of electropherograms are generated which must be analyzed to determine peak areas used for quantification.[1] It has been shown that batch analysis of electropherograms allows for rapid data processing. For example, data processing software (available for download at http://kennedygroup.lsa.umich.edu/downloads/) allows for simultaneous analysis of hundreds of electropherograms to allow for data analysis on the time-scale of the rapid separations. In this strategy hundreds of electropherograms can be aligned based on a common peak, corrected for baseline drift, and peaks of interest can be defined.[71]
3. Targets
3.1. Enzymes
As previously mentioned, CE and MCE are amenable to performing enzymatic assays. Large screens of over 10,000 compounds have only been achieved using commercial microchip systems based on sipping from MWP.[8] The advent of droplet microfluidics interfaced to MCE may prove to have enough throughput, miniaturization, and robustness to provide a step forward for electrophoresis-based screening[15,16,53,54,58–62]; however, this has yet to be proven. The standard approach to enzyme assay by CE is to incubate enzyme with substrate and then separate substrate and product. The incubation may be performed in a MWP. Reduction in volume may be achieved by mixing on chip or in droplet samples as mentioned above. Other strategies, unique to CE, have also been demonstrated for analysis of enzyme modulation. These alternate methods include electrophoretically mediated microanalysis (EMMA), transverse diffusion of laminar flow profiles (TDLFP), and immobilized enzyme reactors (IMERs). Of the techniques, only “mix and separate” has been used for large scale screening with other techniques being demonstrated on small numbers of compounds (Table 1). Examples of screening using these techniques are described below.
Table 1.
Representative CE enzyme assay screens and demonstrations
| Target | Assay Type | Compounds Assayed | Z-score | Reference |
|---|---|---|---|---|
| tyrosine phosphatase | commercial microchip platform | 12,648 | 0.61 | [8] |
| Sirt5 | segmented flow coupled to MCE | 1,280 | 0.8 | [16] |
| glycerol kinase | EMMA | 1 | N.D. | [88] |
| aminopeptidase N | EMMA | 30* | N.D. | [86] |
| neuraminidase | EMMA | 24* | N.D. | [83] |
| β-N-acetylhexosaminidase | TDLFP | 1 | N.D. | [92] |
| four human kinases: GSK3β, DYRK1A, CDK5/p25, CDK1/cylcin B | TDLFP | 13 | N.D. | [93] |
| adenosine deaminase and xanthine oxidase | CE-based IMERs | 20* | 0.82, 0.74 | [101] |
| L-glutamic dehydrogenase | CE-based IMERs | 26* | 0.95 | [100] |
| glucose-6-phosphate dehydrogenase | CE-based IMERs | 6 | N.D. | [97] |
| alkaline phosphatases | CE-based IMERs | 3 | N.D. | [96] |
| acetylcholinesterase | CE-based IMERs | 46* | 0.9 | [95] |
| angiotensin-converting enzyme | CE-based IMERs | 34* | N.D. | [94] |
Not determined (N.D.); electrophoretically mediated microanalysis (EMMA); transverse diffusion of laminar flow profiles (TDLFP); capillary electrophoresis-based immobilized enzyme reactors (CE-based IMERs)
includes crude product, natural extract, or chinese traditional herb
Sirtuins are deacetylases implicated in a number of human diseases including age-related diseases and various cancers making them biologically interesting targets.[72] Both fluorogenic and label free methods have been previously developed to screen modulators of sirtuin activity; however, many of these had drawbacks in regards to high-throughput screening applications.[73] One fluorogenic assay that has been used to assess the activity of Sirtuins 1, 2, and 3 involves the deacylation of a substrate peptide followed by a trypsin digest that releases a fluorescent molecule.[74] Although this format has high throughput, the design of these substrate peptides has resulted in false positive hits.[75] HPLC and MS assays that directly detect substrates and products have been developed; but, their application to high throughput screening campaigns is limited by relatively large sample consumption and analysis time.[76,77]
CE assays for measuring activities of various sirtuins have been reported.[7,16,78–82] An on-chip assay of SIRT1 inhibition by three known inhibitors was demonstrated with 10 min of on-chip reaction time.[78] In another study, a MCE SIRT5 assay was developed using a novel succinylated peptide substrate (GGQSLK[succ]FGKG) labeled with 5-carboxyfluorescein (5-FAM) at the N-terminus as the substrate for LIF detection. Assays were carried out using the previously discussed droplet system shown in Figure 2D.[16] In principle, the entire reaction can be performed in the droplet, as has been demonstrated with fluorescence[60] or MS[35] detection, further reducing sample requirements.
Several other CE assays that minimize sample consumption have been reported for small numbers of compounds. Performing enzymatic reactions within the capillary can minimize enzyme requirements. In EMMA, reactants are sequentially injected onto the capillary and mixed based on their differential electrophoretic mobilities allowing for the enzymatic reaction to occur on-line (Figure 5A).[12,83–88] In comparison to traditional enzyme screening assays, lower IC50 values have been determined using EMMA. Lower observed inhibition has been attributed to the decreased incubation time typical in EMMA.[86] EMMA was also demonstrated for a two substrate enzyme, glycerol kinase, with four reactant plugs: incubation buffer, enzyme and two distinct substrate plugs, mixed in-capillary.[88] Small-scale screening of Chinese herbs and other crude products against enzymatic targets by EMMA with UV detection demonstrated the utility of CE in screening of complex test compound mixtures, where potential optical interference is reduced compared to other optical platforms.[83–87] While EMMA requires very little enzyme per assay optimal conditions for the enzymatic reaction may be easier to achieve in well plate format. Off-line reactions can be completed in parallel, often with automated liquid handling, by carrying out the enzymatic reaction within the capillary, however, on-line incubation limits the throughputs achievable.
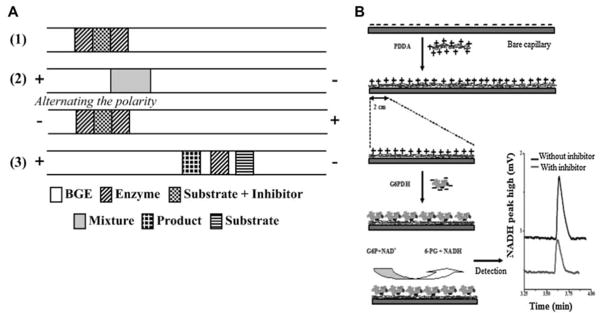
Figure 5.
Schematic of an EMMA strategy for enzyme inhibitor screening where (1) is depiction of sequential injection of plugs (2) polarity switching for mixing between plugs (3) separation of enzyme, substrate and product (A).[12] Schematic of CE-IMER for GAPDH.[97] Reprinted with permission from reference [12] and [97]. Copyright 2011 American Chemical Society and Elsevier.
