Abstract
Esophageal cancer is one of the most common gastrointestinal malignant diseases and there is still no effective treatment. The incidence of esophageal cancer in the world is relatively high and on the increase year by year. Thus, the elaboration on the carcinogenesis of esophageal cancer and the identification of new biomarkers and therapeutic targets is quite beneficial to optimizing the current therapeutic regimen for treating such deadly disease. More and more evidence has shown that non-coding RNAs play an important role in the development and progression of multiple human cancers, including esophageal cancer. microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) are two functional kinds of non-coding RNAs that have been well investigated. They exert tumor suppressive or promoting effect by specifically regulating the expression of certain downstream target genes, which is tumor specific. It is also proved that miRNAs and lncRNAs level in tissue and plasma from esophageal cancer patients are closely correlated with the survival and disease progression, which could be used as a prognostic factor and therapeutic target for esophageal cancer.
Keywords: esophageal cancer, lncRNAs, miRNAs, biomarker, therapeutic target
INTRODUCTION
Esophageal cancer is one of the most common gastrointestinal cancers and the incidence is on the increase in recent years [1, 2]. Patients may have unspecific symptoms like wasteness, nausea, weight loss, and the like at early stage, which makes it difficult to be diagnosed. Although great progress has been made on the carcinogenesis and therapy for esophageal cancer, there is still no effective treatment for such deadly disease and the survival remains very poor [3, 4].
Non-coding RNAs are defined as one kind of transcripts that do not encode any protein [5]. So far, several types of non-coding RNAs have been identified by RNA sequencing and bioinformatics analysis. Among them, long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) have attracted great attentions from researchers all over the world. Most miRNAs are deregulated in multiple human diseases including cancers and cancer-related diseases such as cardiovascular disorders [6]. lncRNAs are a new class of non-coding RNAs that are longer than 200 nucleotides [7], and miRNAs are shorter non-coding RNAs with the length of 21-23 nucleotides [8]. It has been proved that lncRNAs could regulate the expression of downstream genes by mediating chromatin, transcriptional and post-transcriptional modification [9], and miRNAs could post-transcriptionally repress the translation of target genes by binding to the untranslated region (UTR) [10]. Plenty of evidence has verified that multiple lncRNAs and miRNAs participate in the development and progression of esophageal cancer, which could exert regulatory effect on a series of biological processes including cell proliferation, cell migration and invasion, and the like [11, 12]. lncRNAs and mRNAs can have crosstalk and work together in regulatory loops [13]. Thus, in this paper we overviewed the recent research advances on the molecular functions of lncRNAs and miRNAs in esophageal cancer, and further discussed their potential roles of early diagnosis and prognostic biomarkers and therapeutic targets, which could help deepen the current understanding and enlighten the future investigations.
NON-CODING RNAS EXPRESSION PROFILING IN ESOPHAGEAL CANCER
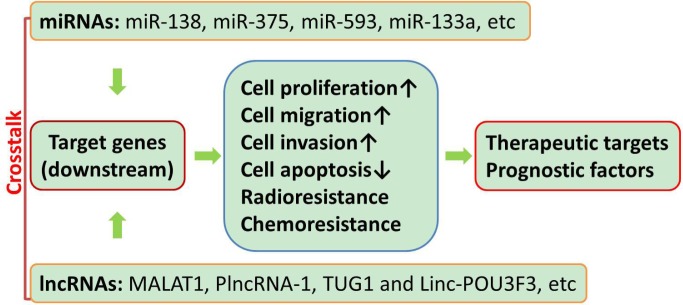
The role of miRNAs and lncRNAs in the pathogenesis of human cancers and other diseases has been elaborated, and there is well-established findings that supports that miRNAs and lncRNAs are very important regulators within human body [14, 15]. They can be either tumor suppressors or tumor promoters [16]. For example, in a cohort of 102 patients who were pathologically diagnosed with esophageal carcinoma, hsa-miR-451a, hsa-miR-144-3p and hsa-miR-144-5p expression in tumor tissues were significantly lower than those in adjacent non-tumor tissues (P < 0.05) [17], which were detected by stem-loop reverse transcription-quantitative polymerase chain reaction and bioinformatics tools. Pearson correlation analysis demonstrated that the expression levels of individual miR-144/451 cluster members were correlated with each other, except for hsa-miR-144-3p and hsa-miR-4732-3p. Further analysis demonstrated that hsa-miR-144-5p expression was highly correlated with hsa-miR-4732-5p and hsa-miR-451a expression, and low hsa-miR-144-3p and hsa-miR-144-5p expression was shown to be the independent risk factors for the onset of esophageal carcinoma. Other miRNAs like miR-138, miR-375, miR-593, miR-133a were down-regulated in esophageal cancer tissue, serving as tumor suppressors, while miR-34b, miR-16, miR-208, miR-423, miR-21, miR-31, miR-223 and miR-373 could have oncogenic actions [18–23] (Figure 1). In the same way, lncRNAs also have dual functions. Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) were down-regulated, while PlncRNA-1, TUG1 and Linc-POU3F3 were up-regulated in esophageal cancer [15, 24–26]. In order to better clarify the expression profiling of non-coding RNAs in esophageal cancer, the database on such patients and tissue collection should be expanded.
