Abstract
Dysfunctional T cells can mediate autoimmunity, but the inaccessibility of autoimmune tissues and the rarity of autoimmune T cells in the blood hinder their study. We describe a method to enrich and harvest autoimmune T cells in vivo by using a biomaterial scaffold loaded with protein antigens. In model antigen systems, we found that antigen-specific T cells become enriched within scaffolds containing their cognate antigens. When scaffolds containing lysates from an insulin-producing β-cell line were implanted subcutaneously in autoimmune diabetes–prone NOD mice, β-cell–reactive T cells homed to these scaffolds and became enriched. These T cells induced diabetes after adoptive transfer, indicating their pathogenicity. Furthermore, T-cell receptor (TCR) sequencing identified many expanded TCRs within the β-cell scaffolds that were also expanded within the pancreata of NOD mice. These data demonstrate the utility of biomaterial scaffolds loaded with disease-specific antigens to identify and study rare, therapeutically important T cells.
Introduction
Many autoimmune diseases are mediated in part by T cells; however, very few disease-initiating autoimmune T cells have been identified either in humans or in model organisms (1–3). A large part of the challenge in identifying and studying autoimmune T cells is their rarity in the blood and, therefore, their inaccessibility. Some estimates suggest that perhaps 1 in ≤105 T cells in the blood may be relevant to ongoing autoimmune diseases (4). Analysis of circulating T cells is further confounded by the inability to freeze or culture these cells without affecting their functional capacities. Although autoimmune T cells are more abundant in tissues undergoing autoimmune attack (5), these tissues are generally inaccessible for routine studies. For example, T-cells drive β-cell destruction and cause type 1 diabetes (T1D) (6), but pancreatic tissue is generally unattainable from patients with T1D. Thus, the ongoing autoimmune T-cell responses in patients with T1D have been exceedingly difficult, if not impossible, to follow with confidence. Methods for enriching rare autoimmune T cells would enable autoimmune T-cell identification and study during disease progression as well as the testing of immune tolerance–promoting drugs.
Antigen-specific T cells can enter into inflamed tissues and proliferate upon T-cell receptor (TCR) engagement with their corresponding antigens. We therefore developed a method for the subcutaneous enrichment of autoimmune T cells by using antigen-loaded biomaterial scaffolds. Biomaterials are routinely used to control the delivery of biomolecules. We previously described the fabrication of biomaterial scaffolds to mimic infectious environments (7). When these scaffolds were loaded with tumor antigens and cytokine adjuvants, they promoted potent T-cell responses and tumor eradication. The ability of these materials to augment immune cell trafficking and deliver antigens suggests that they may be used to enrich antigen-specific T cells in vivo. We hypothesized that controlled antigen release by macroporous scaffolds could be used to recruit and harvest antigen-specific T cells in vivo. Biomaterial scaffolds were fabricated to mimic inflammatory autoimmune lesions through the controlled presentation of the broad set of antigens from β-cell lysates. We tested whether the presentation of scaffold-loaded antigens by recruited antigen-presenting cells would lead to the recruitment and expansion of autoimmune T cells.
Research Design and Methods
Cell Culture
NIT-1 cells, a pancreatic β-cell line, were obtained from American Type Culture Collection (catalog ATCC CRL-2055). They were cultured in complete DMEM/F12 containing 10% FBS.
Mice
C57BL/6 mice, OT-I C57BL/6 mice, OT-II/GFP C57BL/6 mice, female NOD mice, female NOD.SCID mice, NOD-BDC2.5 mice (8), and NOD8.3 mice (9) (The Jackson Laboratory) were used. All experiments involving animals were approved by the Institutional Animal Care and Use Committees of Harvard University and the Joslin Diabetes Center (JDC) (Boston, MA). To monitor diabetes progression in control NOD mice and NOD.SCID mice, blood glucose measurements were performed by using a standard blood glucose monitor (OneTouch) on tail vein blood. Blood measurements were taken weekly, and mice with blood glucose levels >250 mg/dL for 2 consecutive weeks were considered diabetic.
Scaffold Fabrication
A detailed protocol for scaffold fabrication is included in the Supplementary Data. Scaffolds were made by mixing antigens with poly(dl-lactide-co-glycolide) (PLG) microspheres (Degradex PLGA, LG30K; Phosphorex) before processing with gas foaming and particulate leaching. PLG microspheres (18 mg/scaffold) were mixed with either sonicated NIT-1 cell lysate at 1 × 107 cell equivalents/scaffold (∼3.6 mg protein in PBS) or ovalbumin (OVA) protein (5 mg/scaffold in double-distilled H2O). The mixture was vortexed until homogeneous and left at room temperature for 15 min. The solution was vortexed again and snap-frozen in liquid nitrogen. The mixture was then lyophilized and mixed with 200 mg of the porogen, NaCl, or 130 mg sucrose (sieved to a particle size 250–425 μm) and compression molded into discs by using a Carver Model 3850 manual press at 1,500 psi for 1 min (disc diameter 1cm, width 0.2 cm). The resulting discs were placed in a CO2 pressure cylinder, and the pressure was slowly brought up to 800 psi. The scaffolds were equilibrated in 800 psi CO2 for 16 h, and subsequently, the pressure was released over 1–2 min to induce the polymer particles to expand and fuse into an interconnected structure. The NaCl or sucrose was leached from the scaffolds by immersion in water for 4 h, yielding scaffolds that were 85–94% porous. For scanning electron microscopy, scaffolds were sputter coated with gold before visualization.
Scaffold Protein Release Assay
To measure the in vitro release kinetics of either OVA or β-cell lysate release, the scaffolds were placed in 3 mL of PBS in an incubator (37°C). At various time points, the PBS release media were collected and replaced with fresh media. The amount of protein antigen released into PBS over time was determined by a Pierce Micro BCA Protein Assay Kit (Thermo Fisher Scientific).
Scaffold Implantation
Mice were anesthetized by using a continuous flow of an isoflurane/oxygen mixture. A subcutaneous incision (1.5 cm) along the spine was made, and then either one or two subcutaneous pockets (roughly twice the size of the scaffolds) were created by using a pair of scissors. One scaffold was placed in each subcutaneous pocket. The incision along the back was closed with wound clips.
Immunization
C57BL/6 mice were immunized in footpads with ∼0.5 mg of OVA emulsified 1:1 in Complete Freund’s Adjuvant (Sigma). Twenty-one days later, mice were boosted with ∼0.5 mg of OVA emulsified in Complete Freund’s Adjuvant. Scaffolds were implanted 28 days after the second immunization with OVA. Postoperative analgesia (buprenorphine 0.05 mg/kg subcutaneous) was administered to mice undergoing surgery.
Extraction and Flow Cytometry of Cells From Scaffolds and Pancreata
Single-cell suspensions from pancreata and scaffolds were prepared by dicing followed by digestion in DMEM with 1% BSA with collagenase IV (1 mg/mL) and DNase I (10 units/mL) for 20 min at 37°C. Alternatively, scaffolds were digested with collagenase II (250 units/mL in PBS) for 20 min at 37°C. Cells were then filtered through a 70-μm mesh. In some experiments, as noted, T cells were isolated through negative selection (Pan T Cell Isolation Kit II, mouse, 130-095-130; Miltenyi Biotec). Cells were treated with Fc block (anti-mouse CD16/32) and stained with the following antibodies (hybridoma name, fluorochrome, and manufacturer in parentheses): CD45 (30-F11, PE/Cy5; BioLegend), CD11b (M1/70, PE/Cy7; BioLegend), CD11c (HL3, APC/Cy7; BD Biosciences), Gr.1 (8C5, AF680; made in-house), CD19 (6D5, PE/Cy5.5; Invitrogen), CD3ε (145-2C11, PE, BioLegend), CD4 (GK1.5, FITC, made in-house), CD8 (53-6.7, PB; BioLegend), TCR (GL3, Biotin [BioLegend] and Qdot605 streptavidin conjugate [Invitrogen]), PE-conjugated OVA class I tetramer (Beckman Coulter), and propidium iodide. Stained cells were analyzed by using a BD LSRII flow cytometer (BD Biosciences) or sorted on a FACSAria cell sorter (BD Biosciences). The flow cytometry data were analyzed using FlowJo software (Tree Star). The numbers of CD4+ and CD8+ T cells in the scaffolds were determined by spiking each sample with a set number of Accudrop beads and subsequently calculating the absolute cell number by using the results from the flow cytometry data or, alternatively, a hemacytometer. Cell yields from scaffolds ranged from 1 × 105 to 5 × 106 cells.