In-line assays can also be performed with mixing of small volume pressure-injected plugs occurring by TDLFP.[89] Compared to EMMA, this method does not require prior knowledge of differential enzyme and substrate mobilities. The utility of this strategy first demonstrated with an assay of a farnesyltransferase target[90] and was later demonstrated for measuring inhibition of other enzymatic targets with UV detection.[91–93]
CE-based IMERs have also been demonstrated as useful for inhibition screening. Immobilized enzyme reactors can be fabricated within capillaries for on-line enzyme assays, saving enzyme in comparison to bulk assays (Figure 5B). CE-based IMERs for a wide variety of enzymes have been reported for characterizing enzyme inhibitors.[94–101] One study demonstrated the possibilities for multiplexing this CE-based IMER approach by fabricating an IMER of both immobilized adenosine deaminase and xanthine oxidase which was applied to screening of 20 natural extracts with a Z′-scores of 0.82 and 0.74, respectively. The separation was less than 3 min and the on-line incubation was 2.5 min. [101]
3.2. Affinity Interactions
Noncovalent binding of a partner can induce a mobility shift allowing electrophoretic separation. Many different assay schemes have been demonstrated for detecting and quantifying affinity interactions by MCE.[102–104] Noncovalent interactions assayed by CE include protein-nucleic acid [9,103,105], protein-peptide [70,68,106], protein-protein [3,107–110], nucleic acid-small molecule [111,112] and protein-small molecule interactions [113–117]. Of these, a “mix and separate” approach, sometimes called affinity probe CE [118], is perhaps the most amenable for high throughput screening. If the kinetics of dissociation are slow in comparison to the separation time, distinct free protein and protein complex peaks are observed enabling quantification of the bound to free ratio. Rapid separations are therefore preferable for both throughput and maintaining the complex of interest.
Affinity probe CE (APCE) on a microchip has been used to study protein-nucleic acid interactions.[9,104,105] In one example, the protein-nucleic acid interaction between human immunodeficiency virus 1 transactivator of transcription (Tat) and transactivation-responsive RNA (TAR) was studied using a commercial microchip platform. Inhibition of the Tat-TAR complex was demonstrated using a known inhibitor and dose dependent inhibition of the Tat-TAR complex peak was observed, suggesting the potential application of this platform to screening of affinity complexes.[9]
Protein-protein interactions (PPIs) represent a large class of targets that were, until recently, considered intractable. It can be difficult to predict small molecule modulators of these interactions as they often occur with high affinities over large, flat surfaces. Recent success in targeting these interactions has led to an increased interest in screening against them.[119,120] CE has potential as a useful screening technique because of the capability of high efficiency separation of large molecules.
CE has been used in studies of amyloid aggregation[121–123], protein-peptide interactions[70,68,106], and full length PPIs [3,107–110]. Most of these studies have been proof of concept assays; however, one assay did investigate a screen of 3,443 compound library against a target PPI. In this study, a fluorescently labeled heat shock protein 70 (Hsp70) in complex with its co-chaperone Bcl2-associated athanogene 3 (Bag3) was separated by CE. The resulting screen yielded a 1.4% hit rate. A 3.4% hit rate was achieved for the same screen using flow cytometry protein interaction assay (FCPIA). The lower hit rate in the CE assay was attributed to the identification of detection interfering compounds that may be false positives in other assays, such as fluorescent test compounds in the screening library and unexpected aggregation of proteins which could be readily identified by visual inspection of the electropherograms (Figure 1). Comparable Z′-scores of 0.78 for CE and 0.86 for FCPIA were found for these parallel screening platforms. The CE assay was not optimized for throughput however, as it used a commercial, single-channel CE system, and could only perform 220 assays/day. At this throughput CE is potentially more useful as a secondary screening platform.[3] In principle, such assays could be converted to parallel CE or MCE formats for higher throughput.
Although the APCE approach is most amenable to screening, CE-frontal analysis (CE-FA) has been used to measure affinity interactions. In CE-FA, relatively large volumes of equilibrated binding partner mixtures are injected. The large injection volume allows for maintenance of interaction during the separation because the free and complex zones are largely overlapping during the separation. The interaction between apoptosis regulatory, B-cell lymphoma 2 (Bcl-2) family proteins Bcl-XL and BH3-interaction domain (Bid) was studied using a fluorophore-labeled peptide form of Bid. Quantification was achieved based on the plateau height of the free ligand. A high Z′-factor of 0.86 was determined for this method and a screen using sample pooling of 60 compounds with 10 compounds per sample was demonstrated with 10 min/sample separation times.[68]
A number of obstacles still exist for developing CE methods for screening affinity interactions; the protein complex may dissociate during the separation, many interactions induce only a small shift in mobility, and proteins tend to adsorb to the wall of the fused silica capillary which, depending on severity can cause shifts in migration time as well as loss of signal. The small mobility shift and protein adsorption issues must be overcome using conditions that maintain the non-covalent target interactions. One way to simplify method development for interacting proteins, therefore, is to covalently cross-link interacting proteins prior to electrophoresis. Once the proteins are cross-linked, the separation conditions can be optimized without concern of disrupting the interaction. For example, formaldehyde cross-linking followed by SDS-capillary gel electrophoresis separation (denaturing conditions) was shown to be effective for characterizing interaction affinities and small molecule inhibitors of PPI,[110] suggesting potential utility in screening.
Capillary coatings are useful for reducing protein adsorption. A number of recent studies have successfully characterized capillary coatings several of which are compatible with physiological pH and avoid adsorption that was observed on bare-fused silica capillary.[124–128] Unfortunately, a coating that prevents adsorption of one protein may not be as effective at preventing adsorption of a different protein[126,128] and replacing dynamic coatings between separations decreases throughput. Recently, a permanent coating for protein separation was observed to have an electroosmotic flow RSD of only 0.5% over one month.[128]
Assays for small molecule ligand interactions with proteins, peptides, and nucleic acids have been demonstrated by monitoring the mobility shift of the large molecule in the presence of small molecule.[111–117] Small molecule mobility shift assays can be challenging, with some binding events only inducing small changes in large molecule mobility.[114] Still, fragment-based screening has been demonstrated by affinity CE (CEfrag).[116] Successful fragment library screening relies on the ability to detect small amounts of inhibition as well as assay compatibility with high concentrations of test compound. A screen of heat shock protein 90α (Hsp90α) by a mobility shift assay with competitive inhibition was successful applied to a fragment library screen. In this method a small molecule affinity probe that is known to bind to the target molecule is used as an indicator of binding. When the affinity probe small molecule is unbound, it has a different migration time then when it is bound to its target. When another molecule competes with the affinity probe for target binding a mobility shift is observed and a hit is identified. For Hsp90α, using radicicol, a molecule known to interact with Hsp90α as the affinity probe, weaker affinity hits (>500 μM IC50 values) were identified using CE with UV detection versus with a fluorescence polarization assay in a screen of 609 compounds. The throughput of this screen was 100 samples/instrument/day using a 4-capillary instrument and a Z factor of 0.6 was determined.[117]
4. Conclusion
CE and MCE are capable of rapid analysis of small sample volumes making them natural tools for screening. The direct detection of product and resolving power allow for identification of assay interference, providing a powerful advantage relative to fluorescent assays that rely on a single channel optical readout. State of the art screening by CE is embodied in robust commercial microchip systems that sip samples from MWP and inject and separate on parallel arrays of channels. Such systems have been successfully used for a variety of enzyme assays.
