Abstract
Cancer treatments composed of immune checkpoint inhibitors and oncogene-targeted drugs might improve cancer management, but there has been little investigation of their combined potential as yet. To estimate the fraction of cancer cases that might benefit from such combination therapy, we conducted an exploratory study of cancer genomic data sets to determine the proportion with somatic mutation profiles amenable to either immunotherapy or targeted therapy. We surveyed 13,349 genomic profiles from public databases for cases with specific mutations targeted by current agents or a burden of exome-wide non-synonymous mutations (NsM) that exceeds a proposed threshold for response to checkpoint inhibitors. Overall, 8.9% of cases displayed profiles that could benefit from combination therapy, which corresponded to approximately 11.2% of US annual incident cancer cases. Frequently targetable mutations were in PIK3CA, BRAF, NF1, NRAS and PTEN. We also noted a high burden of NsM in cases with targetable mutations in SMO, DDR2, FGFR1, PTCH1, FGFR2 and MET. Our results indicate that a significant proportion of solid tumor patients are eligible for immuno-targeted combination therapy, and they suggest prioritizing specific cancers for trials of certain targeted and checkpoint inhibitor drugs.
Keywords: cancer, targeted therapy, immunotherapy, checkpoint inhibitor, TCGA, non-synonymous mutations
INTRODUCTION
The addition of checkpoint inhibitors (1) to the anticancer therapeutic armamentarium represents an important development and begs the question of how, and in what way, can these agents be wisely utilized in combination with one or more of the already established approaches, including molecularly targeted therapy (2). The potential to combine has strong scientific merit but clinical trials are needed (3). In principle, targeted drugs can induce rapid tumor death leading to the release of neoantigens, which, in turn, could have an impact on immune pathways, and thus, enhance the efficacy of checkpoint inhibitor treatment (4). On the other hand, checkpoint inhibitor treatment could reduce the selection for drug-resistant clones (3,4). In this context, we sought to consider the scope of cancer cases that could benefit from combination therapy and in doing so, provide genomic evidence that could influence the design of new trials needed to establish efficacy for combination therapy.
The rational design of combinations of targeted therapy with immunotherapy based on somatic genomic profiles offers a prioritization scheme for new clinical trials, perhaps minimizing the pursuit of clinical trials less likely to succeed (5). Assessment of somatic mutation profiles from large public databases provides an estimate of the prevalence of both targetable somatic mutations (6) and the burden of somatic nonsynonymous mutations (NsM), used as a surrogate for overall neoantigen load, which, in turn, correlates with clinical utility of checkpoint inhibitors for melanoma and lung adenocarcinoma (7–11). A higher burden of NsM increases the likelihood of response to immunotherapy, and, recently, we and others (12–15) have suggested that an NsM burden of greater than 192 could correlate with response to immunotherapy (16). Here, we analyze NsM count and mutations in genes with available targeted therapy across the 13,349 tumor samples drawn from public databases and infer which available targeted therapies are candidates for combination with checkpoint inhibitor therapy in future clinical trials. Future clinical trials will be necessary to determine the appropriate use of combination therapy in individual patients.
Materials and Methods
Data from 8,181 tumors from the Cancer Genome Atlas (TCGA) (17) Genome Data Analysis Center (November 2, 2015) were accessed and analyzed using the Broad Institute GDAC Firehose (18), a pipeline that includes MuTect for mutation calling and MutSig v2.0 (19) for aggregating mutation counts and rates (Supplementary Table S1). In addition, we examined copy-number and amplification data in 6074 cases from TCGA. For replication, 5,168 non-TCGA samples from the International Cancer Genome Consortium (ICGC) (20) and TumorPortal (TP) (21) were analyzed: 4,373 samples from ICGC (https://dcc.icgc.org/, September 17, 2016) and 795 (22–28) from TumorPortal (http://www.tumorportal.org/, March 9, 2016) (21). In our prior work (16), we observed the distribution of cases in TCGA with a burden exceeding the threshold of 192 NsM, using an empiric metric reported for checkpoint immunotherapy (13–15) and classified fractions of cancers that could potentially benefit from immunotherapy. For the targeted gene therapy response analysis, actionable mutations were selected from the largest precision medicine cancer trial, the NCI-MATCH trial (https://clinicaltrials.gov/ct2/show/NCT02465060, Supplementary Table S2): AKT1, BRAF, DDR2, EGFR, ERBB2, FGFR1, FGFR2, FGFR3, GNA11, GNAQ, KIT, MET, MLH1, MSH2, NF1, NF2, NRAS, PIK3CA, PTCH1, PTEN, and SMO. MLH1 and MSH2 are DNA repair genes that, when mutated, can lead to generation of neoepitopes (29,30). In a checkpoint inhibitor colorectal cancer trial, mismatch-repair deficiency patients showed better response rate (31). Consequently, we used MLH1 and MSH2 as positive controls. A Fisher’s exact test was applied to compare frequencies in the number of samples classified with or without potential checkpoint inhibitor response across gene mutation status. A conditional logistic regression was applied to the discovery (TCGA) and replication (ICGC and TP) sets to evaluate the association between those with ≥ 192 NsM (as a binary outcome) and target gene mutations. Conditional logistic regression results from discovery and replication sets were combined in a standard meta-analysis.