Adoptive Transfer of OT-II/GFP T Cells
Single-cell suspensions from spleens of OT-II/GFP C57BL/6 mice were prepared by mashing the spleens through a 70-μm mesh. Red blood cells were lysed with ammonium-chloride-potassium buffer. The single-cell suspensions were stained with PE-Cy7–conjugated antibodies to B220 (RA3-6B2), CD11b (M1/70), Gr.1 (RB6-8C5), and CD11c (N418), and negative selection was performed by using anti-Cy7 microbeads according to the manufacturer’s protocol (Miltenyi). Purified OT-II/GFP T cells (3 × 106/mouse), were retro-orbitally transplanted into C57BL/6 mice, and during the same procedure, OVA and control scaffolds were implanted on opposing flanks. After 7 days, the scaffolds were extracted and digested with collagenase IV, as described above, and the cells were stained with anti-CD3 (17A2, BV605; BioLegend), anti-B220 (RA3-6B2, PE/Cy5; BioLegend), anti-CD8a (53-6.7, PE/Cy7; BioLegend), and anti-CD4 (GK1.5, PB; BioLegend). Live cells were gated based on forward and side scatter and propidium iodide exclusion.
In Vivo Proliferation
To determine whether T cells proliferate in the scaffolds, OT-II/GFP cells were processed as previously described, labeled with 10 μmol/L CellTrace Violet (Thermo Fisher Scientific) (10), and purified by using magnetic beads as previously described. Enriched OT-II/GFP cells (12 × 106/mouse) were adoptively transferred into C57BL/6. OVA and control scaffolds were implanted during the same procedure. After 8 days, the scaffolds, spleens, and draining lymph nodes were harvested. The single-cell suspensions were stained with antibodies to TCR Vβ5.1, CD19, CD4, CD3, and CD8. Live cells were gated on the basis of forward and side scatter and propidium iodide exclusion. Cells were further gated on either GFP+ cells or unlabeled host cells and then on CD19−, CD3+, CD4+, and TCR+ cells. Levels of CellTrace Violet fluorescence were used to determine cellular proliferation.
Enzyme-Linked ImmunoSpot Analysis
Enzyme-Linked ImmunoSpot (ELISPOT) assays in 96-well plates (S2EM004M99; Fisher Scientific) were performed according the manufacturer’s guidelines. Antibodies used were anti-interleukin 2 (IL-2) (551876; BD Biosciences) and biotinylated anti-IL-2 (551876; BD Biosciences). Assays were performed in serum-free AIM V Medium (12055-091; Life Technologies). In each well, T cells and antigen-presenting cells (with or without antigen or cell lines, as indicated in each figure legend) were incubated at 37°C (5% CO2) overnight. The number of T cells from the β-cell scaffolds and the control scaffolds ranged from 2,000 to 10,000 per well (unless specifically stated in the figure legend). Plates were developed by incubating with 1:1,000 streptavidin-alkaline phosphatase enzyme conjugate (554065; BD Biosciences) for 45 min and developed using SIGMAFAST BCIP/NBT substrate (B5655; Sigma).
Histology
Scaffolds and pancreata were removed from mice, fixed in 4% paraformaldehyde, paraffin-embedded, sectioned, and stained with hematoxylin-eosin. For each group of mice implanted with β-cell or control scaffolds, 63–113 islets were scored. Islets were scored according to islet infiltration (score 0: no infiltration; score 1: leukocytes only bordering islets; score 2: leukocytes penetrating 25% of islet mass; score 3: leukocytes penetrating 75% of islet mass; score 4: end-stage insulitis, <20% of islet area remaining).
Adoptive Transfer for Diabetes Development
To determine whether T cells extracted from β-cell scaffolds could drive diabetes development, β-cell scaffolds and control scaffolds were implanted into 15-week-old NOD mice and removed 2 weeks later. As described above, single-cell suspensions were extracted from each scaffold through collagenase digestion and cell filtration. The total cells retrieved from β-cell and control scaffolds averaged 3.3 × 106 and 1.5 × 106 total cells, respectively. Scaffold-derived cells (2 × 106) were transferred into each NOD.SCID recipient through retro-orbital injection (n = 9 for β-cell scaffold recipients, n = 6 for control scaffold recipients). A subset of NOD.SCID mice (n = 6) also received 2 × 106 splenocytes isolated from the spleens of NOD mice.
TCR Sequencing
Mice were killed and immediately subjected to cardiac perfusion with 10 mmol/L EDTA/PBS. Cells were extracted from scaffolds, spleens, and pancreata as described above. CD4+ T cells were sorted into 2% FBS/HEPES. The TCR β-chain CDR3 regions were amplified and sequenced by Adaptive Biotechnologies. The analysis of TCR β-family members and clonality was performed by using immunoSEQ online software (Adaptive Biotechnologies). Clonality is a measure of TCR diversity within a pool [clonality = 1 − (entropy)/log2(number of productive unique reads)], where entropy is a measure of the distribution of individual sequences. To determine the sequence overlap between scaffolds and pancreata, we pooled the pancreas sequences and used them as the reference for comparison with the TCR sequences from either the β-cell scaffolds or the control scaffolds.
Transduction of NIT-1 Cells
To detect β-cell–specific T cells in ELISPOT assays, we generated a modified NIT-1 cell line that expressed high levels of murine CD86 and either MHC class I or MHC class II. To induce MHC class I, NIT-1 cells were transduced with lentivirus encoding the transcription factor NLRC5. Similarly, to induce MHC class II, NIT-1 cells were transduced with lentivirus encoding CIITA. CD86 was also encoded in a lentiviral vector. After lentiviral transduction, we sorted NIT-1 cells expressing high levels of MHC class I and CD86 (NIT-KD) as well as NIT-1 expressing high levels MHC class II and CD86 (NIT-G7). The NLRC5, CIITA, and CD86 cDNAs were generated by PCR and were inserted into a modified lentiviral pCDH vector (System Biosciences). Lentiviruses were generated in 293T cells by calcium phosphate transfection of lentiviral plasmids plus pCMV delta R8.2 (plasmid 12263; Addgene) and pMD2.G (12259; Addgene), which were gifts from D. Trono (EPFL, Lausanne, France).
Statistical Analysis
Statistical analysis was performed with GraphPad Prism software (La Jolla, CA). Where indicated, values were expressed as mean ± SD. Statistical significance of differences between the groups were analyzed by a two-tailed Student t test, and the exact P values are reported in the figures. P < 0.05 was considered significant. Kaplan-Meier curve statistical analysis was done by using the log-rank test.
Results
Antigen-Specific T Cells Home to OVA-Containing Scaffolds
We engineered porous, antigen-loaded, PLG matrices by using a high-pressure CO2 gas-foaming and particulate-leaching technique (11). As a model antigen, we incorporated OVA into the PLG matrix. The OVA-loaded scaffolds are highly porous (Fig. 1A), allowing space for T-cell infiltration and expansion. These scaffolds also sustained OVA release in vitro for >20 days (Fig. 1B).