Continued development of CE has revealed routes to potentially higher-throughput, lower sample consumption, and new types of assays. Microfluidic sample manipulation, especially droplet systems interfaced to MCE, has shown potential for high throughput, robust screening on nanoliter scale. Besides enzyme assay, affinity assays have been demonstrated that enable CE to be used for a wider range of targets. Although many new assays and formats using CE have been demonstrated, reports of using this new technology for screening large compound libraries are limited.
5. Expert Opinion
Although CE has demonstrated potential as a screening technique, high throughput screening is currently dominated by MWPs used with optical plate readers. This dominance is due to extremely high throughput and entrenchment of existing technology. Furthermore, the expertise of those researchers who develop screens often lies in chemical biology and engineering fluorescent or similar optical changes into desired biological assays rather than CE separations. Nevertheless, current commercial MCE platforms, such as the LabChip system, occupy an important niche in enabling screening when adequate fluorescent assays are challenging to develop, or as a secondary screening tool. Importantly, MCE and CE have demonstrated utility in identifying optical interferences including aggregation and fluorescent test compounds and lower test compound hit rates have been attributed to the identification of these interferences. The viability of CE has been shown with high Z-scores, demonstrating assay robustness on a variety of platforms. These advantages demonstrate the utility of further developing CE for screening.
While the applicability of CE to enzyme screening of diverse targets has been demonstrated using a variety of platforms, assays for affinity binding have yet to see widespread use in MCE screening. CE affinity assays offer significant advantages including sensitivity, ease of method development, multiplexing and information content. However, protein absorption to capillary wall and complex dissociation can complicate method development and have hindered progress in investigating affinity interaction targets by CE. As PPI and protein-nucleic acid interactions continue to grow as drug targets, affinity assays seem to be a promising area in need of further research to fully understand the utility of CE in these assays.
Although the current, commercially available, systems provide a robust platform for screening, they also impose limitations that do not fully support the possible high speeds of separation and therefore potential throughput of CE or MCE. Commercial instruments interface to MWP, containing microliters of each sample. While this is comparable to other screening technologies, interfacing to a MWP does not take advantage of the potential to analyze picoliter to nanoliter sample volumes in CE or MCE. Continued research into CE and MCE has identified promising in-line and microfluidic methods that use less sample volume and achieve higher throughputs. MCE has demonstrated advantages for improving throughput; therefore, the marriage of droplet microfluidics to MCE, so that entire assays are performed on a nanoliter scale, might be the most promising development. The coupling of this droplet technology with other techniques such as MS could also be furthered explored to take advantage of the power of the droplet technology and further the information content of the assay. Current droplet platforms have not demonstrated the robustness necessary for large scale screening efforts by CE and MCE. Further research should emphasize development of robust, integrated, multiplexed droplet and MCE system capable of screening > 104 compounds routinely. Ultimately, such a system, based on MCE and droplet microfluidic technology, would open the door to next generation screening that is not limited by the well plate volume and need for expensive robotics. It would also fully unlock the power of CE screening.
Table 2.
Representative CE affinity interaction screens
| Target | Assay type | Compounds Assayed | Z-score | Reference |
|---|---|---|---|---|
| Tat-TAR | Commercial microchip platform | 1 | N.D. | [9] |
| Hsp70-Bag3 | APCE | 3,443 | 0.78 | [3] |
| Bcl-XL-Bid | CE-FA | 105* | 0.86 | [68] |
| Hsp90α | CEfrag | 609 | 0.6 | [117] |
Not determined (N.D.); affinity probe capillary electrophoresis (APCE); capillary electrophoresis-frontal analysis (CE-FA); fragment screening using capillary electrophoresis (CEfrag)
includes crude product, natural extract, or chinese traditional herb
Article Highlights.
Capillary electrophoresis (CE) and microchip electrophoresis (MCE) offer rapid, low volume separations, well suited for drug discovery and screening.
Few large-scale screens have been reported using CE; many of these have been performed on commercial microchip platform.
Sub-second CE separations are possible in some cases and the throughput of CE can also be improved by parallelization and multiplexing.
A variety of assays are accessible by CE including enzyme assays and biomolecular interactions.
Novel strategies for reducing sample consumption have been reported including droplet microfluidics and on-line reactions demonstrating a future potential of low sample consumption by CE.
Acknowledgments
The authors thank James Grinias for helpful discussion.
Funding:
The authors are supported by National Institutes of Health grant R01GM102236.