Figure 1. Diagram of the molecular functions of miRNAs and lncRNAs in esophageal cancer.
Multiple miRNAs and lncRNAs could regulate cell proliferation, migration, invasion, apoptosis, radioresistance and chemoresistance via modulating the expressions of target genes in esophageal cancer.
NON-CODING RNAS MOLECULAR FUNCTIONS IN ESOPHAGEAL CANCER
The molecular contributions of non-coding RNAs in the carcinogenesis of esophageal cancer have been widely determined (Table 1). lncRNAs and miRNAs could specifically regulate the expression of certain downstream target genes. Consequently, the dysregulation of these genes are able to play an important role in various physical activity and biological processes and multiple signaling transduction pathways are reported to be involved. Besides, miRNA polymorphisms could disturb the miRNA targetome which was defined as the profile of all the target genes [27], evidenced by in silico experimental data. These miRNAs variants may function in a different manner, which lead to the development of esophageal cancer [28]. However, functional experiments need to be conducted to further confirm it. Based on the recent findings, the molecular functions of lncRNAs and miRNAs in esophageal cancer were summarized below.
Table 1. Important non-coding RNAs in human esophageal cancer.
| Non-coding RNAs | Functions |
|---|---|
| lncRNA MALAT1 | Enhance cell proliferation, G2/M cell cycle arrest, cell migration and invasion |
| lncRNA PEG10 | Increase cell proliferation and migration |
| lncRNA TP73-AS1 | Decrease cell apoptosis and induce chemoresistance |
| lncRNA CASC9 | Increase cell migration and invasion |
| lncRNA H19 | Promote cell proliferation and invasion and induce epithelial-to-mesenchymal transition |
| miR-373 | Enhance cell proliferation, G1-phase cell proportion, migration and invasion |
| miR-26b | Enhance cell proliferation, cell-cycle transition and migration |
| miR-100 | Inhibit cell proliferation, migration and invasion and suppress tumor growth |
| miR-98 | Restore radiosensitivity |
| miR-124 | Increase cell apoptosis after radiotherapy |
Cell proliferation regulation
lncRNAs (AFAP1-AS1, UCA1, HOTAIR) were found to be dysregulated in cisplatin-resistant esophageal cancer cells compared with the corresponding parent cells [29]. AFAP1-AS1 was significantly up-regulated in tumor tissues compared with adjacent normal tissues. High AFAP1-AS1 level was closely associated with lymph node metastasis, distant metastasis, advanced tumor stage and chemoresponse. lncRNA SPRY4-IT1 level was up-regulated in human advanced esophageal squamous cell carcinoma tissue, which could enhance the cell viability by activating ZNF703 [30]. MALAT1 is a highly conserved long non-coding RNA, and its oncogenic role has been reported in multiple human cancers. MALAT1 could enhance cell proliferation, G2/M cell cycle arrest, cell migration and invasion of esophageal cancer cells by up-regulating p21 and p27 expression and down-regulating B-MYB expression. miR-101, miR-217 and MALAT1 siRNA have common downstream genes like MIA2, HNF4G, ROBO1, CCT4 and CTHRC1, and miR-101 or miR-217 and MALAT1 were negatively correlated in 42 pairs of esophageal cancer tissue samples and adjacent normal tissues [31]. lncRNA PEG10 was overexpressed in esophageal cancer tissues, which was significantly correlated with lymph node metastases. lncRNA PEG10 silencing could decrease cell proliferation and migration, induce cell apoptosis in two esophageal cancer EC9706 and KYSE150 cells [32].
miR-100 was significantly down-regulated in esophageal cancer, and miR-100 overexpression in esophageal cancer cells significantly inhibited cell proliferation, migration and invasion and suppressed tumor growth via targeting CXCR7 [33]. miRNAs can also regulate sphere formation cell proliferation in esophageal cancer. STAT3 and miR-181b mutually play an important role in controlling cell proliferation and cell apoptosis resistance of sphere formation cells in esophageal cancer in a positive feedback loop [34]. STAT3 can directly increase miR-181b transcription, which targets CYLD gene and miR-181b can enhance the p-STAT3 activity.
Cell cycle regulation
Kyoto Encyclopedia of Genes and Genomes pathway analysis indicated that miR-144/451 cluster could regulate the cell cycle, which validates it to be a promising biomarker for early detection of esophageal carcinoma [17]. miR-26b expression was abnormally up-regulated in esophageal cancer cells and human tissue, whereas no significant change was found on miR-26a expression. miR-26b down-regulation could suppress cell proliferation, cell-cycle transition and migration by targeting TRAF5 [35]. MALAT1 silencing could inhibit proliferation-enhanced apoptosis, cell migration/invasion, and reduce colony formation and induce G2/M arrest [24]. MALAT1 overexpression was associated with a decreased survival rate and MALAT1 can be used as a potential therapeutic target for human esophageal squamous cell carcinoma.
