Abstract
Semiconductor nanotechnology has created the ultimate analytical tool: a nanopore with single molecule sensitivity. This tool offers the intriguing possibility of high- throughput, low cost sequencing of DNA with the absolute minimum of material and preprocessing. The exquisite single molecule sensitivity obviates the need for costly and error-prone procedures like polymerase chain reaction amplification. Instead, nanopore sequencing relies on the electric signal that develops when a DNA molecule translocates through a pore in a membrane. If each base pair has a characteristic electrical signature, then ostensibly a pore could be used to analyze the sequence by reporting all of the signatures in a single read without resorting to multiple DNA copies. The potential for a long read length combined with high translocation velocity should make resequencing inexpensive and allow for haplotyping and methylation profiling in a chromosome.
The aim of genomic science is to predict biological behavior using the encyclopedic information stored in the form of DNA within each cell. The draft sequence of the human genome that emerged in early 2001 was the first step toward this overarching goal.1,2 A daunting challenge that has emerged since then is the detection of the subtle genetic variations in a population. And so the majority of the work ahead involves resequencing genomes with an already known base sequence.
Following Shendure’s analysis, for resequencing the error rate has to be less than the expected variation in the sequence.3 Since human chromosomes differ by approximately 1 in every 1000 base pairs (bp), an error rate of 1/100 kb would be needed to ensure confidence. In the current state-of-the-art sequencing techniques, the accuracy of a raw read is 99.7% and so to cover the diploid human genome (6 billion base-pair), 40 billion raw bases will have to be read. Currently, a run in an Applied Biosystems SOLiD (sequencing by oligo ligation) sequencer requires 5 days and produces 3–4 Gb of sequence data with an average read length of 25–35 bp, costing $5.81 per Mb.4 Applied Biosystems estimates that their SOLiD sequencer will be able to sequence an entire human genome for only $10,000 in just 2 weeks. A faster instrument with longer reads will be cheaper still.
In this review, we will narrowly focus on the prospects for resequencing a single molecule of DNA using a nanopore and the medicine that might be derived from them. More comprehensive reviews describing recent developments in nanopore detection are given elsewhere.5,6 Single molecule DNA sequencing represents the ultimate in sequencing technology, which extracts the maximum amount of information from a minimum of material. And of all the emerging technologies,3 sequencing a single molecule of DNA with a nanopore is the most revolutionary. It is revolutionary because it combines the potential for long read lengths (>5 kb) with high speed (1 bp/10 ns), while obviating the need for costly procedures such as polymerase chain reaction (PCR) amplification due to the single molecule sensitivity. The nanopore sequencing concept uses a new approach to detection, which is reminiscent of Coulter’s original idea of using objects within a constricted current path to alter the electrical resistance.7 Nanopore sequencing relies on the electric signal that develops when DNA translocates through a pore in a membrane. DNA is a highly charged polyanion. By applying an electric field to the membrane, individual DNA molecules can be forced to move through the pore. If each base has a characteristic electrical signature, then potentially a pore could be used to analyze the sequence by reporting all of the signatures in a single read without resorting to multiple copies of DNA.