Footnotes
Declaration of Interest:
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
References
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1••.Perrin D, Frémaux C, Shutes A. Capillary microfluidic electrophoretic mobility shift assays: application to enzymatic assays in drug discovery. Expert Opin Drug Discov. 2009;15:51–63. doi: 10.1517/17460440903493431. Review of use of commercial MCE platform for screening. [DOI] [PubMed] [Google Scholar]
- 2•.Perrin D, Martin T, Cambet Y, et al. Overcoming the Hurdle of Fluorescent Compounds in Kinase Screening: A Case Study. Assay Drug Dev Technol. 2006;4:185–196. doi: 10.1089/adt.2006.4.185. Direct comparison of MCE with other methods for kinase screening. [DOI] [PubMed] [Google Scholar]
- 3.Rauch JN, Nie J, Buchholz TJ, et al. Development of a capillary electrophoresis platform for identifying inhibitors of protein-protein interactions. Anal Chem. 2013;85:9824–9831. doi: 10.1021/ac4023082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perrin D, Frémaux C, Scheer A. Assay Development and Screening of a Serine/Threonine Kinase in an On-Chip Mode Using Caliper Nanofluidics Technology. J Biomol Screen. 2006;11:359–368. doi: 10.1177/1087057106286653. [DOI] [PubMed] [Google Scholar]
- 5.Pommereau A, Pap E, Kannt A. Two Simple and Generic Antibody-Independent Kinase Assays: Comparison of a Bioluminescent and a Microfluidic Assay Format. J Biomol Screen. 2004;9:409–416. doi: 10.1177/1087057104264175. [DOI] [PubMed] [Google Scholar]
- 6.Dunne J, Reardon H, Trinh V, et al. Comparison of On-Chip and Off-Chip Microfluidic Kinase Assay Formats. Assay Drug Dev Technol. 2004;2:121–129. doi: 10.1089/154065804323056468. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y, Gerber R, Wu J, et al. High-throughput assays for sirtuin enzymes: A microfluidic mobility shift assay and a bioluminescence assay. Anal Biochem. 2008;378:53–59. doi: 10.1016/j.ab.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 8.Perrin D, Frémaux C, Besson D, et al. A Microfluidics-Based Mobility Shift Assay to Discover New Tyrosine Phosphatase Inhibitors. J Biomol Screen. 2006;11:996–1004. doi: 10.1177/1087057106294094. [DOI] [PubMed] [Google Scholar]
- 9•.Fourtounis J, Falgueyret JP, Elie Sayegh C. Assessing protein-RNA interactions using microfluidic capillary mobility shift assays. Anal Biochem. 2011;411:161–163. doi: 10.1016/j.ab.2010.11.042. Demonstration of assesing protein-nucleic acid interactions on a commercial, screening compatable, MCE platform. [DOI] [PubMed] [Google Scholar]
- 10.Shanmuganathan M, Britz-McKibbin P. High quality drug screening by capillary electrophoresis: A review. Anal Chim Acta. 2013;773:24–36. doi: 10.1016/j.aca.2013.01.061. [DOI] [PubMed] [Google Scholar]
- 11.Jorgenson JW, Lukacs KD. Zone Electrophoresis in Open-Tubular Glass Capillaries. Anal Chem. 1981;53:1298–1302. [PubMed] [Google Scholar]
- 12.Hai X, Wang X, El-Attug M, et al. In-capillary screening of matrix metalloproteinase inhibitors by electrophoretically mediated microanalysis with fluorescence detection. Anal Chem. 2011;83:425–430. doi: 10.1021/ac1027098. [DOI] [PubMed] [Google Scholar]
- 13.Moore AW, Jorgenson JW. Study of zone broadening in optically gated high-speed capillary electrophoresis. Anal Chem. 1993;65:3550–3560. doi: 10.1021/ac00072a004. [DOI] [PubMed] [Google Scholar]
- 14.Tao L, Thompson JE, Kennedy RT. Optically Gated Capillary Electrophoresis of o-Phthalaldehyde/β-Mercaptoethanol Derivatives of Amino Acids for Chemical Monitoring. Anal Chem. 1998;70:4015–22. doi: 10.1021/ac980324u. [DOI] [PubMed] [Google Scholar]
- 15.Guetschow ED, Steyer DJ, Kennedy RT. Subsecond Electrophoretic Separations from Droplet Samples for Screening of Enzyme Modulators. Anal Chem. 2014;86:10373–10379. doi: 10.1021/ac502758h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16••.Guetschow ED, Kumar S, Lombard DB, Kennedy RT. Identification of sirtuin 5 inhibitors by ultrafast microchip electrophoresis using nanoliter volume samples. Anal Bioanal Chem. 2016;3:721–731. doi: 10.1007/s00216-015-9206-0. Screen of sirtuin 5 inhibitors using droplet microfluidics coupled to MCE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacobson SC, Hergenroder R, Koutny LB, Ramsey JM. High-speed Separations. 1994;66:1114–1118. [Google Scholar]
- 18.Zhong X, Zhang Z, Jiang S, Li L. Recent advances in coupling capillary electrophoresis based separation techniques to ESI and MALDI MS. 2011;4:1214–1225. doi: 10.1002/elps.201300451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roche ME, Oda RP, Machacek D, et al. Enhanced throughput with capillary electrophoresis via continuous-sequential sample injection. Anal Chem. 1997;69:99–104. [Google Scholar]
- 20.Jacobson SC, Koutny LB, Hergenröder R, et al. Microchip Capillary Electrophoresis with an Integrated Postcolumn Reactor. Anal Chem. 1994;66:3472–3476. [Google Scholar]
- 21.Xu H, Ewing AG. High-throughput enzyme assay on a multichannel microchip using optically gated sample introduction. Electrophoresis. 2005;26:4711–4717. doi: 10.1002/elps.200500620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monnig CA, Jorgenson JW. On-column sample gating for high-speed capillary zone electrophoresis. Anal Chem. 1991;63:802–807. doi: 10.1021/ac00008a013. [DOI] [PubMed] [Google Scholar]
- 23.Lemmo AV, Jorgenson J. Transverse flow gating interface for the coupling of microcolumn LC with CZE in a comprehensive two-dimensional system. Anal Chem. 1993;65:1576–1561. [Google Scholar]
- 24.Li Q, Zhu Y, Zhang N-Q, Fang Q. Automatic Combination of Microfluidic Nanoliter-Scale Droplet Array with High-Speed Capillary Electrophoresis. Sci Rep. 2016;6:26654. doi: 10.1038/srep26654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fang X, Fang P, Pan J, Fang Q. A Compact Short-Capillary Based High-Speed Capillary Electrophoresis Bioanalyzer. Electrophoresis. 2016;37:2376–2383. doi: 10.1002/elps.201600195. [DOI] [PubMed] [Google Scholar]
- 26.Fishman HA, Amudi NM, Lee TT, et al. Spontaneous Injection in Microcolumn Separations. Anal Chem. 1994;66:2318–2329. doi: 10.1021/ac00086a018. [DOI] [PubMed] [Google Scholar]
