Abstract
The replication frequency of plasmid R1 is determined by the availability of the initiator protein RepA. Synthesis of RepA is negatively controlled by an antisense RNA, CopA, which forms a duplex with the upstream region of the RepA mRNA, CopT. We have previously shown that the in vitro formation of the CopA-CopT duplex follows second-order kinetics and occurs in at least two steps. The first step is the formation of a transient (kissing) complex, which is subsequently converted to a persistent duplex. Here, we investigate the details of the reaction scheme and determine the rate constants of the pathway from the free RNAs to the complete duplex. Using a shortened CopA RNA (CopI) we have been able to determine the association and dissociation rate constants (k1,k-1) for the kissing complex (which are inferred to be the same for CopI-T and CopA-T), and measured the hybridization rate constant k2 (for CopA-T k2 is at least 1000-fold greater than for CopI-T). The analysis of CopA derivatives of mutant and wild-type origin shows that the rate of formation of the kissing complex is rate-limiting for the overall pairing reaction between CopA and CopT, both in vitro and in vivo. The biological implications of the kinetically irreversible RNA-RNA binding reaction scheme are discussed.
Full text
PDF

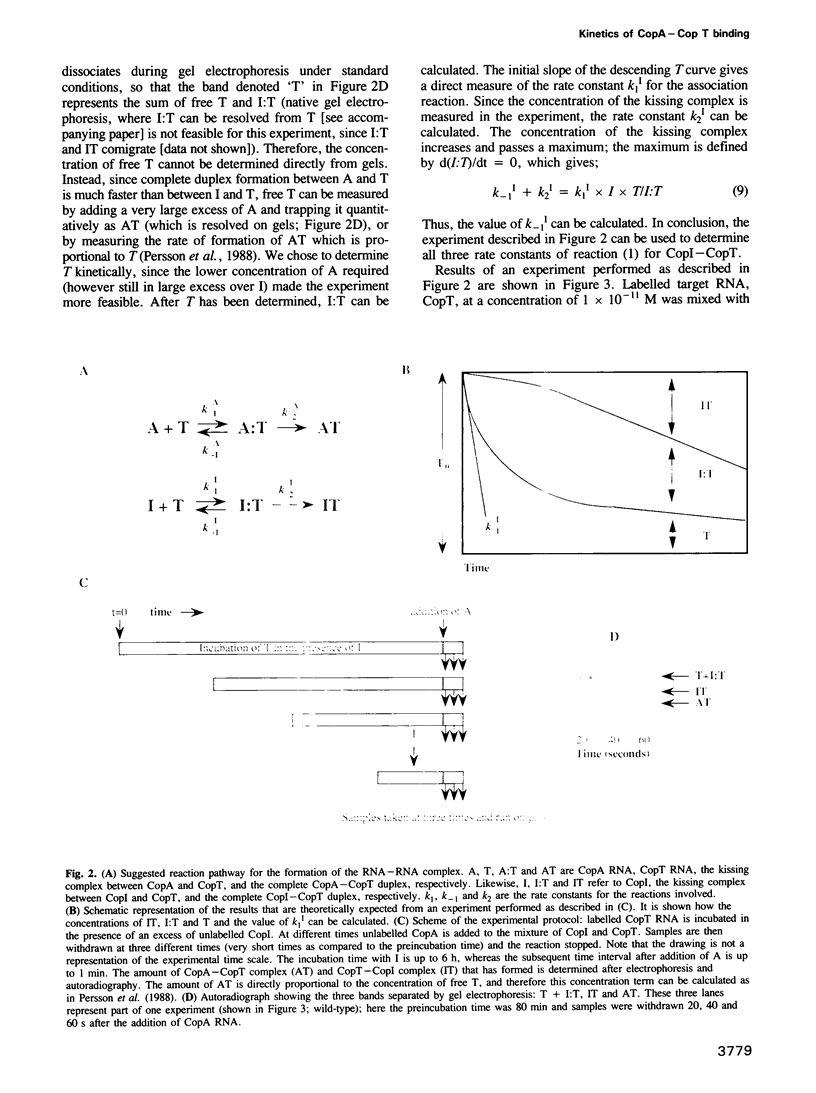
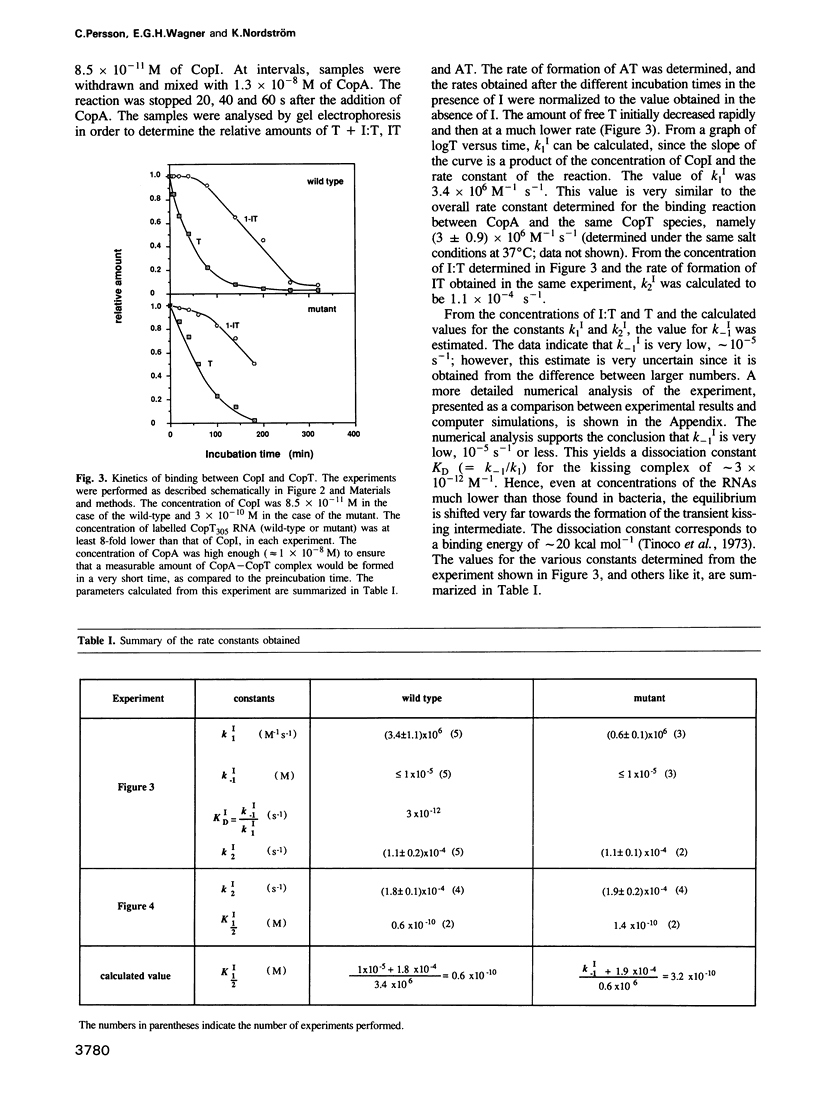





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brady G., Frey J., Danbara H., Timmis K. N. Replication control mutations of plasmid R6-5 and their effects on interactions of the RNA-I control element with its target. J Bacteriol. 1983 Apr;154(1):429–436. doi: 10.1128/jb.154.1.429-436.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig M. E., Crothers D. M., Doty P. Relaxation kinetics of dimer formation by self complementary oligonucleotides. J Mol Biol. 1971 Dec 14;62(2):383–401. doi: 10.1016/0022-2836(71)90434-7. [DOI] [PubMed] [Google Scholar]