One compelling application for fast and cheap resequencing is mutation sequencing where recent work has shown that the majority of human cancers do not always have mutations in the same locations or even the same genes.8 Moreover, the mutations and genotype of the individual have been shown to be important in determining a drug’s effectiveness. Genotyping reveals how an individual differs from a group or species. A single nucleotide polymorphism (SNP) is the most common type of genetic variation—more than 9 million SNPs have already been identified in the human genome.9 A SNP is a variation in a single nucleotide of the DNA sequence that gives rise to different genotypes called alleles. The average frequency of SNPs in the human genome is about 1 per 1000 bp, but they are not uniformly distributed within a chromosome—the two-thirds of SNPs are in noncoding regions of the genome where they act as markers for evolutionary genomics. On the other hand, SNPs found in coding regions can adversely affect protein structure forming the molecular basis for disease, making them important markers in the study of disease genetics and pharmacogenomics.10,11
Following Misra’s thorough review of the subject,11 we find that the assays that have been developed for genotyping can be categorized as: enzymatic cleavage, hybridization, ligation, and primer extension, or some combinations of these—all of them require a PCR amplification step. PCR amplification of a target region of DNA containing the SNP is used to introduce specificity and increase the number of molecules for easy detection. For example, restriction fragment length polymorphism (RFLP) uses enzymes that recognize, bind to, and cleave a specific cognate sequence for detecting genetic variations. While RFLP does not require probes, it has limited throughput and is applicable to a limited number of SNPs. On the other hand, the hybridization strategy has been implemented on high-throughput platforms using microarrays because it does not require any enzymatic reactions to discriminate alleles. Hybridization uses the thermal stability of complementary target-probe strands relative to mismatched strands to discriminate alleles. For example, the GeneChip® uses an array containing a myriad of allele-specific probes, each consisting of 25-mer oligonucleotides immobilized on a glass slide. SNP-containing regions are amplified by PCR from genomic DNA, and then cleaved, tagged, and hybridized to the probe array. Multiple probes that differ at a single position can then be used for analyzing each SNP. One array can hold millions of probes for genotyping SNPs.
SNP genotyping assays such as BeadARRAY® and MassExtend® and pyrosequencing rely on this same kind of multiplexing (detecting up to 105 SNPs per assay) to improve throughput and lower the cost. The cost per genotype can be as low as 1–10 [simble] But almost all of these assays still require PCR amplification, which is expensive, problematic, and ultimately limits the throughput. Despite relentless development, further improvements and new methods are required to provide a cost-effective, routine, genome-wide SNP analyses in large sample collections. Moreover, the current technology is focused on detecting single SNPs whereas the genetic origin of a disease is not always associated with just one SNP.12 Sometimes a combination of SNPs, usually present on the same chromosome, can interact to adversely affect protein expression. Currently, the main limitation to haplotyping is that each SNP is assayed separately and hence it is not possible to accurately infer the exact haplotype map from SNP assays alone.12–14 Instead the haplotype or phase of a set of SNPs is usually inferred computationally from unphased data.14 Detecting SNPs in a single genomic DNA molecule could circumvent this limitation entirely—a read length extending over the length of a chromosome would automatically set the phase of a set of SNPs.
Resequencing can provide other clues about health, besides the bare sequence. An individual organism can express the same genes differently depending on the epigenetic profile. Epigenetic information is not encoded in the DNA sequence, but instead reflects environmental factors affecting gene expression. The DNA sequence is generally written with four chemical bases called adenine (A), guanine (G), cytosine (C), and thymine (T), but the methylation of cytosine is a covalent modification of DNA that produces what amounts to a fifth DNA base, 5-methylcytosine.15 Methylation of cytosine occurs when a methyl group is transferred from S-adenosylmethionine to the C-5 position of the pyrimidine ring by a family of DNA methyltransferases and it usually occurs in the context of the sequence CpG (i.e., 5′–CG–3′ or cytosine directly followed by a guanine). Methylation does not interfere with the Watson/Crick pairing—the methyl group is positioned in the major groove of the DNA. Yet, the pattern of methylation controls protein binding to target sites on the DNA affecting changes in gene expression and in chromatin organization. In eukaryotes, the information encoded in the methylation pattern is used to silence genes, among other things. Physiologically, gene silencing orchestrates a myriad of biological processes such as differentiation, imprinting, and the persistent repression of large chromosomal domains such as the × chromosome in female mammals. Pathologically, silencing can lead to cancer.16
Similar to SNPs, the DNA methylation profile has been touted as a molecular diagnostic—especially for the early detection of cancer. CpG dinucleotides are unevenly distributed across the human genome—vast stretches of DNA are deficient in CpG and these are interspersed by CpG clusters often referred to as CpG islands. CpG islands are defined as being longer than 500 bp with a GC content greater than 55%. Most methylated cytosine residues are found in CpG dinulceotides located outside CpG islands primarily in repetitive sequences,17 while the CpG islands are generally hypomethylated, showing a decreased level of methylated DNA at either an individual CpG or group relative to a reference derived from normal tissue. But methylation of CpG islands in normal tissue increases with age, while the genomic content of 5-methylcytosine generally declines. This same pattern is found in cancer cells but it is more pronounced there.