- 27.Hassan SU, Morgan H, Zhang X, Niu X. Droplet Interfaced Parallel and Quantitative Microfluidic-Based Separations. Anal Chem. 2015;87:3895–3901. doi: 10.1021/ac504695w. [DOI] [PubMed] [Google Scholar]
- 28.Du W, Li L, Nichols KP, Ismagilov RF. SlipChip. Lab Chip. 2009;9:2286. doi: 10.1039/b908978k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ward T, Faivre M, Abkarian M, Stone HA. Microfluidic flow focusing: Drop size and scaling in pressure versus flow-rate-driven pumping. Electrophoresis. 2005;26:3716–3724. doi: 10.1002/elps.200500173. [DOI] [PubMed] [Google Scholar]
- 30.Song H, Tice JD, Ismagilov RF. A Microfluidics System for Controlling Reaction Networks in Time. Angew Chemie - Int Ed. 2003;1:767–772. doi: 10.1002/anie.200390203. [DOI] [PubMed] [Google Scholar]
- 31.Garstecki P, Fuerstman MJ, Stone HA, Whitesides GM. Formation of droplets and bubbles in a microfluidic T-junction-scaling and mechanism of break-up. Lab Chip. 2006;6:437–446. doi: 10.1039/b510841a. [DOI] [PubMed] [Google Scholar]
- 32.Anna SL, Bontoux N, Stone HA. Formation of dispersions using “flow focusing” in microchannels. Appl Phys Lett. 2003;82:364–366. [Google Scholar]
- 33.Chabert M, Dorfman KD, de Cremoux P, et al. Automated microdroplet platform for sample manipulation and polymerase chain reaction. Anal Chem. 2006;78:7722–7728. doi: 10.1021/ac061205e. [DOI] [PubMed] [Google Scholar]
- 34.Gielen F, Van Vliet L, Koprowski BT, et al. A fully unsupervised compartment-on-demand platform for precise nanoliter assays of time-dependent steady-state enzyme kinetics and inhibition. Anal Chem. 2013;85:4761–4769. doi: 10.1021/ac400480z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35••.Sun S, Slaney TR, Kennedy RT. Label Free Screening of Enzyme Inhibitors at Femtomole Scale Using Segmented Flow Electrospray Ionization Mass Spectrometry. Anal Chem. 2012;84:5794–5800. doi: 10.1021/ac3011389. Demonstration of nanoliter volume enzyme assay for screening. [DOI] [PubMed] [Google Scholar]
- 36.Sun S, Kennedy RT. Droplet Electrospray Ionization Mass Spectrometry for High Throughput Screening for Enzyme Inhibitors. Anal Chem. 2014;86:9309–9314. doi: 10.1021/ac502542z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shestopalov I, Tice JD, Ismagilov RF. Multi-step synthesis of nanoparticles performed on millisecond time scale in a microfluidic droplet-based system. Lab Chip. 2004;4:316. doi: 10.1039/b403378g. [DOI] [PubMed] [Google Scholar]
- 38.Song H, Li HW, Munson MS, et al. On-chip titration of an anticoagulant argatroban and determination of the clotting time within whole blood or plasma using a plug-based microfluidic system. Anal Chem. 2006;78:4839–4849. doi: 10.1021/ac0601718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Casadevall i Solvas X, Srisa-Art M, deMello AJ, Edel JB. Mapping of fluidic mixing in microdroplets with 1 microsecond time resolution using fluorescence lifetime imaging. Anal Chem. 2010;82:3950–6. doi: 10.1021/ac100055g. [DOI] [PubMed] [Google Scholar]
- 40.Liau A, Kamik R, Majumdar A, Cate JHD. Mixing crowded biological solutions in milliseconds. Anal Chem. 2005;77:7618–7625. doi: 10.1021/ac050827h. [DOI] [PubMed] [Google Scholar]
- 41.Song H, Bringer MR, Tice JD, et al. Experimental test of scaling of mixing by chaotic advection in droplets moving through microfluidic channels. Appl Phys Lett. 2003;83:4664–4666. doi: 10.1063/1.1630378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Niu X, Gulati S, Edel JB, deMello AJ. Pillar-induced droplet merging in microfluidic circuits. Lab Chip. 2008;8:1837–1841. doi: 10.1039/b813325e. [DOI] [PubMed] [Google Scholar]
- 43.Christopher GF, Bergstein J, End NB, et al. Coalescence and splitting of confined droplets at microfluidic junctions. Lab Chip. 2009;9:1102–1109. doi: 10.1039/b813062k. [DOI] [PubMed] [Google Scholar]
- 44.Fidalgo LM, Abell C, Huck WTS. Surface-induced droplet fusion in microfluidic devices. Lab Chip. 2007;7:984–986. doi: 10.1039/b708091c. [DOI] [PubMed] [Google Scholar]
- 45.Mazutis L, Baret J-C, Griffiths AD. A fast and efficient microfluidic system for highly selective one-to-one droplet fusion. Lab Chip. 2009;9:2665–2672. doi: 10.1039/b903608c. [DOI] [PubMed] [Google Scholar]
- 46.Zagnoni M, Cooper JM. On-chip electrocoalescence of microdroplets as a function of voltage, frequency and droplet size. Lab Chip. 2009;9:2652–2658. doi: 10.1039/b906298j. [DOI] [PubMed] [Google Scholar]
- 47.Adamson DN, Mustafi D, Zhang JXJ, et al. Production of arrays of chemically distinct nanolitre plugs via repeated splitting in microfluidic devices. Lab Chip. 2006;6:1178–1186. doi: 10.1039/b604993a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Link DR, Anna SL, Weitz DA, Stone HA. Geometrically mediated breakup of drops in microfluidic devices. Phys Rev Lett. 2004;92:054503. doi: 10.1103/PhysRevLett.92.054503. [DOI] [PubMed] [Google Scholar]
- 49.Nie J, Kennedy RT. Sampling from nanoliter plugs via asymmetrical splitting of segmented flow. Anal Chem. 2010;82:7852–7856. doi: 10.1021/ac101723x. [DOI] [PubMed] [Google Scholar]
- 50.Choi J-H, Lee S-K, Lim J-M, et al. Designed pneumatic valve actuators for controlled droplet breakup and generation. Lab Chip. 2010;10:456–61. doi: 10.1039/b915596a. [DOI] [PubMed] [Google Scholar]
- 51.Li L, Boedicker JQ, Ismagilov RF. Using a multijunction microfluidic device to inject substrate into an array of preformed plugs without cross-contamination: Comparing theory and experiments. Anal Chem. 2007;79:2756–2761. doi: 10.1021/ac062179n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Slaney TR, Nie J, Hershey ND, et al. Push-Pull Perfusion Sampling with Segmented Flow for High Temporal and Spatial Resolution in Vivo Chemical Monitoring. 2011;83:5207–5213. doi: 10.1021/ac2003938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roman GT, Wang M, Shultz KN, et al. Sampling and electrophoretic analysis of segmented flow streams using virtual walls in a microfluidic device. Anal Chem. 2008;80:8231–8238. doi: 10.1021/ac801317t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang M, Roman GT, Perry ML, Kennedy RT. Microfluidic chip for high efficiency electrophoretic analysis of segmented flow from a microdialysis probe and in vivo chemical monitoring. Anal Chem. 2009;81:9072–9078. doi: 10.1021/ac901731v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fidalgo LM, Whyte G, Bratton D, et al. From microdroplets to microfluidics: Selective emulsion separation in microfluidic devices. Angew Chemie - Int Ed. 2008;47:2042–2045. doi: 10.1002/anie.200704903. [DOI] [PubMed] [Google Scholar]