To estimate the standardized response rate, the percentage of cancer patients that could potentially benefit from combined targeted and immunotherapy in the United States, we reweighted the number of tumors with predicted benefit from combination therapy for each gene in TCGA, ICGC and TP to the population distribution of incident cancer cases by subtype in the United States using 2016 American Cancer Society (ACS) statistics (32).
The p-value threshold when accounting for multiple comparisons (N=21 candidate genes) is 0.0023. All statistical analyses report p-values from two-tailed tests and were performed using R v3.0.1.
RESULTS
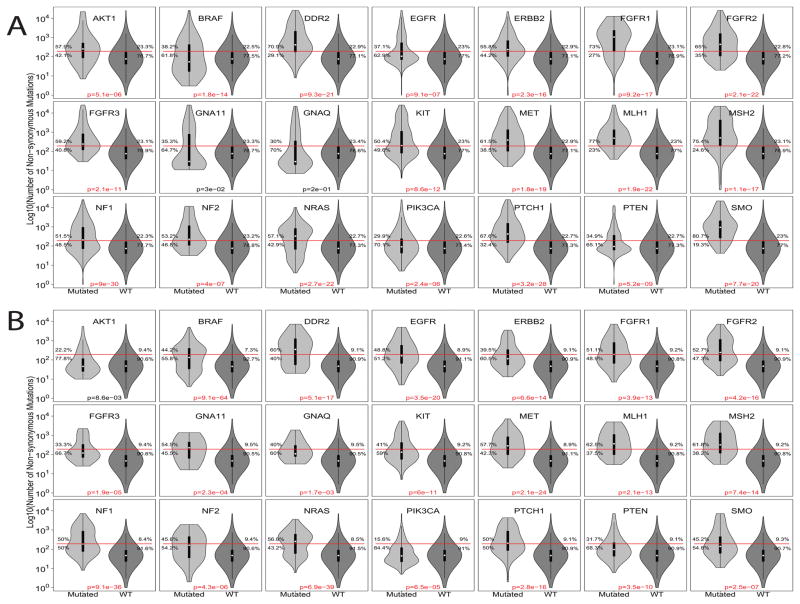
We replicated our previous assessment of the landscape of TCGA (16) using samples for 17 tumor types from ICGC and TP (Supplementary Figure S1). Overall, we observed that the highest fraction of cases with NsM greater than 192 in melanoma, lung adenocarcinoma and squamous-cell carcinoma, colon, bladder and gastric cancer, which could be prioritized for immunotherapy trials. In TCGA, we compared the NsM distribution of cancers either harboring or lacking a targetable mutation: 19 out of the 21 selected genes were observed to have a higher proportion of samples with NsM counts above the 192 threshold (Figure 1A). Tumors with mutations in these 19 genes showed a similar distribution in the replication set (ICGC and TP, Figure 1B), except for AKT1. GNA11, and GNAQ, which were not significant in the discovery set, but were significant in the replication set. MLH1 and MSH2, the positive control genes, were significant in both TCGA and the ICGC and TP sets.
Figure 1.
Violin plot for the number of non-synonymous mutations (log10) across 21 target genes combining all tumor types on (A) TCGA as discovery set and (B) ICGC plus TP as replication set. Red line: 192 non-synonymous mutations threshold. P-value refers to fisher’s test comparing proportion of samples regarding gene mutation and 192 NsM threshold.