Cell migration and invasion regulation
lncRNA CASC9 was significantly up-regulated in esophageal cancer tissues. CASC9 knockdown could significantly suppress cell migration and invasion, and increased CASC9 expression was associated with cell differentiation in vitro, indicating that CASC9 may be identified as a new promising biomarker for poor prognosis and a potential therapeutic target for treating esophageal cancer [36]. lncRNA LOC100130476 up-regulation could suppress cell proliferation and invasion, and hypermethylation of CpG sites in exon 1 could down-regulate LOC100130476 expression in esophageal cancer, which could predict the clinical TNM stage and pathological differentiation [37]. lncRNA H19 was significantly increased and correlated with tumor invasion depth and metastasis. H19 overexpression could promote cell proliferation and invasion and induce epithelial-to-mesenchymal transition, whereas H19 knockdown could inhibit cell proliferation and invasion and reverse epithelial-to-mesenchymal transition in vitro [38]. miR-373 has different functions among different tumor types. miR-373 expression was up-regulated in esophageal cancer tissues and patients’ plasma. miR-373 up-regulation could enhance cell proliferation, G1-phase cell proportion, migration and invasion, while miR-373 silencing could decrease cell proliferation, G1-phase cell proportion, migration and invasion and induce cell apoptosis by directly targeting TIMP3. Overexpression of miR-373 in ECA109 caused a reduction of TIMP3 mRNA and protein, whereas suppression of miR-373 in KYSE410 led to an increase of TIMP3 mRNA and protein [39].
Cell apoptosis regulation
lncRNA TP73-AS1 and its target gene BDH2 were both up-regulated and closely correlated with the tumor location and TNM stage in esophageal cancer tissues. lncRNA TP73-AS1 knockdown inhibited BDH2 expression in EC9706 and KYSE30 cells, which could further inhibit esophageal cancer cell proliferation and induce cell apoptosis via the caspase-3 dependent apoptotic pathway both in vitro and in vivo. BDH2 modulation could partially rescue the effect of lncRNA TP73-AS1 on cell proliferation and apoptosis [40]. lncRNA AFAP1-AS1 was hypomethylated and overexpressed in Barrett's esophagus and esophageal cancer. AFAP1-AS1 silencing by siRNA could inhibit cell proliferation, migration and invasion, suppress colony-forming ability and induce cell apoptosis, but the protein-coding counterpart AFAP1 was not affected [41]. Based on miRNAs profiling data, miR-20b, miR-498 and miR-196 were predicted to be involved in cell apoptosis and autophagy, which are shown to be key regulators of multiple cellular signaling pathways in esophageal cancer [42].
Chemosensitivity regulation
Chemoresistance is the key to the achievement of effective esophageal cancer therapy. miRNA-127-3p, which specifically compromised the homologous recombination repair and significantly increased DNA double strand breaks in cells, could statistically increase the chemosensitivity of esophageal cancer cells to a novel phenanthroline-dione derivative in vivo by mechanistically impairing the recruitment of RAD51 to the damage sites [43]. miR-221 was overexpressed in 5-fluorouracil resistant esophageal cancer cells and human esophageal cancer tissue. miR-221silencing could reduce cell proliferation, increase apoptosis and chemosensitivity, and inactivate the Wnt/β-catenin pathway mediated by alteration in DKK2 expression both in vitro and in vivo [44]. BDH2 or lncRNA TP73-AS1 knockdown enhanced the chemosensitivity of esophageal cancer cells to and cisplatin. Our results suggest that lncRNA TP73-AS1 may be a novel prognostic biomarker that could serve as a potential therapeutic target for the treatment of esophageal cancer [40]. circRNAs dys-regulation, which mainly refer to serum and plasma RNAs were found in human radioresistant esophageal cancer cells, and CircRNA_001059 and circRNA_000167 were the two largest nodes to form a comprehensive and functional circRNA/miRNA co-expression network [45]. LOC285194 expression was significantly down-regulated in esophageal cancer. Low LOC285194 expression was correlated with tumor size, TNM stage, lymph node metastases and distant metastases. Univariate and multivariate analysis demonstrated that LOC285194 down-regulation could predict poor chemotherapy response and survival status, evidenced by the fact that patients with low LOC285194 level had a decreased disease free survival and overall survival [46].
Radiosensitivity modulation
Aberrant miRNAs expression is also responsible for impairing radiosensitivity. miR-98 was down-regulated in radioresistant esophageal cancer cell line, and miR-98 mimic could restore the sensitivity to radiotherapy in esophageal cancer cells by up-regulating miR-98, which could decrease cell proliferation and migration and induce cell apoptosis by directly binding to the promoter of BCL-2 gene [47]. miR-124 expression was reduced in esophageal cancer tissue, and miR-124 overexpression could increase the percentage of apoptotic cells following radiotherapy by targeting CDK4 in esophageal cancer TE-1 cells [48].
POTENTIAL BIOMARKERS AND THERAPEUTIC TARGETS FOR HUMAN ESOPHAGEAL CANCER
Diagnostic and prognostic biomarkers
Detection of miRNAs and lncRNAs could be introduced as the potential biomarkers for early diagnosis and prognosis [49–51]. A microarray analysis has ever identified miR-574-3p, miR-106b, miR-1303, miR-1203, miR-1909, miR-204, miR-371-3p and miR-886-3p, which were differentially expressed between the patients with and without tumor relapse after surgery. Higher expression of miR-574-3p was associated with non-relapse and favorable overall survival, which was a predictor for clinical outcome in patients with esophageal cancer after surgery [18]. Univariate and multivariate analysis revealed that high pretreatment plasma miR-21 level was an independent risk factor for chemoresistance in esophageal cancer, which was greatly correlated with high histopathological response [52]. miR-21 level was significantly elevated in esophageal cancer, while miR-375 was down-regulated. miR-21 overexpression could reduce the radiosensitivity and increase the tumor relapse. The combination of miR-21 and miR-375 level was identified to be a biomarker for early diagnosis and prognosis of esophageal cancer [19].