- Danbara H., Brady G., Timmis J. K., Timmis K. N. Regulation of DNA replication: "target" determinant of the replication control elements of plasmid R6-5 lies within a control element gene. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4699–4703. doi: 10.1073/pnas.78.8.4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi Y., Tomizawa J. Complex formed by complementary RNA stem-loops and its stabilization by a protein: function of CoIE1 Rom protein. Cell. 1990 Jan 26;60(2):199–209. doi: 10.1016/0092-8674(90)90736-x. [DOI] [PubMed] [Google Scholar]
- Givskov M., Molin S. Copy mutants of plasmid R1: effects of base pair substitutions in the copA gene on the replication control system. Mol Gen Genet. 1984;194(1-2):286–292. doi: 10.1007/BF00383529. [DOI] [PubMed] [Google Scholar]
- Green P. J., Pines O., Inouye M. The role of antisense RNA in gene regulation. Annu Rev Biochem. 1986;55:569–597. doi: 10.1146/annurev.bi.55.070186.003033. [DOI] [PubMed] [Google Scholar]
- Grosjean H., Söll D. G., Crothers D. M. Studies of the complex between transfer RNAs with complementary anticodons. I. Origins of enhanced affinity between complementary triplets. J Mol Biol. 1976 May 25;103(3):499–519. doi: 10.1016/0022-2836(76)90214-x. [DOI] [PubMed] [Google Scholar]
- Kittle J. D., Simons R. W., Lee J., Kleckner N. Insertion sequence IS10 anti-sense pairing initiates by an interaction between the 5' end of the target RNA and a loop in the anti-sense RNA. J Mol Biol. 1989 Dec 5;210(3):561–572. doi: 10.1016/0022-2836(89)90132-0. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström K., Molin S., Light J. Control of replication of bacterial plasmids: genetics, molecular biology, and physiology of the plasmid R1 system. Plasmid. 1984 Sep;12(2):71–90. doi: 10.1016/0147-619x(84)90054-4. [DOI] [PubMed] [Google Scholar]
- Perelson A. S., Brendel V. Kinetics of complementary RNA-RNA interaction involved in plasmid ColE1 copy number control. J Mol Biol. 1989 Jul 20;208(2):245–255. doi: 10.1016/0022-2836(89)90386-0. [DOI] [PubMed] [Google Scholar]
- Persson C., Wagner E. G., Nordström K. Control of replication of plasmid R1: kinetics of in vitro interaction between the antisense RNA, CopA, and its target, CopT. EMBO J. 1988 Oct;7(10):3279–3288. doi: 10.1002/j.1460-2075.1988.tb03195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson C., Wagner E. G., Nordström K. Control of replication of plasmid R1: structures and sequences of the antisense RNA, CopA, required for its binding to the target RNA, CopT. EMBO J. 1990 Nov;9(11):3767–3775. doi: 10.1002/j.1460-2075.1990.tb07590.