- 56.Angelescu DE, Mercier B, Sless D, Schroetter R. Microfluidic capillary separation and real-time spectroscopic analysis of specific components from multiphase mixtures. Anal Chem. 2010;82:2412–2420. doi: 10.1021/ac902698m. [DOI] [PubMed] [Google Scholar]
- 57.Niu XZ, Zhang B, Marszalek RT, et al. Droplet-based compartmentalization of chemically separated components in two-dimensional separations. Chem Commun (Camb) 2009;41:6159–6161. doi: 10.1039/b918100h. [DOI] [PubMed] [Google Scholar]
- 58.Pei J, Nie J, Kennedy RT. Parallel electrophoretic analysis of segmented samples on chip for high-throughput determination of enzyme activities. Anal Chem. 2010;82:9261–9267. doi: 10.1021/ac101755y. [DOI] [PubMed] [Google Scholar]
- 59.Niu X, Pereira F, Edel JB, deMello AJ. Droplet-interfaced microchip and capillary electrophoretic separations. Anal Chem. 2013;85:8654–8660. doi: 10.1021/ac401383y. [DOI] [PubMed] [Google Scholar]
- 60.Miller OJ, El A, Mangeat T, et al. High-resolution dose – response screening using droplet-based microfluidics. Proc Natl Acad Sci. 2012;109:378–83. doi: 10.1073/pnas.1113324109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edgar JS, Pabbati CP, Lorenz RM, et al. Capillary electrophoresis separation in the presence of an immiscible boundary for droplet analysis. Anal Chem. 2006;78:6948–6954. doi: 10.1021/ac0613131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.DeLaMarre MF, Shippy SA. Development of a Simplified Microfluidic Injector for Analysis of Droplet Content via Capillary Electrophoresis. Anal Chem. 2014;86:10193–10200. doi: 10.1021/ac502272q. [DOI] [PubMed] [Google Scholar]
- 63.Dishinger JF, Kennedy RT. Multiplexed Detection and Applications for Separations on Parallel Microchips. Electrophoresis. 2008;29:3296–3305. doi: 10.1002/elps.200800067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang XC, Quesada MA, Mathies RA. Capillary array electrophoresis using laser-excited confocal fluorescence detection. Anal Chem. 1992;64:967–972. doi: 10.1021/ac00042a021. [DOI] [PubMed] [Google Scholar]
- 65.Ma L, Gong X, Yeung ES. Combinatorial screening of enzyme activity by using multiplexed capillary electrophoresis. Anal Chem. 2000;72:3383–3387. doi: 10.1021/ac000145o. [DOI] [PubMed] [Google Scholar]
- 66.Emrich CA, Tian H, Medintz IL, Mathies RA. Microfabricated 384-lane capillary array electrophoresis bioanalyzer for ultrahigh-throughput genetic analysis. Anal Chem. 2002;74:5076–5083. doi: 10.1021/ac020236g. [DOI] [PubMed] [Google Scholar]
- 67•.Pei J, Dishinger JF, Roman DL, et al. Microfabricated channel array electrophoresis for characterization and screening of enzymes using RGS-G protein interactions as a model system. Anal Chem. 2008;80:5225–5231. doi: 10.1021/ac800553g. Demonstration of parallelized MCE separations for screening. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu M, Liu C, Zhou M, Li Q, et al. Screening of Small-Molecule Inhibitors of Protein-Protein Interaction with Capillary Electrophoresis Frontal Analysis. Anal Chem. 2016;88:8050–8057. doi: 10.1021/acs.analchem.6b01430. [DOI] [PubMed] [Google Scholar]
- 69.Xue Q, Wainright A, Gangakhedkar S, Gibbons I. Multiplexed enzyme assays in capillary electrophoretic single-use microfluidic devices. Electrophoresis. 2001;22:4000–4007. doi: 10.1002/1522-2683(200110)22:18<4000::AID-ELPS4000>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 70.Yang PL, Whelan RJ, Mao YW, et al. Multiplexed detection of protein-peptide interaction and inhibition using capillary electrophoresis. Anal Chem. 2007;79:1690–1695. doi: 10.1021/ac061936e. [DOI] [PubMed] [Google Scholar]
- 71.Shackman JG, Watson CJ, Kennedy RT. High-throughput automated post-processing of separation data. J Chromatogr A. 2004;1040:273–282. doi: 10.1016/j.chroma.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 72.Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li Y, Liu T, Liao S, et al. A mini-review on Sirtuin activity assays. Biochem Biophys Res Commun. 2015;467:459–466. doi: 10.1016/j.bbrc.2015.09.172. [DOI] [PubMed] [Google Scholar]
- 74.Wegener D, Wirsching F, Riester D, Schwienhorst A. A Fluorogenic Histone Deacetylase Assay Well Suited for High-Throughput Activity Screening. Chem Biol. 2003;10:61–68. doi: 10.1016/s1074-5521(02)00305-8. [DOI] [PubMed] [Google Scholar]
- 75.Pacholec M, Bleasdale JE, Chrunyk B, et al. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J Biol Chem. 2010;285:8340–8351. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Roessler C, Nowak T, Pannek M, et al. Chemical probing of the human sirtuin 5 active site reveals its substrate acyl specificity and peptide-based inhibitors. Angew Chemie - Int Ed. 2014;53:10728–10732. doi: 10.1002/anie.201402679. [DOI] [PubMed] [Google Scholar]
- 77.Rye PT, Frick LE, Ozbal CC, Lamarr WA. Advances in Label-Free Screening Approaches for Studying Sirtuin-Mediated Deacetylation. J Biomol Screen. 2011;16:1217–1226. doi: 10.1177/1087057111420291. [DOI] [PubMed] [Google Scholar]
- 78.Ohla S, Beyreiss R, Scriba GKE, et al. An integrated on-chip sirtuin assay. Electrophoresis. 2010;31:3263–3267. doi: 10.1002/elps.201000220. [DOI] [PubMed] [Google Scholar]
- 79.Fan Y, Ludewig R, Imhof D, Scriba GKE. Development of a capillary electrophoresis-based assay of sirtuin enzymes. Electrophoresis. 2008;29:3717–3723. doi: 10.1002/elps.200800361. [DOI] [PubMed] [Google Scholar]
