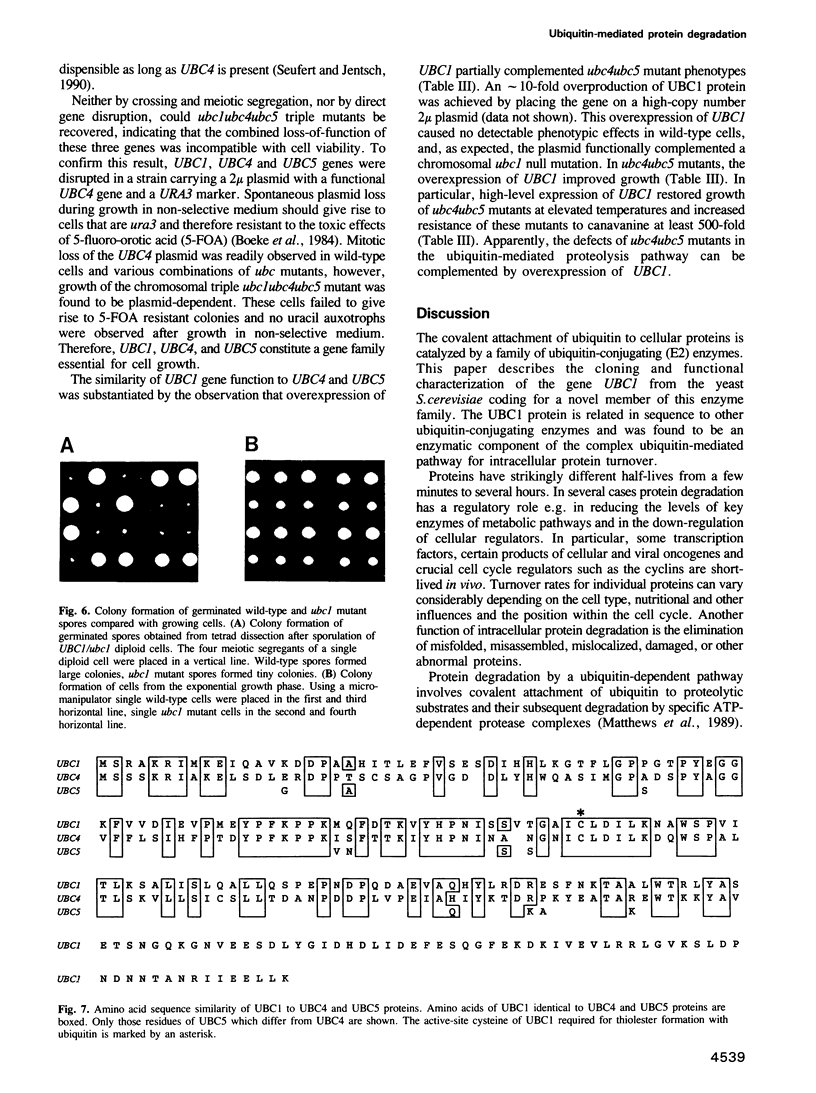
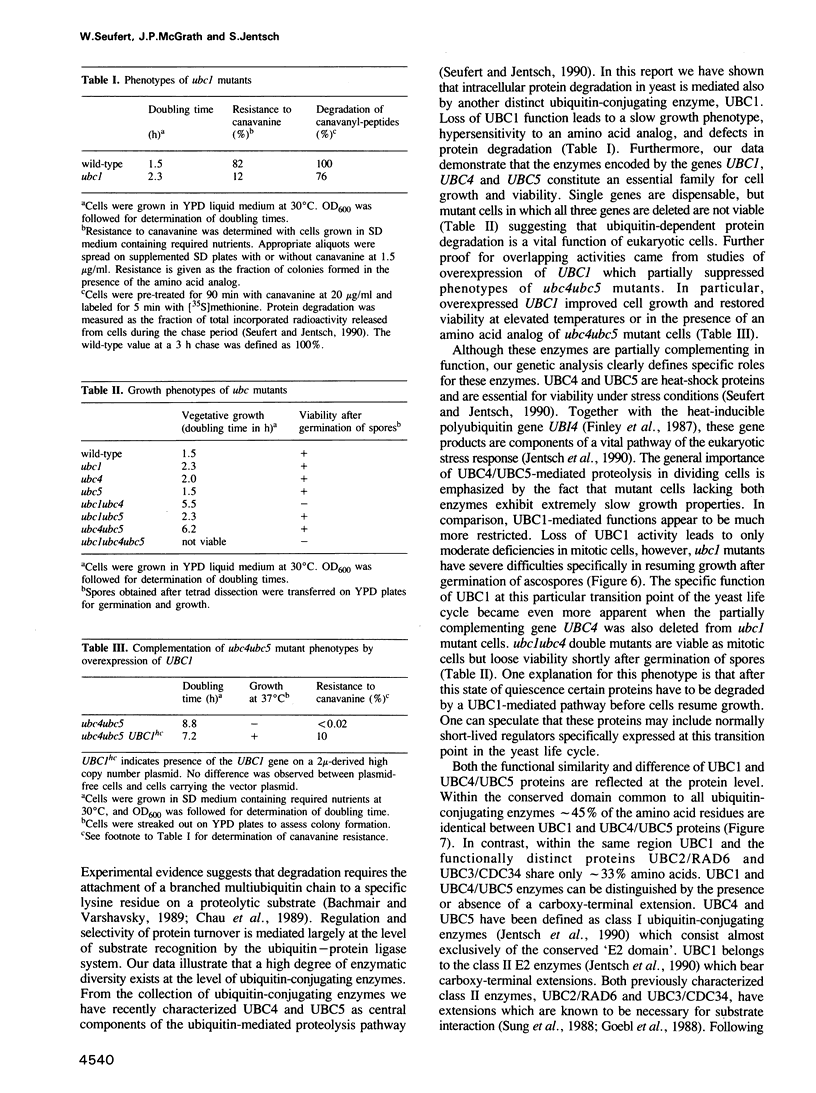
Abstract
The covalent attachment of ubiquitin to cellular proteins is catalyzed by members of a family of ubiquitin-conjugating enzymes. These enzymes participate in a variety of cellular processes, including selective protein degradation, DNA repair, cell cycle control, and sporulation. In the yeast Saccharomyces cerevisiae, two closely related ubiquitin-conjugating enzymes, UBC4 and UBC5, have recently been shown to mediate the selective degradation of short-lived and abnormal proteins. We have now identified a third distinct member of this class of ubiquitin-conjugating enzymes, UBC1. UBC1, UBC4 and UBC5 are functionally overlapping and constitute an enzyme family essential for cell growth and viability. All three mediate selective protein degradation, however, UBC1 appears to function primarily in the early stages of growth after germination of spores. ubc1 mutants generated by gene disruption display only a moderate slow growth phenotype, but are markedly impaired in growth following germination. Moreover, yeast carrying the ubc1ubc4 double mutation are viable as mitotic cells, however, these cells fail to survive after undergoing sporulation and germination. This specific requirement for UBC1 after a state of quiescence suggests that degradation of certain proteins may be crucial at this transition point in the yeast life cycle.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmair A., Varshavsky A. The degradation signal in a short-lived protein. Cell. 1989 Mar 24;56(6):1019–1032. doi: 10.1016/0092-8674(89)90635-1. [DOI] [PubMed] [Google Scholar]
- Ball E., Karlik C. C., Beall C. J., Saville D. L., Sparrow J. C., Bullard B., Fyrberg E. A. Arthrin, a myofibrillar protein of insect flight muscle, is an actin-ubiquitin conjugate. Cell. 1987 Oct 23;51(2):221–228. doi: 10.1016/0092-8674(87)90149-8. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., LaCroute F., Fink G. R. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Brosius J., Lupski J. R. Plasmids for the selection and analysis of prokaryotic promoters. Methods Enzymol. 1987;153:54–68. doi: 10.1016/0076-6879(87)53047-6. [DOI] [PubMed] [Google Scholar]