The efficacy of a methylation assay is ultimately determined by its sensitivity and specificity. The absolute sensitivity refers to the minimal quantity of a pure methylated target DNA that the assay is able to detect, whereas the smallest fraction of methylated DNA that the assay could detect in the presence of an excess of unmethylated DNA is sometimes referred to as specificity. Methylation is not preserved by conventional PCR. Practically, all of the strategies used for the detection of DNA methylation patterns rely on methylation-specific PCR (MSP). According to Irizarry,18 the detection schemes can be categorized as: (1) bisulfite DNA sequencing, (2) bisulfite pyrose-quencing and other methods that interrogate specific single-CpG dinucleotides, and (3) microarray-based methods. Bisulfite DNA sequencing relies on the chemical conversion of cytosine to uracil by sodium bisulfite, followed by PCR, which incorporates T for U (uracil) and then DNA sequencing.18 While offering single-base resolution, the cost is high—$10,000 per megabase of sequence data. Consequently, it is not yet suitable for whole genome analysis on multiple samples. Bisulfite pyrosequencing and related methods such as MethyLight19 and COBRA20 are sensitive, specific, and relatively cheap, but unsuitable for the analysis of a whole genome that incorporates ~28 million CpG dinucleotides. Finally, microarray-based methods can interrogate larger numbers of CpGs than the other techniques at a lower cost, but still suffer from lack of specificity and are limited to the recognition of methylated sites by methyl sensitive restriction enzymes.
NANOPORES FOR SINGLE MOLECULE DETECTION
The detection of mutations, or SNPs, or the methylation profile accomplished with an affordable, bench-top nanopore instrument could conceivably personalize genomic medicine. Using prototypes such as a-hemolysin (α-HL) and its mutants or nanopores in solid-state membranes, the prospects for low cost, high-throughput resequencing are currently being examined.5,6 However, single-base resolution on a translocating strand has not been demonstrated yet. High fidelity reads demand stringent control over both the molecular configuration in the pore and the translocation kinetics. Control of the molecular configuration determines how the ions passing through the pore come into contact with the nucleotides in the constriction, while the translocation kinetics affects the time interval in which the same nucleotides are held in the constriction and data are acquired. None of the nanopore prototypes proffered for sequencing has shown any prospect of satisfying both of these specifications at the same time.
Following the seminal work by Kasianowicz et al.,21 there have been recent reports showing evidence that α-HL can be used to resolve single nucleotides on immobilized DNA and single nucleotides cleaved from DNA, which suggests that sequencing is possible.22,23 For example, Bayley et al.22 recently engineered α-HL in such a way to improve the signal-to-noise ratio (SNR), i.e., to hold a nucleotide in place for a longer period of time, in order to perform more averaging. By modifying the α-HL such that a cyclodextrin is placed in the β-barrel, the time that the pore is occluded by a single nucleotide can be extended. This allows more accurate measurements of which nucleotide is in the pore based on blockade current. This method was used in combination with an exonuclease to determine the composition of single-stranded DNA (ssDNA) using an exonuclease to cleave off individual deoxyribonucleoside monophosphate (dNMPs), and then to measure them as they are captured by the α-HL nanopore.23 However, this scheme is problematic because the cleaved nucleotides have to be transported from the exonuclease to the pore. Consequently, ensuring that nucleotides do not escape detection and arrive in the same sequence as found in the original DNA strand without the exonuclease outpacing current measurements remains a challenge.