Survival analysis revealed that high lncRNA AFAP1-AS1 level was significantly associated with shorter progression free survival and overall survival and multivariate analysis showed that high AFAP1-AS1 level was found to be an independent risk factor for poor clinical response [29]. Hypermethylation of CpG sites in the exon 1 and low expression of LOC100130476 could predict poor survival [37]. HOTAIR overexpression was correlated with short survival in esophageal cancer regardless of the ethnicities, indicating that high HOTAIR level may be a prognostic factor for esophageal cancer [53]. lncRNA CCAT2 was up-regulated in esophageal cancer tissues and positively correlated with TNM stage and lymph node metastasis. Survival analyses revealed that high CCAT2 expression and MYC amplification were significantly associated with poorer overall survival in such patients. Patients with high CCAT2 expression and MYC amplification had an increased risk of cancer-related death, supporting that CCAT2 is a predictive biomarker and therapeutic target for esophageal cancer [54]. lncRNA PCAT-1 was significantly higher in human esophageal cancer, and high PCAT-1 expression was significantly correlated with advanced clinical stage, lymph node metastasis, and poor prognosis, which is a potential prognostic factor for esophageal cancer [55]. Now, the detection of plasma lncRNAs identified POU3F3, HNF1A-AS1 and SPRY4-IT1 were greatly higher in esophageal cancer patients. Among them, plasma POU3F3 had the highest diagnostic performance for esophageal cancer and the combination of POU3F3 and SCCA had a more excellent diagnostic efficiency, especially for early stage [56]. lncRNA UCA1 was overexpressed in esophageal cancer cells and was prone to predict poor prognosis. Targeting UCA1 could decrease cell proliferation, migration and invasion [57]. A study has ever analyzed the associations of lncRNAs with the risk and prognosis of esophageal cancer in 358 patients from eastern China, and validate the findings in another 326 patients from southern China. lncRNA uc002yug.2 was overexpressed in esophageal cancer, which could exert oncogenic effect by overexpressing RUNX1a and reducing CEBPα gene expression. These data suggest that lincRNA-uc002yug.2 level could be a prognostic factor for the esophageal cancer patients’ survival [58].
lncRNAs can also co-function with the adjacent coding genes by forming a lncRNA-mRNA gene pair. For example, lncRNA FOXCUT and its neighboring gene FOXC1. FOXCUT and FOXC1 levels were both up-regulated in esophageal cancer and strongly correlated with poor differentiation and metastasis. Up-regulated FOXCUT or FOXC1 expression could predict a significantly worse prognosis. FOXCUT level was positively correlated with FOXC1 level in esophageal cancer, and the decreased expression of FOXC1 was also observed after being treated by FOXCUT siRNA. In vitro experiments showed that either FOXCUT or FOXC1 silencing could greatly inhibit cell proliferation, colony formation, migration and invasion in esophageal cancer cells. FOXCUT plays a functional role in the development of esophageal cancer and predicts the survival potentially and partially by modulating FOXC1 [59]. Besides, a recent study identified a three lncRNA signature (including ENST00000435885.1, XLOC_013014 and ENST00000547963.1), which was proved to be able to independently predict the outcome of the patients [60].
Therapeutic targets
miR-100 functions as a tumor suppressor in esophageal cancer, which could be potentially applied in treating esophageal cancer [33]. The miR-124/CDK4 axis was an important mechanism in regulating the radiation sensitivity of human esophageal cancer cells, and targeting CDK4 may improve the clinical efficacy of radiotherapy [48]. lncRNA BOKAS was up-regulated and increased WISP1 expression, which could lead to the resistance to radiotherapy in esophageal cancer both in vitro and in vivo. WISP1 was a oncofetal gene in Wnt/β-catenin pathway, and WISP1 level could predict the prognosis of esophageal cancer patients after radiotherapy. Molecular investigation indicated that WISP1 could contribute to radioresistance via repairing irradiation-induced DNA damage and activating PI3K kinase and its function was in a positive feedback loop [61]. Previous researches reported that β-elemene could inhibit cell proliferation of esophageal cancer ECA-109 cells and up-regulate about 85% of the lncRNAs except CDKN2B-AS1. Targeting the β-elemene-mediated lncRNA CDKN2B-AS1 decreased cell proliferation, and increased cell apoptosis by down-regulating the hTERT level [62]. lncRNA HNF1A-AS1 was highly up-regulated in human primary esophageal cancer tissues. HNF1A-AS1 silencing significantly inhibited cell proliferation and anchorage-independent growth, S-phase entry, cell migration and invasion in vitro by preferentially modulating genes that are linked to assembly of chromatin and the nucleosome, which is a key mechanism essential to cell cycle progression. lncRNA HNF1A-AS1 silencing could also inhibit lncRNA H19 expression. Consistent to this finding, HNF1A-AS1 could be a very important therapeutic target for treating esophageal cancer [63]. lncRNA PEG10 up-regulation could suppress cell proliferation, cell invasion and migration in esophageal cancer, which is qualified to be a promising therapeutic target in esophageal cancer [32].