To control for heterogeneity across tumor types, we used a conditional logistic regression analysis (conditioning on tumor type) to assess the impact of the target gene mutation on overall NsM burden (≥192 or not). All genes, except for FGFR3 and GNAQ displayed global significance in the discovery set (TCGA, Supplementary Table S3). In the replication set (ICGC and TP), all genes were significant, except AKT1 (Supplementary Table S4). The discovery and replication sets showed a strong correlation between estimated beta values (Pearson’s r = 0.85; p-value = 1.3 x 10−6; Supplementary Figure S2), suggesting minimal differences in the effect of targetable mutations on the NsM burden. Consequently, we performed a meta-analysis combining results from the discovery and replication sets, which suggest that cases with a mutation in one of the 21 selected target genes also increases the probability of a high NsM (≥192 NsM; Table 1). For the 21 targetable genes, there was no correlation between meta-analysis estimated beta values and gene coding sequence length (Supplementary Figure S3), suggesting minimal influence of gene size.
TABLE 1.
Meta-analysis combining results from the discovery and replication sets
| Gene | p-value | Estimate |
|---|---|---|
| MLH1 | 3.7E-20 | 3.07 |
| MSH2 | 7.1E-18 | 2.81 |
| GNA11 | 2.2E-08 | 2.75 |
| FGFR1 | 1.3E-24 | 2.64 |
| DDR2 | 2.3E-16 | 2.37 |
| NF2 | 1.4E-07 | 2.26 |
| SMO | 4.2E-11 | 2.16 |
| PTCH1 | 3.7E-20 | 2.14 |
| FGFR2 | 4.1E-17 | 2.13 |
| NF1 | 1.1E-32 | 1.94 |
| ERBB2 | 2.2E-22 | 1.93 |
| MET | 5.4E-16 | 1.92 |
| AKT1 | 5.5E-07 | 1.55 |
| GNAQ | 2.0E-03 | 1.54 |
| PTEN | 1.7E-12 | 1.43 |
| EGFR | 4.1E-11 | 1.38 |
| PIK3CA | 1.4E-24 | 1.37 |
| KIT | 1.7E-07 | 1.32 |
| NRAS | 4.3E-08 | 0.95 |
| FGFR3 | 9.1E-04 | 0.94 |
| BRAF | 7.3E-10 | 0.79 |
To evaluate if tumor stage could influence the presence of target genes or NsM burden and, therefore, combination therapy, 5,208 TCGA samples with tumor stage data were used to compare stage I and II (early) samples to stage III and IV (advanced). Although the reduced sample size limited our power, we did not note differences in predicted response to combination therapy between early and advanced tumors (Supplementary Figure S4 and Supplementary Table S5). Concordant with preliminary data showing the therapeutic efficacy of adjuvant and neoadjuvant scenarios (33,34), this analysis suggests that combination therapy could be beneficial for early tumors as well as more advanced cancers.
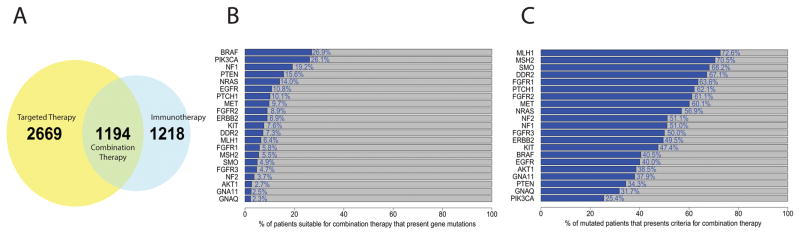
In the combined dataset, 3,863 (28.9%) of the total 13,349 cases showed a mutation in at least one selected target gene and would thus be eligible for targeted therapy. Similarly, 2,412 (18%) cases have a mutation burden exceeding the threshold of 192 NsM, suggesting a greater likelihood for response to immunotherapy. The intersection of the two analyses reveals that 1,194 cases (8.9%) harbor both a targetable mutation and an NsM burden exceeding the threshold for likely response to immunotherapy, thus defining those likely to respond to combination therapy (Figure 2A). Gene mutation frequencies in these cases ranged from 26.9% (BRAF) to 2.3% (GNAQ; Figure 2B and Supplementary Figure S5). Mutations in at least one of the top five mutated genes, BRAF, PIK3CA, NF1, NRAS, and PTEN, accounted for 76.7% overall.
