Abstract
An alkylation repair deficient mutant of Escherichia coli (tag ada), lacking DNA glycosylase activity for removal of alkylated bases, was transformed by a genomic yeast DNA library and clones selected which survived plating on medium containing the alkylating agent methylmethane sulphonate. Three distinct yeast clones were identified which were able to suppress the alkylation sensitive phenotype of the bacterial mutant. Restriction enzyme analysis revealed common DNA fragments present in all three clones spanning 2 kb of yeast DNA. DNA from this region was sequenced and analysed for possible translation of polypeptides with any homology to either the Tag or the AlkA DNA glycosylases of E. coli. One open reading frame of 296 amino acids was identified encoding a putative protein with significant homology to AlkA. DNA containing the open reading frame was subcloned in E. coli expression vectors and cell extracts assayed for alkylbase DNA glycosylase activity. It appeared that such activity was expressed at levels sufficiently high for enzyme purification. The molecular weight of the purified protein was determined by SDS-PAGE to be 35,000 daltons, in good agreement with the 34,340 value calculated from the sequence. The yeast enzyme was able to excise 7-methylguanine as well as 3-methyladenine from dimethyl sulphate treated DNA, confirming the related nature of this enzyme to the AlkA DNA glycosylase from E. coli.
Full text
PDF

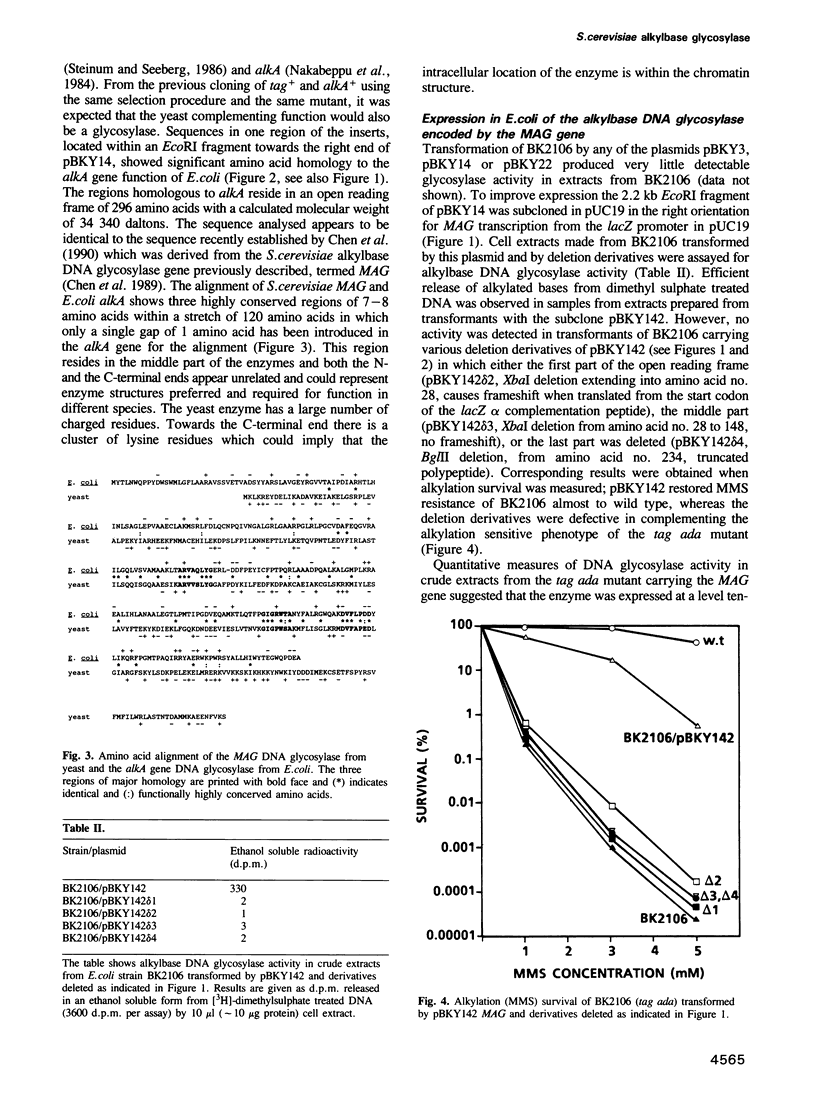



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjelland S., Seeberg E. Purification and characterization of 3-methyladenine DNA glycosylase I from Escherichia coli. Nucleic Acids Res. 1987 Apr 10;15(7):2787–2801. doi: 10.1093/nar/15.7.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtscher H. J., Cooper A. J., Couto L. B. Cellular responses to DNA damage in the yeast Saccharomyces cerevisiae. Mutat Res. 1988 Jul;194(1):1–8. doi: 10.1016/0167-8817(88)90050-8. [DOI] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Chen J., Derfler B., Maskati A., Samson L. Cloning a eukaryotic DNA glycosylase repair gene by the suppression of a DNA repair defect in Escherichia coli. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7961–7965. doi: 10.1073/pnas.86.20.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Derfler B., Samson L. Saccharomyces cerevisiae 3-methyladenine DNA glycosylase has homology to the AlkA glycosylase of E. coli and is induced in response to DNA alkylation damage. EMBO J. 1990 Dec;9(13):4569–4575. doi: 10.1002/j.1460-2075.1990.tb07910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Struhl K. Yeast mRNA initiation sites are determined primarily by specific sequences, not by the distance from the TATA element. EMBO J. 1985 Dec 1;4(12):3273–3280. doi: 10.1002/j.1460-2075.1985.tb04077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevallier M. R., Bloch J. C., Lacroute F. Transcriptional and translational expression of a chimeric bacterial-yeast plasmid in yeasts. Gene. 1980 Oct;11(1-2):11–19. doi: 10.1016/0378-1119(80)90082-7. [DOI] [PubMed] [Google Scholar]
