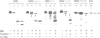Abstract
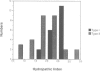
We previously showed that the amino-terminal region of P-450 is responsible not only for targeting to endoplasmic reticulum (ER) membrane but also for stable anchoring to the membrane. In the present study, we introduced several mutations or deletions into the signal-anchor region of the chimeric proteins in which the amino-terminal regions of two forms of cytochrome P-450 were fused to the mature portion of interleukin 2. The amino-terminal acidic amino acid residues were replaced with basic amino acid residues or the hydrophobic core sequences were partially deleted, and these mutant proteins were assayed in vitro for their capacity to be inserted into or translocated across the ER membrane. The proteins that received the former manipulations were processed and the IL-2 portion was translocated across the membrane. In one case, the processing did not occur, thereby enabling the chimeric protein to anchor on the luminal side of the ER. Those that received the latter manipulation were also processed and the IL-2 portion translocated across the ER. These results strongly suggest that the signal-anchor function is determined both by the amino-terminal charged amino acid residues and by the length of the hydrophobic stretch.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Black S. D., Coon M. J. Structural features of liver microsomal NADPH-cytochrome P-450 reductase. Hydrophobic domain, hydrophilic domain, and connecting region. J Biol Chem. 1982 May 25;257(10):5929–5938. [PubMed] [Google Scholar]
- Bos T. J., Davis A. R., Nayak D. P. NH2-terminal hydrophobic region of influenza virus neuraminidase provides the signal function in translocation. Proc Natl Acad Sci U S A. 1984 Apr;81(8):2327–2331. doi: 10.1073/pnas.81.8.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman J., Inukai M., Inouye M. Dual functions of the signal peptide in protein transfer across the membrane. Cell. 1985 Nov;43(1):351–360. doi: 10.1016/0092-8674(85)90040-6. [DOI] [PubMed] [Google Scholar]
- DuBois G. C., Appella E., Armstrong R., Levin W., Lu A. Y., Jerina D. M. Hepatic microsomal epoxide hydrase. Chemical evidence for a single polypeptide chain. J Biol Chem. 1979 Jul 25;254(14):6240–6243. [PubMed] [Google Scholar]
- Ebina Y., Ellis L., Jarnagin K., Edery M., Graf L., Clauser E., Ou J. H., Masiarz F., Kan Y. W., Goldfine I. D. The human insulin receptor cDNA: the structural basis for hormone-activated transmembrane signalling. Cell. 1985 Apr;40(4):747–758. doi: 10.1016/0092-8674(85)90334-4. [DOI] [PubMed] [Google Scholar]
- Fujiki Y., Hubbard A. L., Fowler S., Lazarow P. B. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J Cell Biol. 1982 Apr;93(1):97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garoff H. Using recombinant DNA techniques to study protein targeting in the eucaryotic cell. Annu Rev Cell Biol. 1985;1:403–445. doi: 10.1146/annurev.cb.01.110185.002155. [DOI] [PubMed] [Google Scholar]
- Gilmore R., Walter P., Blobel G. Protein translocation across the endoplasmic reticulum. II. Isolation and characterization of the signal recognition particle receptor. J Cell Biol. 1982 Nov;95(2 Pt 1):470–477. doi: 10.1083/jcb.95.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeuptle M. T., Flint N., Gough N. M., Dobberstein B. A tripartite structure of the signals that determine protein insertion into the endoplasmic reticulum membrane. J Cell Biol. 1989 Apr;108(4):1227–1236. doi: 10.1083/jcb.108.4.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann E., Rapoport T. A., Lodish H. F. Predicting the orientation of eukaryotic membrane-spanning proteins. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5786–5790. doi: 10.1073/pnas.86.15.5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa N., Mihara K., Sato R. Structural analysis of cloned cDNAs for polycyclic hydrocarbon-inducible forms of rabbit liver microsomal cytochrome P-450. J Biochem. 1987 Jun;101(6):1471–1479. doi: 10.1093/oxfordjournals.jbchem.a122017. [DOI] [PubMed] [Google Scholar]
- Krieg P. A., Melton D. A. Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res. 1984 Sep 25;12(18):7057–7070. doi: 10.1093/nar/12.18.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lingappa V. R., Katz F. N., Lodish H. F., Blobel G. A signal sequence for the insertion of a transmembrane glycoprotein. Similarities to the signals of secretory proteins in primary structure and function. J Biol Chem. 1978 Dec 25;253(24):8667–8670. [PubMed] [Google Scholar]
- Lipp J., Dobberstein B. Signal recognition particle-dependent membrane insertion of mouse invariant chain: a membrane-spanning protein with a cytoplasmically exposed amino terminus. J Cell Biol. 1986 Jun;102(6):2169–2175. doi: 10.1083/jcb.102.6.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipp J., Dobberstein B. The membrane-spanning segment of invariant chain (I gamma) contains a potentially cleavable signal sequence. Cell. 1986 Sep 26;46(7):1103–1112. doi: 10.1016/0092-8674(86)90710-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie P. I. Rat liver UDP-glucuronosyltransferase. Sequence and expression of a cDNA encoding a phenobarbital-inducible form. J Biol Chem. 1986 May 5;261(13):6119–6125. [PubMed] [Google Scholar]