- Chau V., Tobias J. W., Bachmair A., Marriott D., Ecker D. J., Gonda D. K., Varshavsky A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989 Mar 24;243(4898):1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- Ciechanover A., Finley D., Varshavsky A. Ubiquitin dependence of selective protein degradation demonstrated in the mammalian cell cycle mutant ts85. Cell. 1984 May;37(1):57–66. doi: 10.1016/0092-8674(84)90300-3. [DOI] [PubMed] [Google Scholar]
- Ciechanover A., Schwartz A. L. How are substrates recognized by the ubiquitin-mediated proteolytic system? Trends Biochem Sci. 1989 Dec;14(12):483–488. doi: 10.1016/0968-0004(89)90180-1. [DOI] [PubMed] [Google Scholar]
- Dunigan D. D., Dietzgen R. G., Schoelz J. E., Zaitlin M. Tobacco mosaic virus particles contain ubiquitinated coat protein subunits. Virology. 1988 Jul;165(1):310–312. doi: 10.1016/0042-6822(88)90691-5. [DOI] [PubMed] [Google Scholar]
- Finley D., Ciechanover A., Varshavsky A. Thermolability of ubiquitin-activating enzyme from the mammalian cell cycle mutant ts85. Cell. 1984 May;37(1):43–55. doi: 10.1016/0092-8674(84)90299-x. [DOI] [PubMed] [Google Scholar]
- Finley D., Ozkaynak E., Varshavsky A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell. 1987 Mar 27;48(6):1035–1046. doi: 10.1016/0092-8674(87)90711-2. [DOI] [PubMed] [Google Scholar]
- Goebl M. G., Yochem J., Jentsch S., McGrath J. P., Varshavsky A., Byers B. The yeast cell cycle gene CDC34 encodes a ubiquitin-conjugating enzyme. Science. 1988 Sep 9;241(4871):1331–1335. doi: 10.1126/science.2842867. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hershko A. Ubiquitin-mediated protein degradation. J Biol Chem. 1988 Oct 25;263(30):15237–15240. [PubMed] [Google Scholar]
- Hochstrasser M., Varshavsky A. In vivo degradation of a transcriptional regulator: the yeast alpha 2 repressor. Cell. 1990 May 18;61(4):697–708. doi: 10.1016/0092-8674(90)90481-s. [DOI] [PubMed] [Google Scholar]
- Jentsch S., McGrath J. P., Varshavsky A. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature. 1987 Sep 10;329(6135):131–134. doi: 10.1038/329131a0. [DOI] [PubMed] [Google Scholar]
- Jentsch S., Seufert W., Sommer T., Reins H. A. Ubiquitin-conjugating enzymes: novel regulators of eukaryotic cells. Trends Biochem Sci. 1990 May;15(5):195–198. doi: 10.1016/0968-0004(90)90161-4. [DOI] [PubMed] [Google Scholar]
- Leung D. W., Spencer S. A., Cachianes G., Hammonds R. G., Collins C., Henzel W. J., Barnard R., Waters M. J., Wood W. I. Growth hormone receptor and serum binding protein: purification, cloning and expression. Nature. 1987 Dec 10;330(6148):537–543. doi: 10.1038/330537a0. [DOI] [PubMed] [Google Scholar]