- Clarke N. D., Kvaal M., Seeberg E. Cloning of Escherichia coli genes encoding 3-methyladenine DNA glycosylases I and II. Mol Gen Genet. 1984;197(3):368–372. doi: 10.1007/BF00329931. [DOI] [PubMed] [Google Scholar]
- Evensen G., Seeberg E. Adaptation to alkylation resistance involves the induction of a DNA glycosylase. Nature. 1982 Apr 22;296(5859):773–775. doi: 10.1038/296773a0. [DOI] [PubMed] [Google Scholar]
- Friedberg E. C. Deoxyribonucleic acid repair in the yeast Saccharomyces cerevisiae. Microbiol Rev. 1988 Mar;52(1):70–102. doi: 10.1128/mr.52.1.70-102.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goth-Goldstein R., Johnson P. L. Repair of alkylation damage in Saccharomyces cerevisiae. Mol Gen Genet. 1990 May;221(3):353–357. doi: 10.1007/BF00259399. [DOI] [PubMed] [Google Scholar]
- Hahn S., Hoar E. T., Guarente L. Each of three "TATA elements" specifies a subset of the transcription initiation sites at the CYC-1 promoter of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8562–8566. doi: 10.1073/pnas.82.24.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaasen I., Evensen G., Seeberg E. Amplified expression of the tag+ and alkA+ genes in Escherichia coli: identification of gene products and effects on alkylation resistance. J Bacteriol. 1986 Nov;168(2):642–647. doi: 10.1128/jb.168.2.642-647.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karran P., Hjelmgren T., Lindahl T. Induction of a DNA glycosylase for N-methylated purines is part of the adaptive response to alkylating agents. Nature. 1982 Apr 22;296(5859):770–773. doi: 10.1038/296770a0. [DOI] [PubMed] [Google Scholar]
- Karran P., Lindahl T., Ofsteng I., Evensen G. B., Seeberg E. Escherichia coli mutants deficient in 3-methyladenine-DNA glycosylase. J Mol Biol. 1980 Jun 15;140(1):101–127. doi: 10.1016/0022-2836(80)90358-7. [DOI] [PubMed] [Google Scholar]
- Lindahl T. DNA repair enzymes. Annu Rev Biochem. 1982;51:61–87. doi: 10.1146/annurev.bi.51.070182.000425. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Sedgwick B., Sekiguchi M., Nakabeppu Y. Regulation and expression of the adaptive response to alkylating agents. Annu Rev Biochem. 1988;57:133–157. doi: 10.1146/annurev.bi.57.070188.001025. [DOI] [PubMed] [Google Scholar]
- Maicas E., Friesen J. D. A sequence pattern that occurs at the transcription initiation region of yeast RNA polymerase II promoters. Nucleic Acids Res. 1990 Jun 11;18(11):3387–3393. doi: 10.1093/nar/18.11.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy T. V., Lindahl T. Methyl phosphotriesters in alkylated DNA are repaired by the Ada regulatory protein of E. coli. Nucleic Acids Res. 1985 Apr 25;13(8):2683–2698. doi: 10.1093/nar/13.8.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabeppu Y., Kondo H., Sekiguchi M. Cloning and characterization of the alkA gene of Escherichia coli that encodes 3-methyladenine DNA glycosylase II. J Biol Chem. 1984 Nov 25;259(22):13723–13729. [PubMed] [Google Scholar]
- Nisson P. E., Lawrence C. W. The isolation and characterization of an alkylating-agent-sensitive yeast mutant, ngs1. Mutat Res. 1986 May;165(3):129–137. doi: 10.1016/0167-8817(86)90047-7. [DOI] [PubMed] [Google Scholar]