- 80.Abromeit H, Kannan S, Sippl W, Scriba GKE. A new nonpeptide substrate of human sirtuin in a capillary electrophoresis-based assay. Investigation of the binding mode by docking experiments. Electrophoresis. 2012;33:1652–1659. doi: 10.1002/elps.201100641. [DOI] [PubMed] [Google Scholar]
- 81.Fan Y, Hense M, Ludewig R, et al. Capillary electrophoresis-based sirtuin assay using non-peptide substrates. J Pharm Biomed Anal. 2011;54:772–778. doi: 10.1016/j.jpba.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 82.Fan Y, Ludewig R, Scriba GKE. 9-Fluorenylmethoxycarbonyl-labeled peptides as substrates in a capillary electrophoresis-based assay for sirtuin enzymes. Anal Biochem. 2009;387:243–248. doi: 10.1016/j.ab.2009.01.038. [DOI] [PubMed] [Google Scholar]
- 83.Jiang T-F, Chong L, Yue M-E, et al. Screening neuraminidase inhibitors from glycosaminoglycan and natural extract by capillary electrophoresis. J Anal Chem. 2016;71:283–288. [Google Scholar]
- 84.Guo LP, Jiang TF, Lv ZH, Wang YH. Screening a-glucosidase inhibitors from traditional Chinese drugs by capillary electrophoresis with electrophoretically mediated microanalysis. J Pharm Biomed Anal. 2010;53:1250–1253. doi: 10.1016/j.jpba.2010.07.041. [DOI] [PubMed] [Google Scholar]
- 85.Zhang WH, Lv ZH, Jiang TF, et al. Screening tyrosinase inhibitors from traditional chinese medicine by capillary electrophoresis with electrophoretically mediated microanalysis. J Food Drug Anal. 2012;20:159–163. [Google Scholar]
- 86•.Wang H, Xu W, Cao J, Wang W. Rapid screening of aminopeptidase N inhibitors by capillary electrophoresis with electrophoretically mediated microanalysis. Electrophoresis. 2015;36:319–325. doi: 10.1002/elps.201400283. Small enzyme screen by EMMA with discussion of discrepancies in inhibitor potencies found by EMMA. [DOI] [PubMed] [Google Scholar]
- 87.Tang Z, Wang ZY, Kang JW. Screening of acetylcholinesterase inhibitors in natural extracts by CE with electrophoretically mediated microanalysis technique. Electrophoresis. 2007;28:360–365. doi: 10.1002/elps.200600327. [DOI] [PubMed] [Google Scholar]
- 88.Nehmé H, Nehmé R, Lafite P, et al. In-capillary reactant mixing for monitoring glycerol kinase kinetics by CE. J Sep Sci. 2013;36:2151–2157. doi: 10.1002/jssc.201300063. [DOI] [PubMed] [Google Scholar]
- 89•.Okhonin V, Liu X, Krylov SN. Transverse diffusion of laminar flow profiles to produce capillary nanoreactors. Anal Chem. 2005;77:5925–5929. doi: 10.1021/ac0508806. First demonstration of TDLFP as an enzyme assay method. [DOI] [PubMed] [Google Scholar]
- 90.Wong E, Okhonin V, Berezovski MV, et al. “Inject-mix-react-separate-quantitate” (IMReSQ) method for screening enzyme inhibitors. J Am Chem Soc. 2008;130:11862–11863. doi: 10.1021/ja804544x. [DOI] [PubMed] [Google Scholar]
- 91.Zhao H, Chen Z. Screening of neuraminidase inhibitors from traditional Chinese medicines by integrating capillary electrophoresis with immobilized enzyme microreactor. J Chromatogr A. 2014;1340:139–145. doi: 10.1016/j.chroma.2014.03.028. [DOI] [PubMed] [Google Scholar]
- 92.Křížek T, Doubnerová V, Ryšlavá H, et al. Offline and online capillary electrophoresis enzyme assays of β-N-acetylhexosaminidase. Anal Bioanal Chem. 2013;405:2425–2434. doi: 10.1007/s00216-012-6607-1. [DOI] [PubMed] [Google Scholar]
- 93.Nehmé H, Nehmé R, Lafite P, et al. Human protein kinase inhibitor screening by capillary electrophoresis using transverse diffusion of laminar flow profiles for reactant mixing. J Chromatogr A. 2013;1314:298–305. doi: 10.1016/j.chroma.2013.08.046. [DOI] [PubMed] [Google Scholar]
- 94.Tang Z, Kang J. Enzyme inhibitor screening by CE with an on-column immobilized enzyme microreactor created by an ionic binding technique. Anal Chem. 2006;78:2514–2520. doi: 10.1021/ac052030w. [DOI] [PubMed] [Google Scholar]
- 95.Tang Z, Wang T, Kang J. Immobilized capillary enzyme reactor based on layer-by-layer assembling acetylcholinesterase for inhibitor screening by CE. Electrophoresis. 2007;28:2981–2987. doi: 10.1002/elps.200700061. [DOI] [PubMed] [Google Scholar]
- 96.Iqbal J. An enzyme immobilized microassay in capillary electrophoresis for characterization and inhibition studies of alkaline phosphatases. Anal Biochem. 2011;414:226–231. doi: 10.1016/j.ab.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 97.Camara MA, Tian M, Guo L, Yang L. Application of capillary enzyme micro-reactor in enzyme activity and inhibitors studies of glucose-6-phosphate dehydrogenase. J Chromatogr B Anal Technol Biomed Life Sci. 2015;990:174–180. doi: 10.1016/j.jchromb.2015.03.019. [DOI] [PubMed] [Google Scholar]
- 98.Wang S, Su P, Yang Y. Online immobilized enzyme microreactor for the glucose oxidase enzymolysis and enzyme inhibition assay. Anal Biochem. 2012;427:139–143. doi: 10.1016/j.ab.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 99.Ji X, Ye F, Lin P, Zhao S. Immobilized capillary tyrosinase microreactor for inhibitor screening in natural extracts by capillary electrophoresis. Talanta. 2010;82:1170–1174. doi: 10.1016/j.talanta.2010.06.029. [DOI] [PubMed] [Google Scholar]
- 100.Zhao S, Ji X, Lin P, Liu YM. A gold nanoparticle-mediated enzyme bioreactor for inhibitor screening by capillary electrophoresis. Anal Biochem. 2011;411:88–93. doi: 10.1016/j.ab.2010.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lin P, Zhao S, Lu X, et al. Preparation of a dual-enzyme co-immobilized capillary microreactor and simultaneous screening of multiple enzyme inhibitors by capillary electrophoresis. J Sep Sci. 2013;36:2538–2543. doi: 10.1002/jssc.201300315. [DOI] [PubMed] [Google Scholar]
- 102.Heegaard NHH. Affinity in Electrophoresis. Electrophoresis. 2009;30:229–239. doi: 10.1002/elps.200900073. [DOI] [PubMed] [Google Scholar]
- 103.Galievsky VA, Stasheuski AS, Krylov SN. Capillary Electrophoresis for Quantitative Studies of Biomolecular Interactions. Anal Chem. 2015;87:157–171. doi: 10.1021/ac504219r. [DOI] [PubMed] [Google Scholar]
- 104.Deeb S, El Wätzig H, El-Hady DA. Capillary electrophoresis to investigate biopharmaceuticals and pharmaceutically-relevant binding properties. TrAC - Trends Anal Chem. 2013;48:112–131. [Google Scholar]