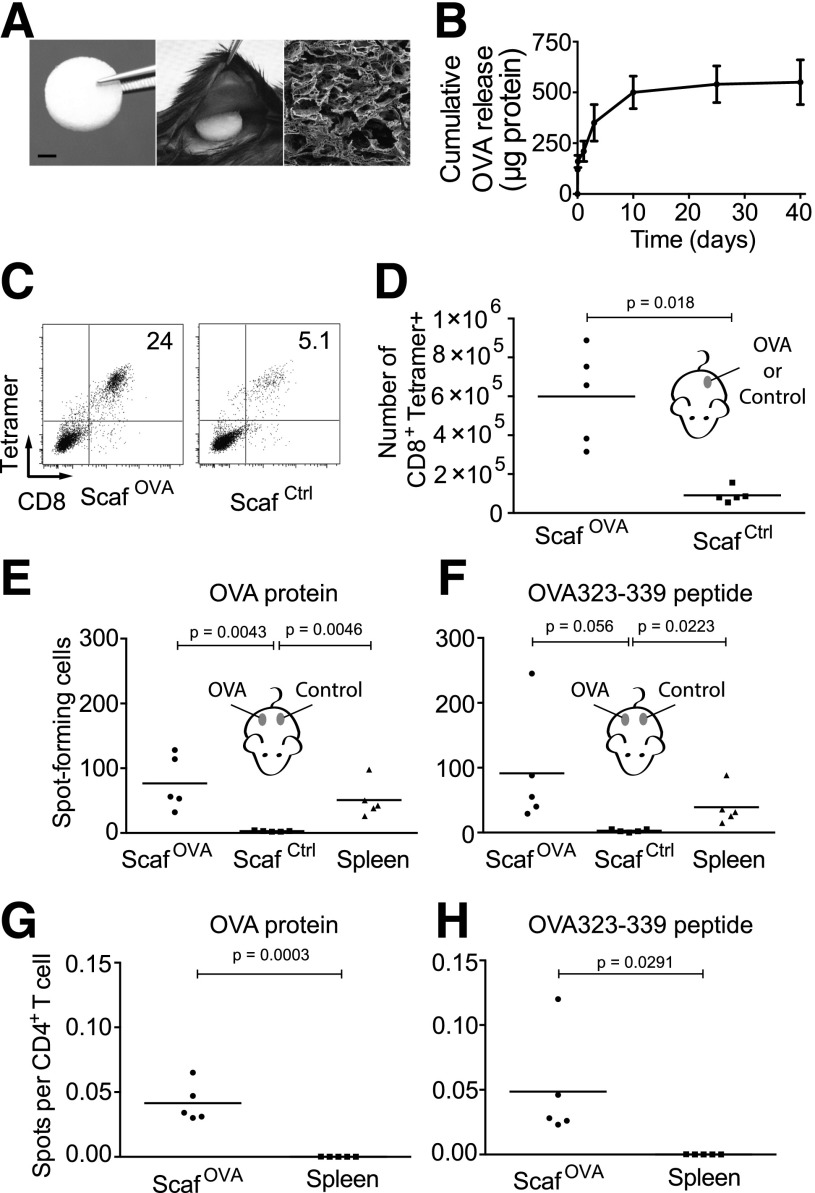
Figure 1.
OVA-specific T cells become enriched in OVA-loaded scaffolds. A: The size (scale bar = 2 mm) and implantation of a PLG scaffold and a scanning electron microscope micrograph of a cross-section of a scaffold. B: The cumulative in vitro release of OVA from PLG scaffolds was analyzed over a period 40 days. C: Flow cytometric plots of OVA-specific, tetramer+, CD8+ T cells in OVA-loaded or control scaffolds at 14 days postimplantation in OT-I mice. Plots show live-gated cells. D: The numbers of OVA tetramer+, CD8+ T cells in OVA and control scaffolds. Each point represents the scaffold T cells from one mouse. E: C57BL/6 mice were immunized twice with OVA, and 28 days postimmunization, each mouse was implanted with OVA and control scaffolds on opposing flanks. After 14 days, cells were extracted from the scaffolds and spleens and were analyzed for IL-2 secretion in an ELISPOT assay in response to 10 μg/mL whole OVA protein or 10 μg/mL OVA323-339 peptide (F). G: The frequency of OVA-specific T cells was determined in the OVA scaffolds and spleens of immunized mice. For spleen samples, 5 × 105 splenocytes from immunized mice were added per well. For OVA scaffolds, one-fourth of the total scaffold-derived cells were added per well, and 2.5 × 105 splenocytes from unimmunized C57BL/6 mice were added to ensure efficient antigen presentation. A portion of splenocytes and scaffolds were analyzed by flow cytometry to determine the CD4+ T-cell frequency, and this was used to determine the frequency of spots per CD4+ T cell in each sample. Each well contained 10 μg/mL whole OVA protein or 10 μg/mL OVA323-339 peptide (H). ScafCtrl, control scaffolds; ScafOVA, OVA-loaded scaffolds.
To determine the ability of antigen-loaded scaffolds to recruit both CD4+ and CD8+ T cells in an antigen-specific manner, we used three different OVA-specific T-cell systems. To determine whether CD8+ T cells become enriched within OVA scaffolds, we implanted either OVA scaffolds or control scaffolds (without protein) subcutaneously (Fig. 1A) in mice expressing a TCR transgene (OT-I) that specifically recognizes an OVA-derived peptide (OVA257-264) presented by MHC class I (12). We found that after 14 days in vivo, the OVA scaffolds contained approximately sevenfold more OVA-specific CD8+ T cells than the control scaffolds, indicating that the implanted scaffolds directed antigen-specific recruitment, retention, and/or expansion of OVA-specific CD8+ T cells (Fig. 1C and D).
Next, CD4+ T-cell enrichment was tested by immunizing wild-type mice with OVA before implantation of OVA and control scaffolds on opposite flanks. OVA scaffolds recruited ∼30-fold more OVA-specific T cells relative to control scaffolds as determined by an IL-2 ELISPOT assay that used either whole OVA protein or the dominant MHC class II–restricted peptide OVA323-339 as antigen sources (Fig. 1E and F and Supplementary Fig. 1A and B). Approximately 1 in 20 CD4+ T cells within OVA scaffolds were antigen-specific, whereas OVA-specific T cells in the spleens of the same animals were present at ∼1 in 10,000, indicating a 500-fold enrichment within the OVA scaffolds (Fig. 1G and H and Supplementary Fig. 1C).
We next investigated the recruitment of naïve, circulating antigen-specific T cells. T cells expressing GFP and a TCR transgene (OT-II), which specifically recognizes the OVA-derived peptide (OVA323-339) presented by MHC class II, were adoptively transferred into wild-type mice (13). Subsequently, one OVA scaffold and one control scaffold were implanted on opposite flanks. After 7 days, the scaffolds were removed, and the frequencies of OT-II/GFP CD4+ T cells were determined in the scaffolds and spleens. Approximately 22% of the CD4+ T cells in OVA scaffolds were OT-II/GFP CD4+ T cells compared with only 2% and 6% of the CD4+ cells in control scaffolds and spleens, respectively (Fig. 2A and B). These results indicate that antigen-loaded PLG scaffolds become preferentially populated by T cells that are specific for the loaded antigens.