CONCLUSIONS
Multiple miRNAs and lncRNAs play an important role in the development and progression of esophageal cancer, which could be identified to be new promising therapeutic targets and prognostic factors. However, the current findings mainly focus on the basic research on the functions of non-coding RNAs in esophageal cancer. Great progress should be made to fully understand the clinical application of miRNAs and lncRNAs in diagnosing and treating such devastating disease. Targeting specific downstream genes by miRNAs and lncRNAs in esophageal cancer will be a promising therapeutic regimen, which should be further validated in future studies.
Footnotes
Authors’ contribution
X.H. and J.W. wrote the paper; Z.R. revised the paper; G.Z. designed the study and revised the paper.
CONFLICTS OF INTERESTS
The author(s) declare no competing financial interests.
REFERENCES
- 1.Pakzad R, Mohammadian-Hafshejani A, Khosravi B, Soltani S, Pakzad I, Mohammadian M, Salehiniya H, Momenimovahed Z. The incidence and mortality of esophageal cancer and their relationship to development in Asia. Ann Transl Med. 2016;4:29. doi: 10.3978/j.issn.2305-5839.2016.01.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan HZ, Lin WJ, Huang JQ, Dai M, Fu JH, Huang QH, Chen WM, Xu YL, Ye TT, Lin ZY, Lin XS, Cai JX, Dong YH, et al. Updated incidence rates and risk factors of esophageal cancer in Nan’ao Island, a coastal high-risk area in southern China. Dis Esophagus. 2016 doi: 10.1111/dote.12468. [DOI] [PubMed] [Google Scholar]
- 3.Patti MG. Updates on the Treatment of Esophageal Cancer. J Laparoendosc Adv Surg Tech A. 2016 doi: 10.1089/lap.2016.29008.mgp. [DOI] [PubMed] [Google Scholar]
- 4.Allemann P, Mantziari S, Wagner D, Digklia A, Ozsahin E, De Bari B, Dorta G, Godat S, Montserrat F, Sempoux C, Brunel C, Demartines N, Schafer M. [Curative treatment for esophageal cancer: results of a multidisciplinary consensus] Rev Med Suisse. 2016;12:1165–9. [PubMed] [Google Scholar]
- 5.de Almeida RA, Fraczek MG, Parker S, Delneri D, O’Keefe RT. Non-coding RNAs and disease: the classical ncRNAs make a comeback. Biochem Soc Trans. 2016;44:1073–8. doi: 10.1042/BST20160089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taibi F, Metzinger-Le Meuth V, Massy ZA, Metzinger L. miR-223: An inflammatory oncomiR enters the cardiovascular field. Biochimica Et Biophysica Acta. 2014;1842:1001–9. doi: 10.1016/j.bbadis.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 7.Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10:925–33. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Evans JR, Feng FY, Chinnaiyan AM. The bright side of dark matter: lncRNAs in cancer. J Clin Invest. 2016;126:2775–82. doi: 10.1172/JCI84421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 11.Hemmatzadeh M, Mohammadi H, Karimi M, Musavishenas MH, Baradaran B. Differential role of microRNAs in the pathogenesis and treatment of Esophageal cancer. Biomed Pharmacother. 2016;82:509–19. doi: 10.1016/j.biopha.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 12.Harada K, Baba Y, Ishimoto T, Shigaki H, Kosumi K, Yoshida N, Watanabe M, Baba H. The role of microRNA in esophageal squamous cell carcinoma. J Gastroenterol. 2016;51:520–30. doi: 10.1007/s00535-016-1161-9. [DOI] [PubMed] [Google Scholar]
- 13.Bayoumi AS, Sayed A, Broskova Z, Teoh JP, Wilson J, Su H, Tang YL, Kim IM. Crosstalk between Long Noncoding RNAs and MicroRNAs in Health and Disease. Int J Mol Sci. 2016;17:356. doi: 10.3390/ijms17030356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Ju J, Ni B, Wang H. The emerging role of miR-506 in cancer. Oncotarget. 2016;7:62778–62788. doi: 10.18632/oncotarget.11294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen WJ, Zhang F, Zhao X, Xu J. LncRNAs and Esophageal Squamous Cell Carcinoma - Implications for Pathogenesis and Drug Development. J Cancer. 2016;7:1258–64. doi: 10.7150/jca.14869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sugihara H, Ishimoto T, Miyake K, Izumi D, Baba Y, Yoshida N, Watanabe M, Baba H. Noncoding RNA Expression Aberration Is Associated with Cancer Progression and Is a Potential Biomarker in Esophageal Squamous Cell Carcinoma. Int J Mol Sci. 2015;16:27824–34. doi: 10.3390/ijms161126060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao Z, Liu R, Liao J, Yang M, Pan E, Yin L, Pu Y. Possible tumor suppressive role of the miR-144/451 cluster in esophageal carcinoma as determined by principal component regression analysis. Mol Med Rep. 2016 doi: 10.3892/mmr.2016.5691. [DOI] [PubMed] [Google Scholar]