- 105.Zou D, Zhang D, Liu S, et al. Interplay of binding stoichiometry and recognition specificity for the interaction of MBD2b protein and methylated DNA revealed by affinity capillary electrophoresis coupled with laser-induced fluorescence analysis. Anal Chem. 2014;86:1775–1782. doi: 10.1021/ac4036636. [DOI] [PubMed] [Google Scholar]
- 106.Yang PL, Mao Y, Lee AWM, Kennedy RT. Measurement of dissociation rate of biomolecular complexes using CE. Electrophoresis. 2009;30:457–464. doi: 10.1002/elps.200800397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pedersen JT, Østergaard J, Houen G, Heegaard NHH. Affinity capillary electrophoresis for identification and investigation of human Gc-globulin (vitamin D-binding protein) and its isoforms interacting with G-actin. Electrophoresis. 2008;29:1723–1733. doi: 10.1002/elps.200700618. [DOI] [PubMed] [Google Scholar]
- 108.Shimura K, Waki T, Okada M, et al. Analysis of protein-protein interactions with a multi-capillary electrophoresis instrument. Electrophoresis. 2006;27:1886–1894. doi: 10.1002/elps.200500239. [DOI] [PubMed] [Google Scholar]
- 109.Chen X, Flynn GC. A high throughput dimer screening assay for monoclonal antibodies using chemical cross-linking and microchip electrophoresis. J Chromatogr B. 2009;877:3012–3018. doi: 10.1016/j.jchromb.2009.07.020. [DOI] [PubMed] [Google Scholar]
- 110.Ouimet CM, Shao H, Rauch JN, et al. Protein Cross-linking Capillary Electrophoresis for Protein-Protein Interaction Analysis. Anal Chem. 2016;88:8272–8278. doi: 10.1021/acs.analchem.6b02126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ouameur AA, Tajmir-Riahi H-A. Structural Analysis of DNA Interactions with Biogenic Polyamines and Cobalt(III)hexamine Studied by Fourier Transform Infrared and Capillary Electrophoresis. J Biol Chem. 2004;279:42041–42054. doi: 10.1074/jbc.M406053200. [DOI] [PubMed] [Google Scholar]
- 112.Ouameur AA, Tajmir-Riahi H-A. Probing tRNA interaction with biogenic polyamines. RNA. 2010;16:1968–1979. doi: 10.1261/rna.1994310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Quaglia M, Carazzone C, Sabella S, et al. Search of ligands for the amyloidogenic protein β 2-microglobulin by capillary electrophoresis and other techniques. Electrophoresis. 2005;26:4055–4063. doi: 10.1002/elps.200500313. [DOI] [PubMed] [Google Scholar]
- 114.Mcdonnell PA, Caldwell GW, Masucci JA. Using capillary electrophoresis / frontal analysis to screen drugs interacting with human serum proteins. Electrophoresis. 1998;19:448–454. doi: 10.1002/elps.1150190315. [DOI] [PubMed] [Google Scholar]
- 115.Liu L, Xu X, Liu Y, et al. Screening HIV-1 fusion inhibitors based on capillary electrophoresis head-end microreactor targeting to the core structure of gp41. J Pharm Biomed Anal. 2016;120:153–157. doi: 10.1016/j.jpba.2015.12.021. [DOI] [PubMed] [Google Scholar]
- 116.Hughes D. Affinity Capillary Electrophoresis Method for Assessing a Biological Interaction of a Ligand/Receptor Pair Such as G Protein Coupled Receptor and its Targets as well as for Drug Screening. 2012/0175255 A1. US. 2012
- 117•.Austin C, Pettit SN, Magnolo SK, et al. Fragment screening using capillary electrophoresis (CEfrag) for hit identification of heat shock protein 90 ATPase inhibitors. J Biomol Screen. 2012;17:868–76. doi: 10.1177/1087057112445785. Demonstration of fragment screening by CE binding assay. [DOI] [PubMed] [Google Scholar]
- 118.Shimura K, Karger BL. Affinity Probe Capillary Electrophoresis: Analysis of Recombinant Human Growth Hormone with a Fluorescent Labeled Antibody Fragment. Anal Chem. 1994;66:9–15. doi: 10.1021/ac00073a004. [DOI] [PubMed] [Google Scholar]
- 119.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 120.Arkin MR, Tang Y, Wells JA. Small-molecule inhibitors of protein-protein interactions: Progressing toward the reality. Chem Biol. 2014;21:1102–1114. doi: 10.1016/j.chembiol.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pryor NE, Moss MA, Hestekin CN. Capillary electrophoresis for the analysis of the effect of sample preparation on early stages of Aβ1 – 40 aggregation. Electrophoresis. 2014;35:1814–1820. doi: 10.1002/elps.201400012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Picou RA, Kheterpal I, Wellman AD, et al. Analysis of Aβ (1-40) and Aβ (1-42) monomer and fibrils by capillary electrophoresis. J Chromatogr B Anal Technol Biomed Life Sci. 2011;879:627–632. doi: 10.1016/j.jchromb.2011.01.030. [DOI] [PubMed] [Google Scholar]
- 123.Picou RA, Schrum DP, Ku G, et al. Separation and detection of individual AB aggregates by capillary electrophoresis with laser-induced fluorescence detection. Anal Biochem. 2012;425:104–112. doi: 10.1016/j.ab.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 124.Liyanage R, Krylova SM, Krylov SN. Minimizing adsorption of histidine-tagged proteins for the study of protein-deoxyribonucleic acid interactions by kinetic capillary electrophoresis. J Chromatogr A. 2013;1322:90–96. doi: 10.1016/j.chroma.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 125.de Jong S, Epelbaum N, Liyanage R, Krylov SN. A semipermanent coating for preventing protein adsorption at physiological pH in kinetic capillary electrophoresis. Electrophoresis. 2012;33:2584–2590. doi: 10.1002/elps.201200153. [DOI] [PubMed] [Google Scholar]
- 126.Tan L, Zheng X, Chen L, Wang Y. Quality testing of human albumin by capillary electrophoresis using thermally cross-linked poly(vinyl pyrrolidone)-coated fused-silica capillary. J Sep Sci. 2014;37:2974–2982. doi: 10.1002/jssc.201400463. [DOI] [PubMed] [Google Scholar]
- 127.You J, Zhao L, Wang G, et al. Quaternized cellulose-supported gold nanoparticles as capillary coatings to enhance protein separation by capillary electrophoresis. J Chromatogr A. 2014;1343:160–166. doi: 10.1016/j.chroma.2014.03.079. [DOI] [PubMed] [Google Scholar]
- 128.Witos J, Karesoja M, Karjalainen E, et al. Surface initiated polymerization of a cationic monomer on inner surfaces of silica capillaries: Analyte separation by capillary electrophoresis versus polyelectrolyte behavior. J Sep Sci. 2013;36:1070–1077. doi: 10.1002/jssc.201200945. [DOI] [PubMed] [Google Scholar]