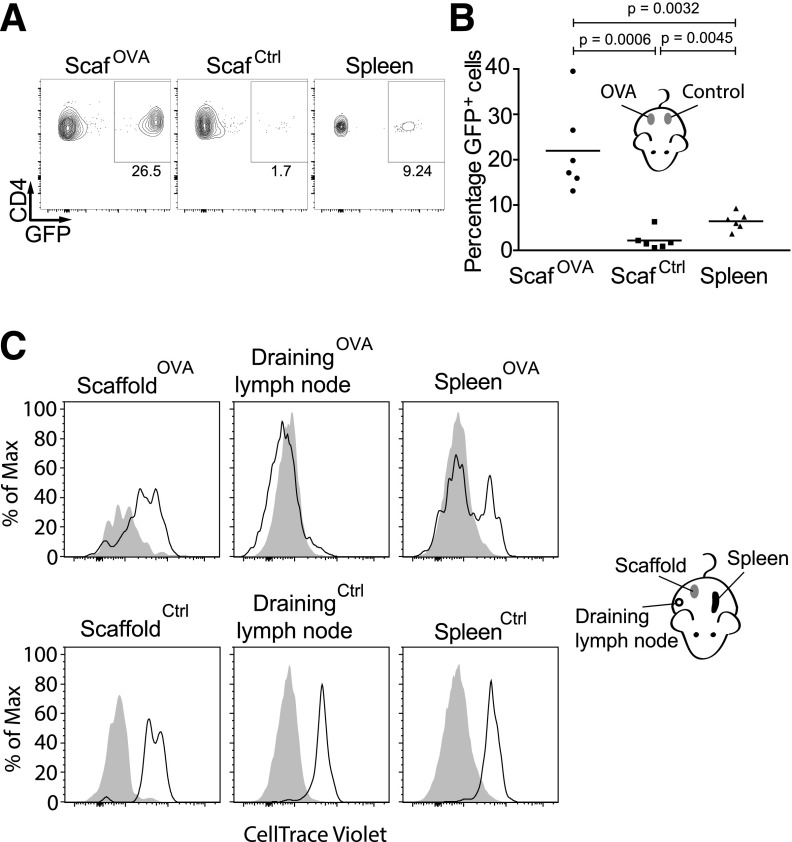
Figure 2.
Antigen-specific enrichment and proliferation of naïve CD4+ T cells in scaffolds. A: C57BL/6 mice were intravenously injected with 3 × 106 OT-II/GFP CD4+ T cells and implanted with OVA and control scaffolds on opposite flanks. After 7 days, scaffold-derived T cells were analyzed by flow cytometry. Representative FACS plots show the frequencies of OT-II/GFP CD4+ T cells in OVA and control scaffolds. Cells are pregated on live CD4+ T cells. B: Scaffold T-cell frequencies were determined in six mice. C: CD4+ T cells were enriched from OT-II/GFP mice and labeled with CellTrace Violet dye. The labeled cells were transferred intravenously into C57BL/6 mice that contained either OVA or control scaffolds implanted subcutaneously. After 8 days, the cells in the scaffolds were analyzed by flow cytometry. The histograms indicate the level of CellTrace Violet dye fluorescence of gated TCR Vβ5.1+ CD4+ GFP+ cells from scaffolds, spleens, and the inguinal lymph nodes of the recipients. Lower levels of fluorescence indicate greater proliferation. Shaded control histograms indicate the background fluorescence of unlabeled host TCR Vβ5.1+ T cells. ScafCtrl, control scaffolds; ScafOVA, OVA-loaded scaffolds.
To determine whether antigen-specific T cells proliferate in the scaffolds or are recruited without proliferation, we labeled naïve OT-II T cells with CellTrace Violet dye, which enables evaluation of proliferation through dye dilution. After adoptive transfer, OVA scaffolds were implanted, and 8 days postimplantation, the proliferation of OT-II T cells was assessed in the scaffolds, spleens, and draining lymph nodes. The dye dilution indicated that OT-II cells proliferated a small amount in the OVA scaffolds, whereas they proliferated extensively in the draining lymph node (Fig. 2C). This experiment suggests that enrichment of antigen-specific T cells within antigen-loaded scaffolds is due to antigen-specific recruitment rather than to intrascaffold proliferation. The extensive proliferation within the draining lymph nodes may reflect leakage of OVA from the scaffolds (Fig. 1A) or might also be due to migration of antigen-presenting cells from the scaffolds to the draining lymph nodes.
β-Cell Lysate-Containing Scaffolds Locally Enrich Autoimmune T Cells in NOD Mice
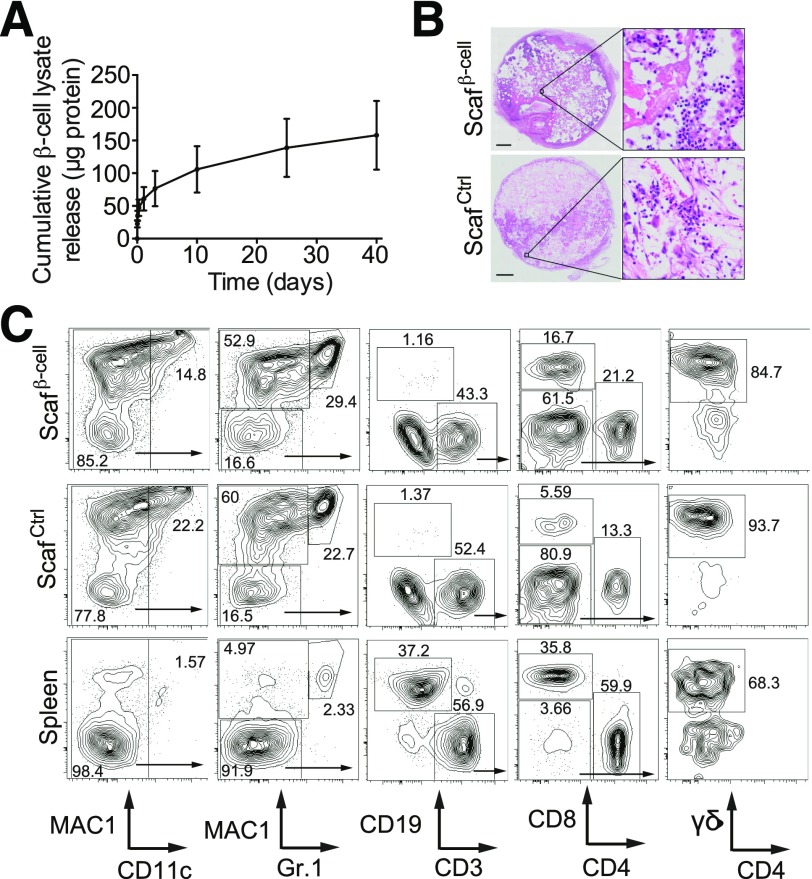
The utility of the scaffolds for enriching antigen-specific T cells in vivo suggests that scaffolds might be useful for enriching rare, autoimmune T cells. NOD mice develop spontaneous autoimmune diabetes. The development of diabetes in NOD mice has many parallels with human T1D, including the major role of autoimmune T cells in driving β-cell destruction (14). Therefore, we tested whether the scaffolds could be used to enrich autoimmune T cells in these mice. Because β-cells in the pancreas are the target of T cells in T1D, we incorporated cell lysates from the NOD-derived β-cell line NIT-1 (15) in the scaffolds (herein, β-cell scaffolds). β-Cell scaffolds released protein for >40 days in a similar fashion to OVA scaffolds (Figs. 1B and 3A). To examine whether β-cell scaffolds can enrich autoimmune T cells, we implanted either β-cell scaffolds or control scaffolds (without protein) subcutaneously in mice and removed the scaffolds for analysis after 14 days. Hematoxylin-eosin staining of the scaffolds after extraction revealed abundant infiltrates of hematopoietic cells both in the β-cell scaffolds and in the control scaffolds (Fig. 3B). Flow cytometry of scaffold-derived cells revealed multiple hematopoietic lineages within the scaffolds, including CD4+, CD8+, and γδ-T cells (Fig. 3C and Supplementary Table 1). Both the β-cell scaffolds and the control scaffolds also contained abundant Mac1+ myeloid lineages (macrophages, granulocytes, and dendritic cells). The recruitment of these lineages to the scaffolds is likely due to the inflammatory properties of the PLG material itself (11) because both the β-cell scaffolds and the control scaffolds recruit similar assortments of cells. The myeloid and dendritic cells likely play important roles in the phagocytosis and presentation of scaffold-derived antigens. Of note, the number of CD4+ and CD8+ T cells in both β-cell scaffolds and control scaffolds increased in parallel with blood glucose levels (Supplementary Fig. 2). This observation suggests that T-cell migration may change during progression of diabetes.