Figure 2.
(A) Venn diagram for all 13,349 samples from discovery and replication set combined showing proportion that could respond to targeted, checkpoint inhibitor and combination therapy in our analysis. (B) Stacked plot shows the percent of patients suitable for combination therapy that presents each studied gene mutation in samples. Patients can be in more than one category since patients can have multiple genes mutated. (C) Stacked plot shows, for each studied gene, the proportion of mutated patients that presents criteria for combination therapy. Patients can be in more than one category since patients can have multiple genes mutated.
We further examined the subset of cases harboring a high NsM burden (≥192) in those with a targetable gene mutation. Figure 2C shows the percentage of samples with mutations of each gene eligible for immune checkpoint inhibitor therapy; more than 60% of cases harboring mutations in SMO, DDR2, FGFR1, PTCH1, FGFR2, and MET presented with a high burden of NsM (≥192). Although less common, they could be prioritized as candidates to study whether combination treatment could be more efficacious.
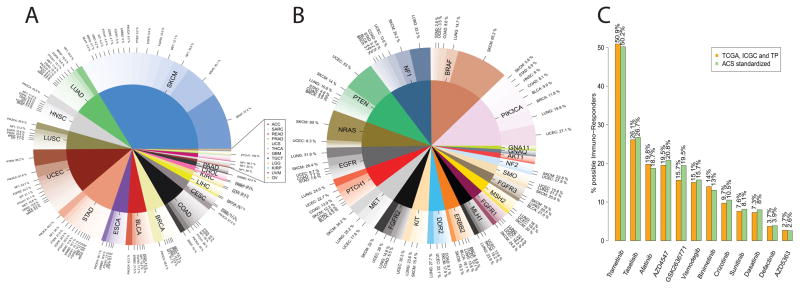
Cancer genomic databases such as TCGA, ICGC and TP do not adequately represent incidence rates of tumor types present in the general population. It is notable that TCGA selected for large tumor biopsies that would provide adequate material for multiple platforms, and thus are oversampled for cases with late stage disease. To estimate how many cancer patients could potentially benefit from combination therapy, we standardized the number of tumors (Figure 3A) to the United States cancer incidence rate for 21 out of 28 tumor types using data from 2016 American Cancer Society (ACS) statistics (32) (Supplementary Table S6). The fraction of 8.9% observed in TCGA, ICGC and TP that could benefit from combination therapy corresponds to 11.2% of cancer patients in the US. Although minor changes in the ranking of targeted genes were present after standardization, the prevalence of targetable mutations in PIK3CA, BRAF, NF1, NRAS, and PTEN would still represent a sizable fraction for possible combination therapy (Figure 3B).
Figure 3.
(A) Plot showing the most common mutations in each tumor type for the samples predicted to respond to combination therapy. These genes can be selected for tumor specific clinical trials for combination therapy. (B) Genes and tumor type standardize to 2016 Cancer Statistics. (C) Percent of samples predicted to benefit from combination therapy that could receive each targeted drug based on the mutation profile. Orange, data from combined databases (TCGA, ICGC and TP). Green, data from 2016 Cancer Statistics Standardized analysis.
Since drugs can target more than one gene, we matched mutated genes to the NCI-MATCH list of targeted drugs (Supplementary Figure S6) to determine which were also enriched for a high burden of NsM (≥192). Of the cases in TCGA, ICGC and TP that could benefit from combination therapy, 50.9% had BRAF, NF1, GNAQ and/or GNA11 mutations which can be targeted by Trametinib. Similarly, Taselisib (targets PIK3CA mutations) could be used in 26.1% of these cases and Afatinib (EGFR and ERBB2 mutations) in 19.8% of cases (Figure 3C).
Point mutations are not the only gene abnormalities that can be used for targeted therapy selection. In the NCI-MATCH trial, copy-number alteration, amplification and translocations are also used for therapy selection, mainly for MET, ERBB2, NF2, and PTEN. TCGA provides copy numbers data for 6074 patients that allowed us to evaluate the impact of these parameters on combination therapy, for these three genes. When considering copy number alterations, an additional 64 patients could benefit from MET-targeted therapy combined with checkpoint inhibitors, 84 for ERRB2, 8 for NF2, and 43 for PTEN (Supplementary Figure S7 and Supplementary Table S7). Considering only TCGA patients with mutation and copy-number data, the proportion of US patients suitable for combination therapy shifts from 12.6% to 15.8% when MET, ERBB2, NF2, and PTEN copy-number and amplification, data are included.