- 18.Okumura T, Kojima H, Miwa T, Sekine S, Hashimoto I, Hojo S, Nagata T, Shimada Y. The expression of microRNA 574-3p as a predictor of postoperative outcome in patients with esophageal squamous cell carcinoma. World J Surg Oncol. 2016;14:228. doi: 10.1186/s12957-016-0985-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lv H, He Z, Wang H, Du T, Pang Z. Differential expression of miR-21 and miR-75 in esophageal carcinoma patients and its clinical implication. Am J Transl Res. 2016;8:3288–98. [PMC free article] [PubMed] [Google Scholar]
- 20.Sun J, Song K, Feng X, Gao S. MicroRNA-367 is a potential diagnostic biomarker for patients with esophageal squamous cell carcinoma. Biochem Biophys Res Commun. 2016;473:363–9. doi: 10.1016/j.bbrc.2016.01.042. [DOI] [PubMed] [Google Scholar]
- 21.Sun L, Dong S, Dong C, Sun K, Meng W, Lv P, Yin H, Ming L, He F. Predictive value of plasma miRNA-718 for esophageal squamous cell carcinoma. Cancer Biomark. 2016;16:265–73. doi: 10.3233/CBM-150564. [DOI] [PubMed] [Google Scholar]
- 22.Phatak P, Byrnes KA, Mansour D, Liu L, Cao S, Li R, Rao JN, Turner DJ, Wang JY, Donahue JM. Overexpression of miR-214-3p in esophageal squamous cancer cells enhances sensitivity to cisplatin by targeting survivin directly and indirectly through CUG-BP1. Oncogene. 2016;35:2087–97. doi: 10.1038/onc.2015.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuzaki J, Suzuki H. Role of MicroRNAs-221/222 in Digestive Systems. J Clin Med. 2015;4:1566–77. doi: 10.3390/jcm4081566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yao W, Bai Y, Li Y, Guo L, Zeng P, Wang Y, Qi B, Liu S, Qin X, Zhao B. Upregulation of MALAT-1 and its association with survival rate and the effect on cell cycle and migration in patients with esophageal squamous cell carcinoma. Tumour Biol. 2016;37:4305–12. doi: 10.1007/s13277-015-4223-3. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Shi X, Yang W, Lu Z, Wang P, Chen Z, He J. Transcriptome profiling of lncRNA and co-expression networks in esophageal squamous cell carcinoma by RNA sequencing. Tumour Biol. 2016 doi: 10.1007/s13277-016-5227-3. [DOI] [PubMed] [Google Scholar]
- 26.Cao B, Song N, Zhang M, Di C, Yang Y, Lu Y, Chen R, Lu ZJ, Guo M. Systematic study of novel lncRNAs in different gastrointestinal cancer cells. Discov Med. 2016;21:159–71. [PubMed] [Google Scholar]
- 27.Vlad A, Rohrs S, Klein-Hitpass L, Muller O. The first five years of the Wnt targetome. Cell Signal. 2008;20:795–802. doi: 10.1016/j.cellsig.2007.10.031. [DOI] [PubMed] [Google Scholar]
- 28.Nariman-Saleh-Fam Z, Bastami M, Somi MH, Samadi N, Abbaszadegan MR, Behjati F, Ghaedi H, Tavakkoly-Bazzaz J, Masotti A. In silico dissection of miRNA targetome polymorphisms and their role in regulating miRNA-mediated gene expression in esophageal cancer. Cell Biochemistry and Biophysics. 2016 doi: 10.1007/s12013-016-0754-5. [DOI] [PubMed] [Google Scholar]
- 29.Zhou XL, Wang WW, Zhu WG, Yu CH, Tao GZ, Wu QQ, Song YQ, Pan P, Tong YS. High expression of long non-coding RNA AFAP1-AS1 predicts chemoradioresistance and poor prognosis in patients with esophageal squamous cell carcinoma treated with definitive chemoradiotherapy. Mol Carcinog. 2016 doi: 10.1002/mc.22454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xue-Liang J, Ming-Dong W, Ya-Bi Z, Wang-Yue W. Upregulated long noncoding RNA SPRY4-IT1 contributes to increased cell viability by activating zinc finger 703 expression in esophageal squamous cell carcinoma. Indian J Cancer. 2015;52(Suppl 3):E164–7. doi: 10.4103/0019-509X.186566. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Li M, Wang Z, Han S, Tang X, Ge Y, Zhou L, Zhou C, Yuan Q, Yang M. Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. J Biol Chem. 2015;290:3925–35. doi: 10.1074/jbc.M114.596866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zang W, Wang T, Huang J, Li M, Wang Y, Du Y, Chen X, Zhao G. Long noncoding RNA PEG10 regulates proliferation and invasion of esophageal cancer cells. Cancer Gene Ther. 2015;22:138–44. doi: 10.1038/cgt.2014.77. [DOI] [PubMed] [Google Scholar]