Figure 3.
β-Cell lysate scaffolds recruit multiple hematopoietic lineages. A: Cumulative release of protein from PLG scaffolds loaded with β-cell lysate was analyzed over a period of 40 days. B: Hematoxylin-eosin staining of sections of β-cell and control scaffolds at day 14 postimplantation in NOD mice. Scale bar = 1 mm. C: FACS plots of hematopoietic lineages within β-cell and control scaffolds and spleens explanted 14 days postimplantation in 14-week-old NOD mice. Frequencies of dendritic cells (CD11c+), granulocytes (Mac1+ Gr.1+), monocytes (Mac1+ Gr.1−), B cells (CD19+), and CD4+, CD8+, and γδ-TCR+ T cells were determined. Plots were pregated on live CD45+ cells, and arrows within plots indicate subgating. Scafβ-cell, β-cell scaffolds; ScafCtrl, control scaffolds.
β-Cell Scaffolds Contain β-Cell–Reactive T Cells
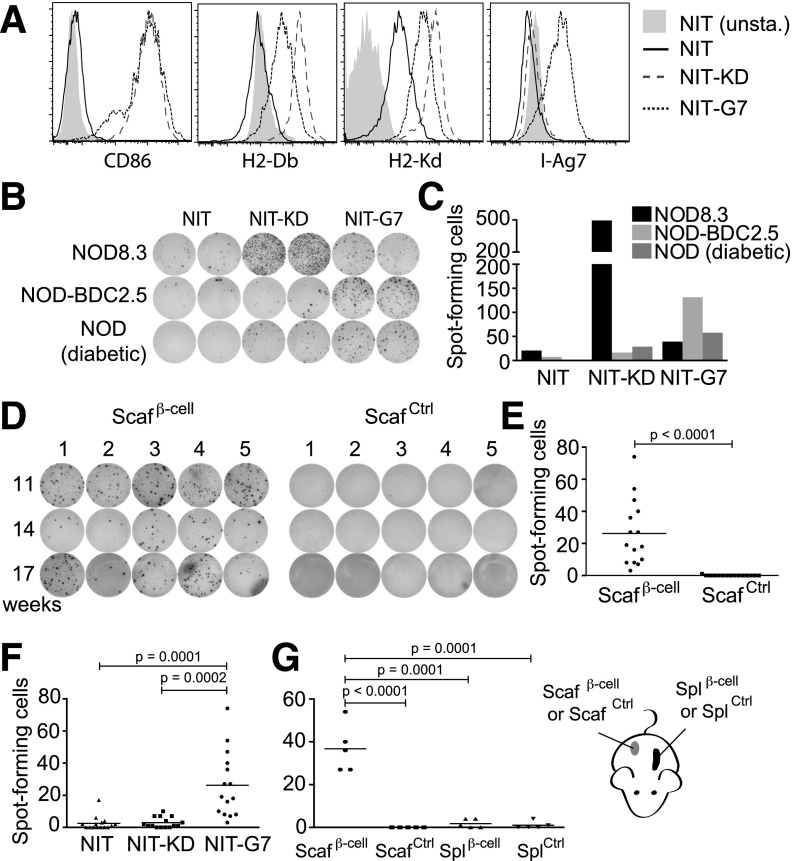
To determine whether the β-cell scaffolds contained T cells that are specific for β-cell antigens, we established an ELISPOT assay to measure β-cell–reactive T cells. We modified the NOD-derived β-cell line NIT-1 to improve its ability to present antigens to T cells. We stably transduced NIT-1 cells with the transcriptional regulators NLRC5 or CIITA, which upregulate the MHC class I and MHC class II pathways (16,17). CIITA was previously shown to upregulate autoantigen presentation on NIT-1 cells (18) (Fig. 4A). We also transduced these cells with the costimulatory molecule B7.2 (CD86). Live NIT-1 cells expressing NLRC5 (NIT-KD) directly stimulated CD8+ T cells from NOD8.3 mice, which recognize an IGRP-derived peptide (Fig. 4B and C). Similarly, live CIITA-expressing NIT-1 cells (NIT-G7) potently stimulated CD4+ T cells from BDC2.5 mice, which recognize a β-cell–specific peptide presented by IA-G7. Therefore, addition of antigen processing and costimulatory molecules to NIT-1 cells enabled them to directly stimulate autoimmune T cells in an ELISPOT assay. We next used the modified NIT-1 β-cells to test whether scaffolds could enrich autoimmune T cells.
Figure 4.
β-Cell scaffolds enrich T cells specific for β-cell proteins. A: NIT-1 cells (NIT) were transduced with CD86 and either NLRC5 (NIT-KD) or CIITA (NIT-G7) and were analyzed by flow cytometry for expression of CD86, MHC class I (H2-Db and H2-Kd), and MHC class II (I-Ag7) or left unstained (NIT unsta.). B: Splenocytes (5 × 105/well) from NOD8.3, NOD-BDC2.5, and diabetic NOD mice were analyzed for IL-2 secretion in an ELISPOT assay in response to live NIT-1, NIT-KD, and NIT-G7 cells (1 × 105/well). T cells from NOD-BDC2.5 and NOD8.3 transgenic mice express TCRs that recognize peptides derived from the β-cell proteins on I-Ag7 and H2-Kd, respectively. C: Quantification of IL-2+ T-cell spots in B by using an ELISPOT analyzer. D: β-Cell and control scaffolds were removed from the backs of 11-, 14-, and 17-week-old NOD mice at 14 days postimplantation. Cells were extracted from the scaffolds and tested for IL-2 production in response to live NIT-G7 cells in an ELISPOT assay. E: IL-2 production was quantified with an ELISPOT analyzer. F: IL-2 production measured in response to live NIT, NIT-KD, and NIT-G7 cells from three cohorts of mice (11, 14, and 17 weeks old). G: T cells (7 × 103/well) from β-cell scaffolds, control scaffolds, or spleens (Spl) from mice implanted with either β-cell scaffolds or control scaffolds were assayed in an IL-2 ELISPOT on live NIT-G7 cells and quantified. Scafβ-cell, β-cell scaffolds; ScafCtrl, control scaffolds.
Mice were implanted with either β-cell scaffolds or control scaffolds, and then T cells within the scaffolds were assayed for their ability to respond to live NIT-G7 cells in an ELISPOT assay. β-Cell scaffolds contained significantly more NIT-G7–reactive T cells than control scaffolds (Fig. 4D and E). The majority of the β-cell–specific T cells in the β-cell scaffolds were restricted to MHC class II, as evidenced by ninefold more T cells reacting to the MHC class II–expressing NIT-G7 than to the MHC class I–expressing NIT-KD cells (Fig. 4F). To determine whether the β-cell–specific T cells were enriched in β-cell scaffolds compared with the spleens and control scaffolds, we assayed the same number of T cells derived from spleens or from scaffolds on NIT-G7. These data revealed 0.5% of NIT-G7–reactive T cells in the β-cell scaffolds compared with 0% in the control scaffolds and 0.02% in the spleens (Fig. 4G). These data show that autoimmune T cells preferentially populate β-cell scaffolds, where they become significantly enriched over both control scaffolds and spleens.