On the other hand, engineered proteinaceous pores offer a platform for numerous biomedical applications, specifically proteometrics. Specific recognition groups can be engineered into the pore with a binding affinity that can be easily tuned to detect the different protein variants secreted by pathogens, for example.24 Protein folding dynamics can also be detected using nanopores like α-HL.25 While α-HL translocates proteins that are unfolded, it also have been shown that the dynamics of bulkier proteins in folded conformations can be monitored for study in their native aqueous environment.26
Recent advances in semiconductor nanofabrication have enabled the creation of nanometer-scale pores in solid-state membranes that mimic proteinaceous pores. In contrast to α-HL, the size and shape of the pore in a solid-state membrane can be controlled on a subnanometer scale, allowing for a specific geometry to be tailored to the purpose. For example, the constriction shown in Figure 1(a) is too small to allow a double stranded DNA (dsDNA) molecule to translocate across the membrane at low voltages (<200 mV), while a ssDNA can easily permeate the pore.27 Solid-state membranes are also resilient in chemical and thermal environments useful for denaturing the DNA and allow for easier integration with other electrical components. Moreover, there is a range of choices for membrane materials; insulators, semiconductors, and metal films28 have all been used. And there is a panoply of methods for fabricating pores: ion track etching in ~10 µm thick poly(ethylene-terephthalate) (PET),29 ion or electron beam sculpting in Si3N4 < 30-nm thick membranes28,30,31 to name a few. Through microfabrication techniques, the solid-state membrane can also be reduced to submicrometer scales, mitigating parasitic capacitance effects and improving electrical performance.32,33
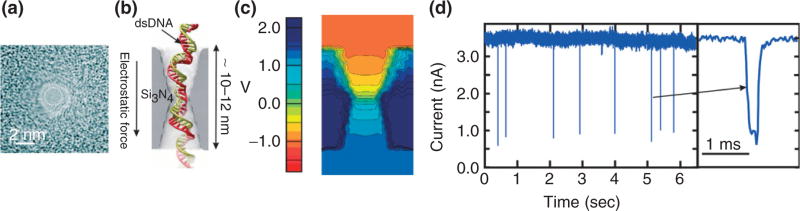
FIGURE 1.
Nanopores in thin membranes used for single molecule detection. (a) A transmission electron image of a pore with a diameter of 2.2 nm in a 15-nm thick Si3N4 membrane. (b) A cross-section through a 2-nm nanopore in a silicon nitride membrane showing a dsDNA in the pore. (c) Distribution of the electrostatic potential in a nanopore at 2.6-V bias across the membrane in 1 M KCl. (d) A measurement of the electrolytic current through the 2.2-nm pore interacting with a λ-DNA with 1.0 V applied across the silicon nitride membrane. The current blockades are observed as DNA translocates through the pore. The figure on the right is an expanded view of a blockade. Figures (b) and (c) courtesy of A. Aksimentiev.
A technique used prevalently to create a nanopore in a membrane is stimulated beam decomposition and sputtering using a tightly focused electron beam of transmission electron microscope (TEM), first reported by Ho et al.34 This method allows high resolution imaging of the geometry during sputtering and is applicable to a wide range of membrane materials such as NiCr, Si3N4, SiO2, Si, and Al2O3.30,34–37 High resolution TEM images taken at different tilt angles allow us to model the pore topography as two intersecting cones each with >15–30° cone angle,34 as illustrated in Figure 1(b). Biconical pores like this focus the electric field in the center of the pore where two cones intersect.38,39 Figure 1(c) is a simulation illustrating the electrostatic voltage distribution in a biconical pore. For this geometry, most of the voltage drop occurs near the constriction in the center of the membrane.
The flexibility inherent to nanofabrication technology holds out the prospect of controlling the both molecular configuration and the translocation kinetics at the same time. The ability to control the membrane thickness, diameter, and the cone angle of a nanopore is especially important in tuning the selectivity of the pore to molecular size and specifying the electrical potential profile inside the pore. In comparison, a-HL can only accommodate the single strand of DNA since it has a diameter less than 1.6 nm and the lipid bilayer cannot sustain high electric fields.