- Matthews W., Driscoll J., Tanaka K., Ichihara A., Goldberg A. L. Involvement of the proteasome in various degradative processes in mammalian cells. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2597–2601. doi: 10.1073/pnas.86.8.2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C. M., Rose I. A. Functional heterogeneity of ubiquitin carrier proteins. J Biol Chem. 1985 Feb 10;260(3):1573–1581. [PubMed] [Google Scholar]
- Seufert W., Jentsch S. Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J. 1990 Feb;9(2):543–550. doi: 10.1002/j.1460-2075.1990.tb08141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegelman M., Bond M. W., Gallatin W. M., St John T., Smith H. T., Fried V. A., Weissman I. L. Cell surface molecule associated with lymphocyte homing is a ubiquitinated branched-chain glycoprotein. Science. 1986 Feb 21;231(4740):823–829. doi: 10.1126/science.3003913. [DOI] [PubMed] [Google Scholar]
- Snyder M., Elledge S., Davis R. W. Rapid mapping of antigenic coding regions and constructing insertion mutations in yeast genes by mini-Tn10 "transplason" mutagenesis. Proc Natl Acad Sci U S A. 1986 Feb;83(3):730–734. doi: 10.1073/pnas.83.3.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P., Prakash S., Prakash L. The RAD6 protein of Saccharomyces cerevisiae polyubiquitinates histones, and its acidic domain mediates this activity. Genes Dev. 1988 Nov;2(11):1476–1485. doi: 10.1101/gad.2.11.1476. [DOI] [PubMed] [Google Scholar]
- Wilkinson K. D., Lee K. M., Deshpande S., Duerksen-Hughes P., Boss J. M., Pohl J. The neuron-specific protein PGP 9.5 is a ubiquitin carboxyl-terminal hydrolase. Science. 1989 Nov 3;246(4930):670–673. doi: 10.1126/science.2530630. [DOI] [PubMed] [Google Scholar]
- Wu R. S., Kohn K. W., Bonner W. M. Metabolism of ubiquitinated histones. J Biol Chem. 1981 Jun 10;256(11):5916–5920. [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yarden Y., Escobedo J. A., Kuang W. J., Yang-Feng T. L., Daniel T. O., Tremble P. M., Chen E. Y., Ando M. E., Harkins R. N., Francke U. Structure of the receptor for platelet-derived growth factor helps define a family of closely related growth factor receptors. Nature. 1986 Sep 18;323(6085):226–232. doi: 10.1038/323226a0. [DOI] [PubMed] [Google Scholar]