β-Cell Scaffold–Derived T Cells Induce Diabetes in NOD.SCID Mice
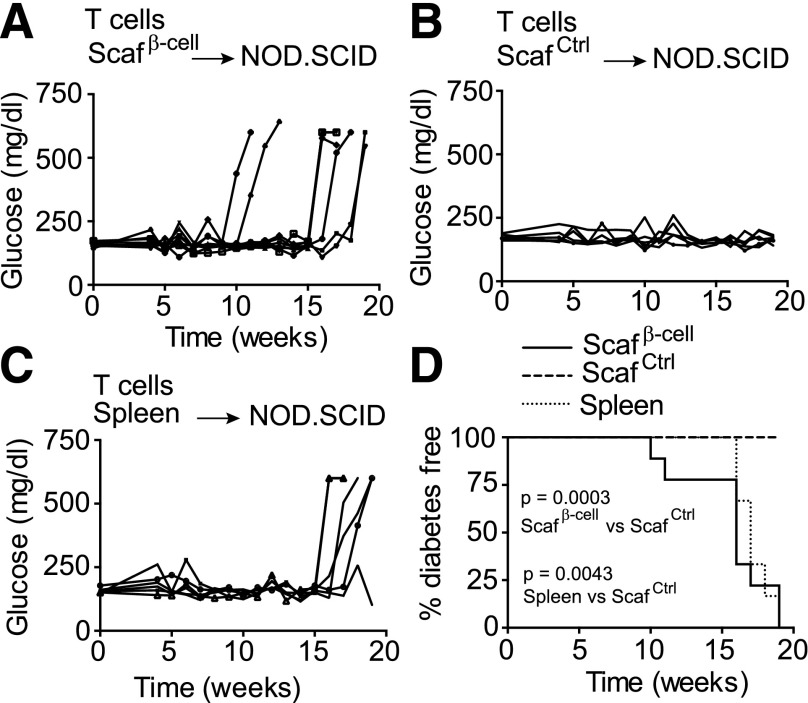
We next tested whether the T cells that were enriched in the β-cell scaffolds were disease relevant and could cause diabetes upon transplantation. NOD.SCID mice lack both T and B cells and can be used to assess the ability of adoptively transferred T cells to induce diabetes (19). β-Cell scaffolds or control scaffolds were implanted in NOD mice for 14 days. Scaffold-derived T cells were then adoptively transferred into NOD.SCID mice, and diabetes development was tracked by weekly glucose measurements. We found that 100% (nine of nine) of recipients of the β-cell scaffold–derived T cells developed diabetes 10–20 weeks after transfer, whereas recipients of T cells from control scaffolds did not (zero of six) (Fig. 5A, B, and D). As controls, NOD.SCID mice were transplanted with splenic T cells; these also developed diabetes (five of six) as predicted (Fig. 5C and D). These results indicate that β-cell scaffolds mimic the antigenic environment of the pancreas and become populated by β-cell–reactive, diabetes-inducing T cells.
Figure 5.
T cells extracted from β-cell lysate scaffolds induce diabetes after adoptive transfer into NOD.SCID mice. β-Cell or control scaffolds were implanted in NOD mice for 14 days, and then scaffold-derived cells were isolated. A–C: T cells from β-cell scaffolds, control scaffolds, and spleens were adoptively transferred into NOD.SCID mice. Blood glucose levels in recipient mice were monitored weekly. D: Kaplan-Meier plot shows diabetes incidence in mice that received β-cell scaffold–derived cells, control scaffold–derived cells, or splenocytes. Mice with two consecutive readings of >250 mg/dL were considered diabetic. Scafβ-cell, β-cell scaffolds; ScafCtrl, control scaffolds.
TCR Sequences Overlap Between β-Cell Scaffold– and Pancreas-Derived T Cells
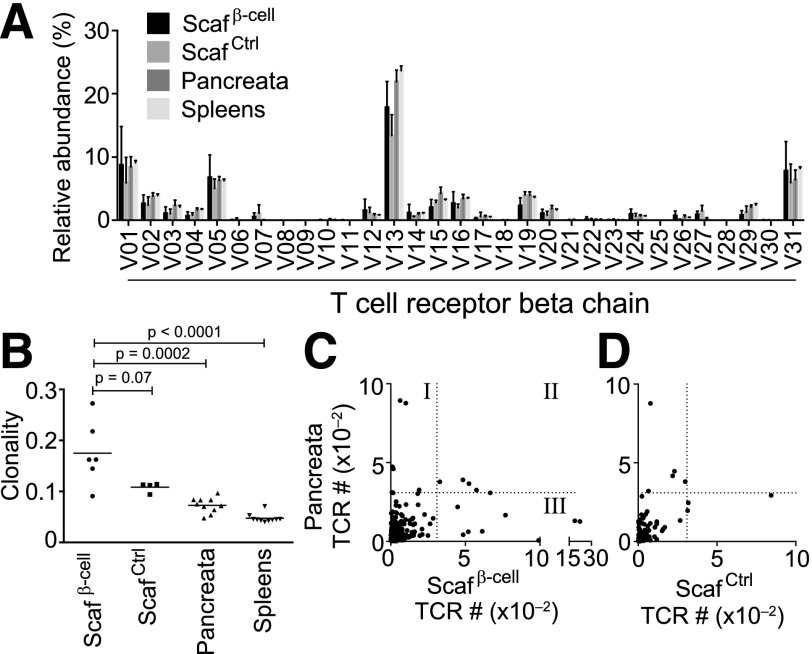
The finding that T cells within β-cell scaffolds induced T1D suggests that this population of scaffold-derived T cells represents the population of T cells within the pancreas. To test this hypothesis, TCR sequences from pancreata and scaffolds were compared. Because the ELISPOT data indicated that reactive T cells in the β-cell scaffolds were primarily MHC class II restricted, TCR analysis focused specifically on CD4+ T cells. We implanted β-cell or control scaffolds in NOD mice for 14 days. Upon scaffold removal, we also harvested the pancreata and spleens. CD4+ T cells were sorted from scaffolds, spleens, and pancreata, and the highly diverse CDR3 regions of their TCRβ chains were sequenced (20). There are 31 TCR Vβ-family members. Initial analysis of the sequencing data revealed a similar distribution of these Vβ-subunits among scaffolds, pancreata, and spleens, consistent with diverse populations of T cells occupying each of these sites (Fig. 6A).
Figure 6.
T cells from β-cell scaffolds have partially overlapping TCR repertoires with pancreas-derived T cells. β-Cell or control scaffolds were implanted in NOD mice for 14 days. Cells were extracted from β-cell and control scaffolds, spleens, and pancreata. CD4+ T cells were sorted, and their TCRβ chains were sequenced. A: Distribution of TCR Vβ-region usage in T cells from β-cell and control scaffolds, spleens, and pancreata. B: TCRβ sequences from CD4+ T-cell populations in β-cell and control scaffolds, pancreata, and spleens were used to calculate the clonality of each population. C: TCRβ protein sequences that overlapped between scaffolds and pancreata. The number of times each sequence was found in pancreata and β-cell scaffolds or control scaffolds (D) are plotted. The dotted lines mark 3 SDs above the mean and represent highly expanded clones in each tissue. Scafβ-cell, β-cell scaffolds; ScafCtrl, control scaffolds.
Because the majority of T-cell clones have unique sequences in their antigen-recognizing CDR3 regions, we used the CDR3 sequences to determine the frequency of T-cell clones that overlapped among pancreata, spleens, and scaffolds. To determine whether individual T-cell clones expanded disproportionately within the scaffolds, we calculated the clonality of the T-cell populations within each site (21). A monoclonal population has a clonality of 1, and a perfectly diverse population has a clonality of 0. By this measurement, the clonality of TCR sequences within the β-cell scaffolds was more than double the clonality of TCRs in pancreata and spleens, strongly suggesting that T cells within the β-cell scaffolds had undergone clonal expansion (Fig. 6B).