Just like for α-HL, a nanopore in a solid-state membrane can be used as a transducer to tease out information about the structure associated with a single molecule from an electrical current signal.25,40 In normal operation, a thin membrane with a nanopore through it separates two chambers each filled with electrolyte. A driving bias, V0, is then applied and a corresponding ionic current, I0, flows through the nanopore. The driving bias causes a charged molecule like DNA in the vicinity to migrate toward the pore and eventually it is captured by the field as the dsDNA shown in Figure 1(b). When a DNA molecule enters the pore, the ionic current through it changes drastically. The effective cross-sectional area of the pore changes, due to the DNA charge and the excluded volume that ions could previously travel through. Figure 1(d) illustrates blockades in the open pore current I0 ~ 3.5 nA associated with the translocation of a λ-DNA molecule through the 2.2 ± 0.2 nm pore. Thus, if it is small enough, single molecules in an electrolytic solution can be transported through a pore one at a time by applying an electric field along the pore axis.
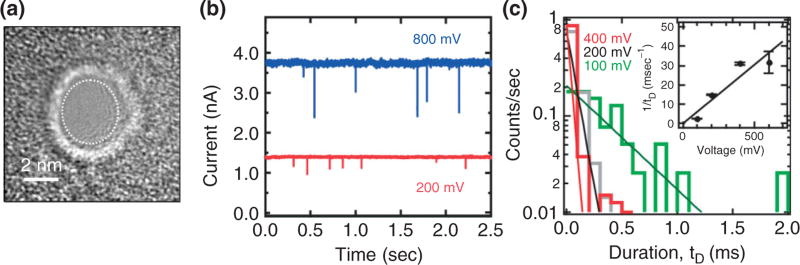
Prior work has indicated that the translocation velocity of DNA through a solid-state pore may be large, exceeding 1 bp/10 ns for the electric fields used in our experiments.41–44 The results for the 3.6 × 3.2 nm nanopore shown in Figure 2(a)–(c) are typical. We find that when λ-DNA is injected into the electrolyte at the negative (cis) electrode and 200 mV is applied across a 31.5 ± 2.0 nm thick nitride membrane with a 3.6 × 3.2 nm nanopore in it, current blockades are observed as illustrated in Figure 2(b). These blockades are supposed to be due to the reduction of the electrolytic current through the pore due to the translocation of DNA. If the blockade duration corresponds with the interval that DNA blocks the pore, then the average transient width tD signifies the time required for 48.502 kb λ-DNA to translocate through the pore, which is tD = 0.0677 ± 0.003 ms for 200 mV, indicating a translocation velocity of 48.5 kb/0.067 ms = 1 bp/1.3 ns consistent with other estimates.41
FIGURE 2.
A nanopore larger than double-stranded DNA in thin membrane. (a) A transmission electron micrograph of a 3.6 × 3.2 nm cross-section nanopore in a 31.5-nm thick Si3N4 membrane. (b) Blockades in the electrolytic current are measured in a 100 mM KCl as a function of membrane bias through the pore shown in (a) as a function of time. (c) Distributions illustrating the frequency as a function of the duration of a current blockade, tD, at 400 mV (red); 200 mV (black) and 100 mV (green). The distribution depends sensitively on the voltage. Inset: The reciprocal of the duration, 1/tD, as a function of the applied voltage. 1/tD seems to depend linearly on the voltage.
This attribute for detecting, identifying, and counting molecules one at a time, ostensibly makes a nanopore the ultimate analytical tool, provided that a signature of the translocation can be identified in the pore current. While there are several base-pair present, the current will be most sensitive to the base-pair in the constriction of the biconically shape pore. Ions passing through the pore are forced into contact with the portion of the molecule in the constriction. At low bias, the electric potential of the nucleotides in the constriction presents an energy barrier to the passage of ions. Because the passage rate is exponentially related to the height of the barrier, differences in the heights for different molecules have a substantial effect on the current–voltage characteristic. Accordingly, control of the distribution of the electric potential in the pore relies on the control over the configuration of the molecule as it translocates. Different transients all associated with the same DNA translocating through the same pore can be generated, depending on the molecular configuration, the pore charge, and electrolyte molarity.36–40,45,46 Presumably, the cone angle, the thickness of the membrane, and the pore diameter will all affect the ultimate resolution. Thus, sequencing demands precise, subnanometer control over the thickness and composition of the membrane and a pore geometry smaller than the double helix.