DISCUSSION
Combining targeted therapy with checkpoint inhibitors has the potential to improve cancer outcomes (3). Our study explores the mutational profiles in existing genomic data and should be viewed as an exercise to estimate possible benefit and prioritization of studies absolutely required to evaluate the efficacy of checkpoint inhibitors and targeted therapy combination. Our results suggest 8.9% of cases drawn from the genomic databases (TCGA, ICGC and TP; n=13,349), which after normalizing for incidence rates in the US is approximately 11.2%, could potentially benefit from combination therapy.
We performed two analytical approaches for prioritization: first we investigated the most common genes mutated in the 8.9% of TCGA, ICGC and TP samples that could benefit from combination therapy. Then we searched for mutated genes known to be targeted that also had a higher burden of NsM. Five genes, namely BRAF, PIK3CA, NF1, NRAS, and PTEN, were mutated in 76.7% of samples that could benefit from combination therapy, suggesting drugs targeting these genes should be prioritized for future clinical trials. From the second approach, patients already known to have SMO, DDR2, FGFR1, PTCH1, FGFR2 and/or MET mutations could represent high priority candidates for combination therapy, since more than 60% of samples with these mutations also have an NsM burden above 192.
The NCI-MATCH trial is based upon targeting known genomic abnormalities, regardless of tumor type, with either FDA-approved drugs or agents approved for testing. Trametinib would be the suitable targeted drug for 50.9% of the patients predicted to benefit from combination therapy. Trametinib targets BRAF (nonV600 alone or V600 with Dabrafenib) and NF1, two of the top 5 mutated genes. Two additional drugs, Taselisib (targets PIK3CA), and Afatinib (targets EFGR and HER2) could each be indicated in one quarter of those with high NsM burden as well. It is notable that 25.5% of cases with ≥192 NsM present with more than one drug-targetable mutation, opening opportunities for other drug combinations for subsequent lines of therapy in case of relapse or in three-way combination (Supplementary Table S5). Still, in a dynamic clinical scenario in which intra-tumor heterogeneity can limit the effect of targeted therapy (35), the possibility of multiple combinations could be important. It is also plausible that heterogeneity could undermine the precision of our model based on a dichotomous assessment of the mutational landscape.
The estimated proportion of patients suitable for combination therapy with immunotherapy and targeted therapy found in this study reflects the current state of directly related to targeted therapies, which is sure to increase in the near future. It is important to point out that other molecular mechanisms, not only mutations can play an important role on cancer biology and may be a biomarker for drug selection. The NCI-MATCH trial selects patients with copy-number alterations of MET, ERBB2, NF2, and PTEN. We are limited by the number of available samples in TCGA with information on copy number for these genes, but including this information in our analysis increases the number of patients who could potentially respond to combination therapy.
Our assessment of possible immunotherapy response is based on available exome-wide NsM counts, which can vary by tumor and stage. A more precise estimate will be possible when new biomarkers, and larger datasets that include information of molecular abnormalities become available. Nonetheless, we did observe replication of our earlier finding for six cancers (16). We recognize that the mutational load, one of 7 parameters important in the recently proposed cancer immunogram (12), may not adequately account for the response. However, the use of large cancer genomic datasets enables a scalable approach that could influence the decision of when and how to study combination therapy including targeted mutations and checkpoint inhibition, as well as perhaps other anticancer modalities.
In conclusion, our survey of somatic mutations in 13,349 tumor samples has provided a foundation for considering how and when to apply combination therapy. Validation with clinical trials is required for testing whether up to one tenth of cases could be candidates for combination therapy, before any individual patient receives these treatments. At the same time, the accumulation of new data on cancer genomes could improve the precision of the estimated prevalence of mutational events that could be useful in driving research into combination therapy.
Supplementary Material
Acknowledgments
Financial support: This study was supported by the Intramural Research Program of the National Institutes of Health.
Footnotes
Conflicts of interest: The authors declare that they have no competing interests.