- Markoff L., Lin B. C., Sveda M. M., Lai C. J. Glycosylation and surface expression of the influenza virus neuraminidase requires the N-terminal hydrophobic region. Mol Cell Biol. 1984 Jan;4(1):8–16. doi: 10.1128/mcb.4.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T., Emi Y., Kawabata S., Omura T. Purification and characterization of three male-specific and one female-specific forms of cytochrome P-450 from rat liver microsomes. J Biochem. 1986 Nov;100(5):1359–1371. doi: 10.1093/oxfordjournals.jbchem.a121842. [DOI] [PubMed] [Google Scholar]
- Meyer D. I., Krause E., Dobberstein B. Secretory protein translocation across membranes-the role of the "docking protein'. Nature. 1982 Jun 24;297(5868):647–650. doi: 10.1038/297647a0. [DOI] [PubMed] [Google Scholar]
- Mihara K., Sato R. Molecular cloning and sequencing of cDNA for yeast porin, an outer mitochondrial membrane protein: a search for targeting signal in the primary structure. EMBO J. 1985 Mar;4(3):769–774. doi: 10.1002/j.1460-2075.1985.tb03695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier S., Van Luc P., Kreibich G., Sabatini D. D., Adesnik M. Signals for the incorporation and orientation of cytochrome P450 in the endoplasmic reticulum membrane. J Cell Biol. 1988 Aug;107(2):457–470. doi: 10.1083/jcb.107.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan D. O., Edman J. C., Standring D. N., Fried V. A., Smith M. C., Roth R. A., Rutter W. J. Insulin-like growth factor II receptor as a multifunctional binding protein. Nature. 1987 Sep 24;329(6137):301–307. doi: 10.1038/329301a0. [DOI] [PubMed] [Google Scholar]
- Nagata K., Matsunaga T., Gillette J., Gelboin H. V., Gonzalez F. J. Rat testosterone 7 alpha-hydroxylase. Isolation, sequence, and expression of cDNA and its developmental regulation and induction by 3-methylcholanthrene. J Biol Chem. 1987 Feb 25;262(6):2787–2793. [PubMed] [Google Scholar]
- Nebert D. W., Gonzalez F. J. P450 genes: structure, evolution, and regulation. Annu Rev Biochem. 1987;56:945–993. doi: 10.1146/annurev.bi.56.070187.004501. [DOI] [PubMed] [Google Scholar]
- Nelson D. R., Strobel H. W. On the membrane topology of vertebrate cytochrome P-450 proteins. J Biol Chem. 1988 May 5;263(13):6038–6050. [PubMed] [Google Scholar]
- Sakaguchi M., Mihara K., Sato R. A short amino-terminal segment of microsomal cytochrome P-450 functions both as an insertion signal and as a stop-transfer sequence. EMBO J. 1987 Aug;6(8):2425–2431. doi: 10.1002/j.1460-2075.1987.tb02521.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi M., Mihara K., Sato R. Signal recognition particle is required for co-translational insertion of cytochrome P-450 into microsomal membranes. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3361–3364. doi: 10.1073/pnas.81.11.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid S. R., Spiess M. Deletion of the amino-terminal domain of asialoglycoprotein receptor H1 allows cleavage of the internal signal sequence. J Biol Chem. 1988 Nov 15;263(32):16886–16891. [PubMed] [Google Scholar]
- Schneider C., Owen M. J., Banville D., Williams J. G. Primary structure of human transferrin receptor deduced from the mRNA sequence. Nature. 1984 Oct 18;311(5987):675–678. doi: 10.1038/311675b0. [DOI] [PubMed] [Google Scholar]
- Spiess M., Lodish H. F. An internal signal sequence: the asialoglycoprotein receptor membrane anchor. Cell. 1986 Jan 17;44(1):177–185. doi: 10.1016/0092-8674(86)90496-4. [DOI] [PubMed] [Google Scholar]
- Szczesna-Skorupa E., Browne N., Mead D., Kemper B. Positive charges at the NH2 terminus convert the membrane-anchor signal peptide of cytochrome P-450 to a secretory signal peptide. Proc Natl Acad Sci U S A. 1988 Feb;85(3):738–742. doi: 10.1073/pnas.85.3.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesna-Skorupa E., Kemper B. NH2-terminal substitutions of basic amino acids induce translocation across the microsomal membrane and glycosylation of rabbit cytochrome P450IIC2. J Cell Biol. 1989 Apr;108(4):1237–1243. doi: 10.1083/jcb.108.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P., Blobel G. Purification of a membrane-associated protein complex required for protein translocation across the endoplasmic reticulum. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7112–7116. doi: 10.1073/pnas.77.12.7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P., Ibrahimi I., Blobel G. Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to in-vitro-assembled polysomes synthesizing secretory protein. J Cell Biol. 1981 Nov;91(2 Pt 1):545–550. doi: 10.1083/jcb.91.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka H., Morohashi K., Sogawa K., Miyata T., Kawajiri K., Hirose T., Inayama S., Fujii-Kuriyama Y., Omura T. Structural analysis and specific expression of microsomal cytochrome P-450(M-1) mRNA in male rat livers. J Biol Chem. 1987 Feb 5;262(4):1706–1711. [PubMed] [Google Scholar]
- Zerial M., Huylebroeck D., Garoff H. Foreign transmembrane peptides replacing the internal signal sequence of transferrin receptor allow its translocation and membrane binding. Cell. 1987 Jan 16;48(1):147–155. doi: 10.1016/0092-8674(87)90365-5. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Analysis of the distribution of charged residues in the N-terminal region of signal sequences: implications for protein export in prokaryotic and eukaryotic cells. EMBO J. 1984 Oct;3(10):2315–2318. doi: 10.1002/j.1460-2075.1984.tb02132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Control of topology and mode of assembly of a polytopic membrane protein by positively charged residues. Nature. 1989 Oct 5;341(6241):456–458. doi: 10.1038/341456a0. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem. 1983 Jun 1;133(1):17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]