- 33.Zhou SM, Zhang F, Chen XB, Jun CM, Jing X, Wei DX, Xia Y, Zhou YB, Xiao XQ, Jia RQ, Li JT, Sheng W, Zeng Y. miR-100 suppresses the proliferation and tumor growth of esophageal squamous cancer cells via targeting CXCR7. Oncol Rep. 2016;35:3453–9. doi: 10.3892/or.2016.4701. [DOI] [PubMed] [Google Scholar]
- 34.Xu DD, Zhou PJ, Wang Y, Zhang L, Fu WY, Ruan BB, Xu HP, Hu CZ, Tian L, Qin JH, Wang S, Wang X, Li YC, et al. Reciprocal activation between STAT3 and miR-181b regulates the proliferation of esophageal cancer stem-like cells via the CYLD pathway. Mol Cancer. 2016;15:40. doi: 10.1186/s12943-016-0521-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Z, Zhao L, Zhao F, Yang G, Wang J. MicroRNA-26b regulates cancer proliferation migration and cell cycle transition by suppressing TRAF5 in esophageal squamous cell carcinoma. Am J Transl Res. 2016;8:1957–70. [PMC free article] [PubMed] [Google Scholar]
- 36.Pan Z, Mao W, Bao Y, Zhang M, Su X, Xu X. The long noncoding RNA CASC9 regulates migration and invasion in esophageal cancer. Cancer Med. 2016 doi: 10.1002/cam4.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo W, Dong Z, Shi Y, Liu S, Liang J, Guo Y, Guo X, Shen S, Shan B. Aberrant methylation-mediated downregulation of long noncoding RNA LOC100130476 correlates with malignant progression of esophageal squamous cell carcinoma. Dig Liver Dis. 2016;48:961–9. doi: 10.1016/j.dld.2016.05.010. [DOI] [PubMed] [Google Scholar]
- 38.Huang C, Cao L, Qiu L, Dai X, Ma L, Zhou Y, Li H, Gao M, Li W, Zhang Q, Han K, Lv H. Upregulation of H19 promotes invasion and induces epithelial-to-mesenchymal transition in esophageal cancer. Oncol Lett. 2015;10:291–296. doi: 10.3892/ol.2015.3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu W, Li M, Chen X, Zhang D, Wei L, Zhang Z, Wang S, Meng L, Zhu S, Li B. MicroRNA-373 promotes migration and invasion in human esophageal squamous cell carcinoma by inhibiting TIMP3 expression. Am J Cancer Res. 2016;6:1–14. [PMC free article] [PubMed] [Google Scholar]
- 40.Zang W, Wang T, Wang Y, Chen X, Du Y, Sun Q, Li M, Dong Z, Zhao G. Knockdown of long non-coding RNA TP73-AS1 inhibits cell proliferation and induces apoptosis in esophageal squamous cell carcinoma. Oncotarget. 2016;7:19960–74. doi: 10.18632/oncotarget.6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu W, Bhagat TD, Yang X, Song JH, Cheng Y, Agarwal R, Abraham JM, Ibrahim S, Bartenstein M, Hussain Z, Suzuki M, Yu Y, Chen W, et al. Hypomethylation of noncoding DNA regions and overexpression of the long noncoding RNA, AFAP1-AS1, in Barrett's esophagus and esophageal adenocarcinoma. Gastroenterology. 2013;144:956–966. doi: 10.1053/j.gastro.2013.01.019. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu BL, Wang D, Bai WJ, Zhang F, Zhao X, Yi Y, Zhang T, Shen WJ, Li EM, Xu LY, Xu JZ. An integrative framework to identify cell death-related microRNAs in esophageal squamous cell carcinoma. Oncotarget. 2016;7:56758–56766. doi: 10.18632/oncotarget.10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhen N, Yang Q, Zheng K, Han Z, Sun F, Mei W, Yu Y. MiroRNA-127-3p targets XRCC3 to enhance the chemosensitivity of esophageal cancer cells to a novel phenanthroline-dione derivative. Int J Biochem Cell Biol. 2016;79:158–167. doi: 10.1016/j.biocel.2016.08.026. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y, Zhao Y, Herbst A, Kalinski T, Qin J, Wang X, Jiang Z, Benedix F, Franke S, Wartman T, Camaj P, Halangk W, Kolligs FT, et al. miR-221 Mediates Chemoresistance of Esophageal Adenocarcinoma by Direct Targeting of DKK2 Expression. Ann Surg. 2016 doi: 10.1097/SLA.0000000000001928. [DOI] [PubMed] [Google Scholar]
- 45.Su H, Lin F, Deng X, Shen L, Fang Y, Fei Z, Zhao L, Zhang X, Pan H, Xie D, Jin X, Xie C. Profiling and bioinformatics analyses reveal differential circular RNA expression in radioresistant esophageal cancer cells. J Transl Med. 2016;14:225. doi: 10.1186/s12967-016-0977-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tong YS, Zhou XL, Wang XW, Wu QQ, Yang TX, Lv J, Yang JS, Zhu B, Cao XF. Association of decreased expression of long non-coding RNA LOC285194 with chemoradiotherapy resistance and poor prognosis in esophageal squamous cell carcinoma. J Transl Med. 2014;12:233. doi: 10.1186/s12967-014-0233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin YY, Chen QJ, Wei Y, Wang YL, Wang ZW, Xu K, He Y, Ma HB. Upregulation of microRNA-98 increases radiosensitivity in esophageal squamous cell carcinoma. J Radiat Res. 2016 doi: 10.1093/jrr/rrw068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang YH, Wang QQ, Li H, Ye T, Gao F, Liu YC. miR-124 radiosensitizes human esophageal cancer cell TE-1 by targeting CDK4. Genet Mol Res. 2016 doi: 10.4238/gmr.15027893. [DOI] [PubMed] [Google Scholar]