- Percival K. J., Klein M. B., Burgers P. M. Molecular cloning and primary structure of the uracil-DNA-glycosylase gene from Saccharomyces cerevisiae. J Biol Chem. 1989 Feb 15;264(5):2593–2598. [PubMed] [Google Scholar]
- Popoff S. C., Spira A. I., Johnson A. W., Demple B. Yeast structural gene (APN1) for the major apurinic endonuclease: homology to Escherichia coli endonuclease IV. Proc Natl Acad Sci U S A. 1990 Jun;87(11):4193–4197. doi: 10.1073/pnas.87.11.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash L., Prakash S. Isolation and characterization of MMS-sensitive mutants of Saccharomyces cerevisiae. Genetics. 1977 May;86(1):33–55. doi: 10.1093/genetics/86.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riazuddin S., Lindahl T. Properties of 3-methyladenine-DNA glycosylase from Escherichia coli. Biochemistry. 1978 May 30;17(11):2110–2118. doi: 10.1021/bi00604a014. [DOI] [PubMed] [Google Scholar]
- Rydberg B., Lindahl T. Nonenzymatic methylation of DNA by the intracellular methyl group donor S-adenosyl-L-methionine is a potentially mutagenic reaction. EMBO J. 1982;1(2):211–216. doi: 10.1002/j.1460-2075.1982.tb01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakumi K., Nakabeppu Y., Yamamoto Y., Kawabata S., Iwanaga S., Sekiguchi M. Purification and structure of 3-methyladenine-DNA glycosylase I of Escherichia coli. J Biol Chem. 1986 Nov 25;261(33):15761–15766. [PubMed] [Google Scholar]
- Samson L., Cairns J. A new pathway for DNA repair in Escherichia coli. Nature. 1977 May 19;267(5608):281–283. doi: 10.1038/267281a0. [DOI] [PubMed] [Google Scholar]
- Sancar A., Sancar G. B. DNA repair enzymes. Annu Rev Biochem. 1988;57:29–67. doi: 10.1146/annurev.bi.57.070188.000333. [DOI] [PubMed] [Google Scholar]
- Seeberg E., Nissen-Meyer J., Strike P. Incision of ultraviolet-irradiated DNA by extracts of E. coli requires three different gene products. Nature. 1976 Oct 7;263(5577):524–526. doi: 10.1038/263524a0. [DOI] [PubMed] [Google Scholar]
- Steinum A. L., Seeberg E. Nucleotide sequence of the tag gene from Escherichia coli. Nucleic Acids Res. 1986 May 12;14(9):3763–3772. doi: 10.1093/nar/14.9.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas L., Yang C. H., Goldthwait D. A. Two DNA glycosylases in Escherichia coli which release primarily 3-methyladenine. Biochemistry. 1982 Mar 16;21(6):1162–1169. doi: 10.1021/bi00535a009. [DOI] [PubMed] [Google Scholar]
- Varshney U., Hutcheon T., van de Sande J. H. Sequence analysis, expression, and conservation of Escherichia coli uracil DNA glycosylase and its gene (ung). J Biol Chem. 1988 Jun 5;263(16):7776–7784. [PubMed] [Google Scholar]
- Yamagata Y., Odawara K., Tomita K., Nakabeppu Y., Sekiguchi M. Crystallization and preliminary X-ray diffraction studies of 3-methyladenine-DNA glycosylase II from Escherichia coli. J Mol Biol. 1988 Dec 20;204(4):1055–1056. doi: 10.1016/0022-2836(88)90063-0. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y., Katsuki M., Sekiguchi M., Otsuji N. Escherichia coli gene that controls sensitivity to alkylating agents. J Bacteriol. 1978 Jul;135(1):144–152. doi: 10.1128/jb.135.1.144-152.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]