To determine whether the T-cell populations within the β-cell scaffolds overlapped with those in the pancreata, we compared CDR3 sequences among those sites. Of note, β-cell scaffolds contained more highly expanded clones (present 3 SDs above the mean) than the control scaffolds (13 vs. 3) (Fig. 6C and D and Supplementary Table 2). Additionally, four highly expanded clones in the β-cell scaffolds (∼31% of highly expanded clones) overlapped with highly expanded clones in the pancreas (Supplementary Table 2). In contrast, control scaffolds were devoid of expanded TCR clones that were highly expanded in the pancreata. Taken together with the previous experiments that show both the β-cell reactivity and the pathogenicity of β-cell scaffold–derived T cells, the expanded TCRβ sequences within the β-cell scaffolds may represent a subset of pathogenic T cells that contribute to autoimmune diabetes.
In Vivo T-Cell Enrichment Does Not Promote Disease Progression
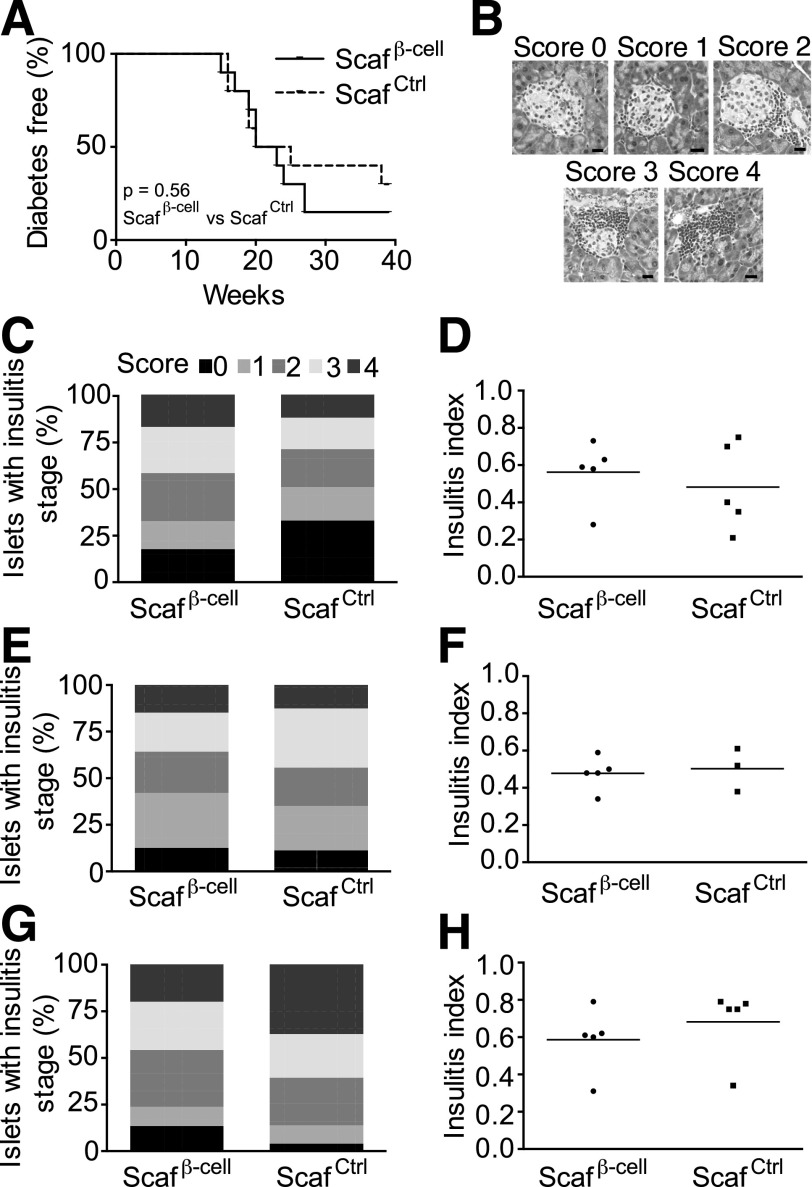
We next asked whether antigen-loaded scaffolds contribute to the progression of autoimmunity because this is a critical safety consideration for potential use of scaffolds in humans as well as an experimental concern for using scaffolds in animals. To address this risk, we implanted β-cell scaffolds or control scaffolds in NOD mice and followed blood glucose levels over time. Mice implanted with β-cell scaffolds or control scaffolds developed diabetes at the same normal rate as untreated NOD mice (Fig. 7A and Supplementary Fig. 3). To further investigate the autoimmune pathogenesis, we quantified mononuclear cell infiltrates in islets at 14 days postimplantation of β-cell or control scaffolds in mice of various ages. The pancreata were harvested, and mononuclear cell infiltrates in the islets were quantified. The insulitis score from 0 to 4 was based on the histopathological features of islets (Fig. 7B). The results revealed no significant differences in insulitis index between β-cell and control scaffolds in any of the cohorts (Fig. 7C–H). These data indicate that implanted scaffolds enrich for autoimmune T cells locally at the scaffold site but do not affect diabetes development or gross immune cell infiltration within the pancreas.
Figure 7.
β-Cell scaffold implantation does not induce diabetes. β-Cell or control scaffolds were implanted in NOD mice at 10 weeks of age, and weekly glucose measurements were taken until the age of 40 weeks. A: A Kaplan-Meier plot shows diabetes incidences in mice that were transplanted with either β-cell or control scaffolds. Diabetes was defined as two consecutive blood glucose readings >250 mg/dL. In each group, 10 mice were followed for diabetes development. B: Representative pancreas sections illustrate islet leukocytic infiltration and the determination of insulitis score. Islets were rated 0–4 according to the level of leukocytic infiltration. NOD mice were implanted with β-cell or control scaffolds, and insulitis scores were determined 14 days after implantation. Scale bars = 20 μm. The first cohort of mice was implanted at 9 weeks, and insulitis scores (C) and insulitis indexes (D) were determined at 11 weeks. The second cohort was implanted at 15 weeks, and insulitis scores (E) and insulitis indexes (F) were determined at 17 weeks. The third cohort was implanted at 22 weeks, and insulitis scores (G) and insulitis indexes (H) were determined at 24 weeks. Insulitis index: I = (0 × number of score 0) + (1 × number of score 1) + (2 × number of score 2) + (3 × number of score 3) + (4 × number of score 4)/(total number of scored islets). From each group of β-cell or control scaffolds, 63–113 islets were scored. Scafβ-cell, β-cell scaffolds; ScafCtrl, control scaffolds.
Discussion
T cells play a major role in T1D, but the study of these T cells has been limited by the inaccessibility of the human pancreas and the rarity of autoimmune T cells in the blood. We found that antigen-specific T cells home to scaffolds that contain their cognate antigens. Moreover, we demonstrate that β-cell scaffolds can be used to safely enrich for the β-cell–reactive T cells that drive autoimmune diabetes progression. These observations suggest that PLG scaffolds loaded with β-cell antigens can act as pancreas surrogates for autoimmune T-cell populations and offer a new method for isolating autoimmune T cells.
PLG scaffolds have the potential to be highly complementary to tetramer, sequencing, and other single-cell approaches for the identification of autoimmune T cells. By preenriching autoimmune T-cell populations in vivo, the measurement of T-cell frequency along with specificity and/or sequence in scaffolds could be a novel approach for monitoring autoimmune disease status. This study did not analyze the exact antigen specificities, although β-cell scaffolds may offer an approach for studying how antigen specificities and targeting changes during the course of autoimmune diabetes. This information may also be used for autoantigen discovery and to identify novel drug targets.
Although the β-cell scaffolds recruit and enrich autoimmune T cells in NOD mice, they do not appear to accelerate diabetes progression or increase islet infiltration, suggesting that β-cell scaffolds are safe from a diabetes standpoint. This finding is somewhat surprising because the response to the scaffold antigens is not limited to the scaffolds themselves but extends to the nearby lymph nodes. In scaffolds loaded with OVA, we found extensive expansion of OVA-specific T cells in the draining lymph nodes. This result suggests that additional signals may be required before autoimmune T cells that expand in a peripheral lymph node are able to exacerbate diabetes progression. Despite the findings of safety in this study, design modifications may be needed before using antigen-loaded scaffolds in patients with autoimmune disease. For example, scaffolds may be designed to immobilize antigens by covalent attachment or other conjugation techniques to better localize antigen presentation and T-cell accumulation.