References
- 1.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–64. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khalil DN, Smith EL, Brentjens RJ, Wolchok JD. The future of cancer treatment: immunomodulation, CARs and combination immunotherapy. Nat Rev Clin Oncol. 2016;13:273–90. doi: 10.1038/nrclinonc.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharma P, Allison JP. Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell. 2015;161:205–14. doi: 10.1016/j.cell.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vanneman M, Dranoff G. Combining immunotherapy and targeted therapies in cancer treatment. Nat Rev Cancer. 2012;12:237–51. doi: 10.1038/nrc3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morgan S, Grootendorst P, Lexchin J, Cunningham C, Greyson D. The cost of drug development: a systematic review. Health Policy. 2011;100:4–17. doi: 10.1016/j.healthpol.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Collins FS, Varmus H. A New Initiative on Precision Medicine. N Engl J Med. 2015;372:793–5. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gubin MM, Zhang X, Schuster H, Caron E, Ward JP, Noguchi T, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515:577–81. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tran E, Turcotte S, Gros A, Robbins PF, Lu Y-C, Dudley ME, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344:641–5. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–4. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castle JC, Kreiter S, Diekmann J, Löwer M, van de Roemer N, de Graaf J, et al. Exploiting the mutanome for tumor vaccination. Cancer Res. 2012;72:1081–91. doi: 10.1158/0008-5472.CAN-11-3722. [DOI] [PubMed] [Google Scholar]
- 11.Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity. 2013;39:1–10. doi: 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 12.Blank CU, Haanen JB, Ribas A, Schumacher TN. CANCER IMMUNOLOGY. The “cancer immunogram”. Science. 2016;352:658–60. doi: 10.1126/science.aaf2834. [DOI] [PubMed] [Google Scholar]
- 13.Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–11. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–99. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–8. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colli LM, Machiela MJ, Myers TA, Jessop L, Yu K, Chanock SJ. Burden of Nonsynonymous Mutations among TCGA Cancers and Candidate Immune Checkpoint Inhibitor Responses. Cancer Res. 2016;76:3767–72. doi: 10.1158/0008-5472.CAN-16-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, Ellrott K, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–20. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Center BITGDA. Analysis-ready standardized TCGA data from Broad GDAC Firehose stddata__2015_06_01 run. Broad Inst MIT Harvard; 2015. [Google Scholar]
- 19.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–8. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.International Cancer Genome Consortium. Hudson TJ, Anderson W, Artez A, Barker AD, Bell C, et al. International network of cancer genome projects. Nature. 2010;464:993–8. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–20. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat J-P, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–9. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat J-P, et al. A Landscape of Driver Mutations in Melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–6. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–60. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, et al. Mapping the Hallmarks of Lung Adenocarcinoma with Massively Parallel Sequencing. Cell. 2012;150:1107–20. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486:405–9. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chabanon RM, Pedrero M, Lefebvre C, Marabelle A, Soria J-C, Postel-Vinay S. Mutational Landscape and Sensitivity to Immune Checkpoint Blockers. Clin Cancer Res. 2016;22:4309–21. doi: 10.1158/1078-0432.CCR-16-0903. [DOI] [PubMed] [Google Scholar]
- 30.Jeggo PA, Pearl LH, Carr AM. DNA repair, genome stability and cancer: a historical perspective. Nat Rev Cancer. 2015;16:35–42. doi: 10.1038/nrc.2015.4. [DOI] [PubMed] [Google Scholar]
- 31.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509–20. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 33.Eggermont AMM, Chiarion-Sileni V, Grob J-J, Dummer R, Wolchok JD, Schmidt H, et al. Adjuvant ipilimumab versus placebo after complete resection of high-risk stage III melanoma (EORTC 18071): a randomised, double-blind, phase 3 trial. Lancet Oncol. 2015;16:522–30. doi: 10.1016/S1470-2045(15)70122-1. [DOI] [PubMed] [Google Scholar]
- 34.Liu J, Blake SJ, Yong MCR, Harjunpää H, Ngiow SF, Takeda K, et al. Improved Efficacy of Neoadjuvant Compared to Adjuvant Immunotherapy to Eradicate Metastatic Disease. Cancer Discov. 2016;6 doi: 10.1158/2159-8290.CD-16-0577. [DOI] [PubMed] [Google Scholar]
- 35.Fisher R, Pusztai L, Swanton C. Cancer heterogeneity: implications for targeted therapeutics. Br J Cancer. 2013;108:479–85. doi: 10.1038/bjc.2012.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.