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pörschke D., Eigen M. Co-operative non-enzymic base recognition. 3. Kinetics of the helix-coil transition of the oligoribouridylic--oligoriboadenylic acid system and of oligoriboadenylic acid alone at acidic pH. J Mol Biol. 1971 Dec 14;62(2):361–381. doi: 10.1016/0022-2836(71)90433-5. [DOI] [PubMed] [Google Scholar]
- Rosen J., Ryder T., Inokuchi H., Ohtsubo H., Ohtsubo E. Genes and sites involved in replication and incompatibility of an R100 plasmid derivative based on nucleotide sequence analysis. Mol Gen Genet. 1980;179(3):527–537. doi: 10.1007/BF00271742. [DOI] [PubMed] [Google Scholar]
- Simons R. W., Kleckner N. Biological regulation by antisense RNA in prokaryotes. Annu Rev Genet. 1988;22:567–600. doi: 10.1146/annurev.ge.22.120188.003031. [DOI] [PubMed] [Google Scholar]
- Stougaard P., Molin S., Nordström K., Hansen F. G. The nucleotide sequence of the replication control region of the resistance plasmid R1drd-19. Mol Gen Genet. 1981;181(1):116–122. doi: 10.1007/BF00339014. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Tomizawa J. Control of ColE1 plasmid replication: initial interaction of RNA I and the primer transcript is reversible. Cell. 1985 Mar;40(3):527–535. doi: 10.1016/0092-8674(85)90201-6. [DOI] [PubMed] [Google Scholar]
- Tomizawa J. Control of ColE1 plasmid replication: the process of binding of RNA I to the primer transcript. Cell. 1984 Oct;38(3):861–870. doi: 10.1016/0092-8674(84)90281-2. [DOI] [PubMed] [Google Scholar]
- Twigg A. J., Sherratt D. Trans-complementable copy-number mutants of plasmid ColE1. Nature. 1980 Jan 10;283(5743):216–218. doi: 10.1038/283216a0. [DOI] [PubMed] [Google Scholar]
- Womble D. D., Rownd R. H. Regulation of IncFII plasmid DNA replication. A quantitative model for control of plasmid NR1 replication in the bacterial cell division cycle. J Mol Biol. 1986 Dec 5;192(3):529–547. doi: 10.1016/0022-2836(86)90274-3. [DOI] [PubMed] [Google Scholar]