- 49.Shah AK, Saunders NA, Barbour AP, Hill MM. Early diagnostic biomarkers for esophageal adenocarcinoma—the current state of play. Cancer Epidemiol Biomarkers Prev. 2013;22:1185–209. doi: 10.1158/1055-9965.EPI-12-1415. [DOI] [PubMed] [Google Scholar]
- 50.Tan C, Qian X, Guan Z, Yang B, Ge Y, Wang F, Cai J. Potential biomarkers for esophageal cancer. Springerplus. 2016;5:467. doi: 10.1186/s40064-016-2119-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wan J, Wu W, Che Y, Kang N, Zhang R. Insights into the potential use of microRNAs as a novel class of biomarkers in esophageal cancer. Dis Esophagus. 2016;29:412–20. doi: 10.1111/dote.12338. [DOI] [PubMed] [Google Scholar]
- 52.Komatsu S, Ichikawa D, Kawaguchi T, Miyamae M, Okajima W, Ohashi T, Imamura T, Kiuchi J, Konishi H, Shiozaki A, Fujiwara H, Okamoto K, Otsuji E. Circulating miR-21 as an independent predictive biomarker for chemoresistance in esophageal squamous cell carcinoma. Am J Cancer Res. 2016;6:1511–23. [PMC free article] [PubMed] [Google Scholar]
- 53.Ma G, Wang Q, Lv C, Qiang F, Hua Q, Chu H, Du M, Tong N, Jiang Y, Wang M, Zhang Z, Wang J, Gong W. The prognostic significance of HOTAIR for predicting clinical outcome in patients with digestive system tumors. J Cancer Res Clin Oncol. 2015;141:2139–45. doi: 10.1007/s00432-015-1980-8. [DOI] [PubMed] [Google Scholar]
- 54.Zhang X, Xu Y, He C, Guo X, Zhang J, Zhang L, Kong M, Chen B, Zhu C. Elevated expression of CCAT2 is associated with poor prognosis in esophageal squamous cell carcinoma. J Surg Oncol. 2015;111:834–9. doi: 10.1002/jso.23888. [DOI] [PubMed] [Google Scholar]
- 55.Shi WH, Wu QQ, Li SQ, Yang TX, Liu ZH, Tong YS, Tuo L, Wang S, Cao XF. Upregulation of the long noncoding RNA PCAT-1 correlates with advanced clinical stage and poor prognosis in esophageal squamous carcinoma. Tumour Biol. 2015;36:2501–7. doi: 10.1007/s13277-014-2863-3. [DOI] [PubMed] [Google Scholar]
- 56.Tong YS, Wang XW, Zhou XL, Liu ZH, Yang TX, Shi WH, Xie HW, Lv J, Wu QQ, Cao XF. Identification of the long non-coding RNA POU3F3 in plasma as a novel biomarker for diagnosis of esophageal squamous cell carcinoma. Mol Cancer. 2015;14:3. doi: 10.1186/1476-4598-14-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li JY, Ma X, Zhang CB. Overexpression of long non-coding RNA UCA1 predicts a poor prognosis in patients with esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7:7938–44. [PMC free article] [PubMed] [Google Scholar]
- 58.Wu H, Zheng J, Deng J, Zhang L, Li N, Li W, Li F, Lu J, Zhou Y. LincRNA-uc002yug.2 involves in alternative splicing of RUNX1 and serves as a predictor for esophageal cancer and prognosis. Oncogene. 2015;34:4723–34. doi: 10.1038/onc.2014.400. [DOI] [PubMed] [Google Scholar]
- 59.Pan F, Yao J, Chen Y, Zhou C, Geng P, Mao H, Fang X. A novel long non-coding RNA FOXCUT and mRNA FOXC1 pair promote progression and predict poor prognosis in esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7:2838–49. [PMC free article] [PubMed] [Google Scholar]
- 60.Li J, Chen Z, Tian L, Zhou C, He MY, Gao Y, Wang S, Zhou F, Shi S, Feng X, Sun N, Liu Z, Skogerboe G, et al. LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut. 2014;63:1700–10. doi: 10.1136/gutjnl-2013-305806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang H, Luo H, Hu Z, Peng J, Jiang Z, Song T, Wu B, Yue J, Zhou R, Xie R, Chen T, Wu S. Targeting WISP1 to sensitize esophageal squamous cell carcinoma to irradiation. Oncotarget. 2015;6:6218–34. doi: 10.18632/oncotarget.3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu Z, Wu H, Li Y, Hou Q, Wang Y, Li S, Xia B, Wu S. beta-Elemene inhibits the proliferation of esophageal squamous cell carcinoma by regulating long noncoding RNA-mediated inhibition of hTERT expression. Anticancer Drugs. 2015;26:531–9. doi: 10.1097/CAD.0000000000000216. [DOI] [PubMed] [Google Scholar]
- 63.Yang X, Song JH, Cheng Y, Wu W, Bhagat T, Yu Y, Abraham JM, Ibrahim S, Ravich W, Roland BC, Khashab M, Singh VK, Shin EJ, et al. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut. 2014;63:881–90. doi: 10.1136/gutjnl-2013-305266. [DOI] [PMC free article] [PubMed] [Google Scholar]