Of note, antigen-specific T cells proliferate far less within scaffolds than within the nearby lymph node, which suggests that environmental signals within the scaffolds, although sufficient to drive antigen-specific T-cell localization, are inadequate for promoting T-cell proliferation. Thus, PLG scaffolds, in addition to their use in enriching antigen-specific T cells, may offer a platform for separately evaluating environmental signals that modulate T-cell expansion and localization.
In addition to T1D, other T-cell–mediated autoimmune diseases also involve tissues that are relatively inaccessible, such as in multiple sclerosis and ankylosing spondylitis. Scaffolds loaded with antigens from nervous system tissue or spine could enable efficient isolation of T cells important to these diseases. In addition to autoimmune T cells, rare T cells specific for virtually any antigen, including tumor antigens, could be enriched by using the scaffolds described here.
PLG is routinely used medically as a resorbable suture material (22). In addition, PLG scaffolds containing tumor cell lysate along with adjuvants are currently being used in a phase I cancer vaccine trial for the treatment of metastatic melanoma (ClinicalTrials.gov identifier NCT01753089), setting a clinical precedent for these antigen-loaded materials. The widespread use of PLG in the clinic and the undetectable effect of β-cell scaffolds on diabetes progression in mice both suggest that antigen-loaded PLG scaffolds could have utility in the clinic to enrich rare, antigen-specific T cells. Antigen-loaded scaffolds offer a new approach for studying the immune cell components of autoimmune disease or tumor biology to advance therapeutic modalities.
Supplementary Material
Article Information
Acknowledgments. The authors thank Aldo Rossini (JDC) for helping to initiate and support this study. They also thank MiJeong Kim (JDC) for helpful advice and David Pober (JDC) for statistical advice. The authors acknowledge the excellent support of Adaptive Biotechnologies, the Joslin Animal Facility, and the Joslin Flow Cytometry Core. The authors also acknowledge technical support from Alexander O’Donovan (JDC), Sara E. Vazquez (JDC), and Vani Manchanda (JDC).
Funding. The JDC Flow Cytometry Core is supported by a National Institute of Diabetes and Digestive and Kidney Diseases grant (P30-DK-036836). This work was supported in part by a Mary K. Iacocca Fellowship provided by the Iacocca Foundation to M.A.T. and in part by fellowships from the Swedish Society of Medicine, The Tegger Foundation, and the Swedish Society for Medical Research to M.A.T. and the Fleisher Family Foundation, the Pittsburgh Foundation, and the Harvard Stem Cell Institute to T.S.
Author Contributions. M.A.T., S.K., S.T.K., S.A.V., T.S., and O.A.A. performed experiments. D.W. provided reagents. M.A.T., F.V., A.L.W., D.J.M., T.S., and O.A.A. provided intellectual contributions. M.A.T., D.J.M., T.S., and O.A.A. analyzed the data and wrote the manuscript. T.S. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db16-0946/-/DC1.
See accompanying article, p. 2066.
References
- 1.Compston A, Coles A. Multiple sclerosis. Lancet 2008;372:1502–1517 [DOI] [PubMed] [Google Scholar]
- 2.Lehuen A, Diana J, Zaccone P, Cooke A. Immune cell crosstalk in type 1 diabetes. Nat Rev Immunol 2010;10:501–513 [DOI] [PubMed] [Google Scholar]
- 3.Cope AP, Schulze-Koops H, Aringer M. The central role of T cells in rheumatoid arthritis. Clin Exp Rheumatol 2007;25(Suppl. 46):S4–S11 [PubMed] [Google Scholar]
- 4.Roep BO. Autoreactive T cells in endocrine/organ-specific autoimmunity: why has progress been so slow? Springer Semin Immunopathol 2002;24:261–271 [DOI] [PubMed]
- 5.Delong T, Wiles TA, Baker RL, et al. Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion. Science 2016;351:711–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stadinski B, Kappler J, Eisenbarth GS. Molecular targeting of islet autoantigens. Immunity 2010;32:446–456 [DOI] [PubMed] [Google Scholar]
- 7.Ali OA, Emerich D, Dranoff G, Mooney DJ. In situ regulation of DC subsets and T cells mediates tumor regression in mice. Sci Transl Med 2009;1:8ra19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katz JD, Wang B, Haskins K, Benoist C, Mathis D. Following a diabetogenic T cell from genesis through pathogenesis. Cell 1993;74:1089–1100 [DOI] [PubMed] [Google Scholar]
- 9.Verdaguer J, Schmidt D, Amrani A, Anderson B, Averill N, Santamaria P. Spontaneous autoimmune diabetes in monoclonal T cell nonobese diabetic mice. J Exp Med 1997;186:1663–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quah BJC, Parish CR. New and improved methods for measuring lymphocyte proliferation in vitro and in vivo using CFSE-like fluorescent dyes. J Immunol Methods 2012;379:1–14 [DOI] [PubMed] [Google Scholar]
- 11.Ali OA, Huebsch N, Cao L, Dranoff G, Mooney DJ. Infection-mimicking materials to program dendritic cells in situ. Nat Mater 2009;8:151–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR. T cell receptor antagonist peptides induce positive selection. Cell 1994;76:17–27 [DOI] [PubMed] [Google Scholar]
- 13.Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol. Immunol Cell Biol 1998;76:34–40 [DOI] [PubMed] [Google Scholar]
- 14.Babad J, Geliebter A, DiLorenzo TP. T-cell autoantigens in the non-obese diabetic mouse model of autoimmune diabetes. Immunology 2010;131:459–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamaguchi K, Gaskins HR, Leiter EH. NIT-1, a pancreatic beta-cell line established from a transgenic NOD/Lt mouse. Diabetes 1991;40:842–849 [DOI] [PubMed] [Google Scholar]
- 16.Chang CH, Guerder S, Hong SC, van Ewijk W, Flavell RA. Mice lacking the MHC class II transactivator (CIITA) show tissue-specific impairment of MHC class II expression. Immunity 1996;4:167–178 [DOI] [PubMed] [Google Scholar]
- 17.Biswas A, Meissner TB, Kawai T, Kobayashi KS. Cutting edge: impaired MHC class I expression in mice deficient for Nlrc5/class I transactivator. J Immunol 2012;189:516–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suri A, Walters JJ, Rohrs HW, Gross ML, Unanue ER. First signature of islet beta-cell-derived naturally processed peptides selected by diabetogenic class II MHC molecules. J Immunol 2008;180:3849–3856 [DOI] [PubMed] [Google Scholar]
- 19.Christianson SW, Shultz LD, Leiter EH. Adoptive transfer of diabetes into immunodeficient NOD-scid/scid mice. Relative contributions of CD4+ and CD8+ T-cells from diabetic versus prediabetic NOD.NON-Thy-1a donors. Diabetes 1993;42:44–55 [DOI] [PubMed] [Google Scholar]
- 20.Marrero I, Hamm DE, Davies JD. High-throughput sequencing of islet-infiltrating memory CD4+ T cells reveals a similar pattern of TCR Vβ usage in prediabetic and diabetic NOD mice. PLoS One 2013;8:e76546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pielou EC. The measurement of diversity in different types of biological collections. J Theor Biol 1966;13:131–144 [Google Scholar]
- 22.Makadia HK, Siegel SJ. Poly lactic-co-glycolic acid (PLGA) as biodegradable controlled drug delivery carrier. Polymers (Basel) 2011;3:1377–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.