The bandwidth and noise performance present severe limitations for high-throughput resequencing with a solid-state nanopore. As a first attempt to reconcile the SNR required for single molecule detection with the bandwidth, Smeets et al.32 have analyzed the electrical characteristics of a nanopore, representing it by an equivalent lumped element circuit consisting of a resistance associated with the pore, Rp, in parallel with the total membrane capacitance (including parasitics), Cm, and a series resistance, Rel, associated predominately with the electrolyte. The transient voltage response across the membrane is determined essentially by: fc ≤ 1/2πRelCm with Cm > 10 pF for a 12–15 nm thick membrane, and Rel ~ 10–100 kΩ for 10–100 mM KCl, so that RelCm ~ 1–10 µs. However, considering the typical velocity of long DNA fragments through a nanopore is ~ 1 bp/10 ns, a response time <10 ns, is required to capture a current transient associated with a single base.
The data shown in Figure 1(c) indicate that the relative change in current associated with DNA blockading a pore is ΔI/I < 0.8, which translates to a ΔI ~ 3 nA for a 2.2-nm diameter pore (in 100 mM KCl). Therefore, for SNR > 2, we need peak-to-peak noise <1.5 nA or a root-mean-square (rms) value of ΔIrms ~ 1.5 nA/8 = 190 pA. If dielectric noise associated with the capacitance predominates as suggested by Smeets et al.,32 then where D is the dielectric loss constant (D ~ 0.2 for our membranes). For a bandwidth Δf ~ 16 MHz corresponding to a sampling time of 10 ns, we estimate that DCm ~ 3 fF is required to detect any current signature, which is about 2500x smaller than the typical membrane capacitance and demands microfabrication.
Besides a current blockade, there are alternative strategies for the detection of DNA sequence in a nanopore. For example, it has been proposed to use optical fluorescence to optically readout the sequence,47 but this scheme suffers the same limitations as second generation sequencing technology: it relies on enzymatic incorporation of a fluorescently labeled nucleotide through a polymerase and then applying techniques that suppress the ambient radiation so that one molecule can be identified as a time. And it is slow to detect fluorescence, which demands multiple pores operating in parallel for high-throughput. Another strategy for sequencing DNA, which involves the detection of the transverse tunneling current using electrodes placed adjacent to the pore as molecule translocates, not only promises more sensitivity, but it also demands more stringent control of the molecule position relative to the probes.48,49 Measuring tunneling current requires two electrodes to be placed within few nanometers of each other and stringent control over the orientation of the molecule. A third approach leverages current advances in semiconductor fabrication by using a nanopore sputtered in a capacitor layers that are separated by thin layer of dielectric (~1 nm) that is somewhat comparable to the base-pair separation of a stretched DNA molecule.50,51 The voltage change induce in capacitor by the presence of the dipole moment of individual base-pair has potential of allowing to record base-pair information of the DNA molecule as it translocates through the pore.
USING A NANOPORE TO DETECT MUTATIONS
B-form dsDNA is a stiff, highly charged polymer with a solvated, helical structure of about 2.6–2.9 nm in diameter, according to neutron scattering, that depends on the sequence and the number of strongly bound water molecules included in the primary hydration shell.52 We have shown that only a small electric field (<0.2 V/10nm) is necessary to force dsDNA through a pore >3 nm in diameter,22,34 but the permeability changes dramatically if the DNA is bound to a restriction endonuclease that is larger than the pore diameter.
We have explored the prospects for using a solid-state nanopore to detect base-pair mutations in dsDNA by measuring the binding of a restriction enzyme.53,54 As illustrated by the snapshots taken from a molecular dynamics (MD) simulation shown in Figure 3(a), this method uses the electric field in the pore to pull on the polyanionic DNA complexed with a restriction endonuclease, shearing the protein from the cognate sites in DNA and rupturing the bond. The signature of a rupture, which is the translocation of DNA across the membrane through the pore, appears to depend dramatically on the voltage as illustrated in Figure 3(b). This method is a derivative of RFLP, which also uses restriction enzymes to recognize a specific sequence in a dsDNA fragment and then cleaves the strand at a site in the sequence at or near to it, creating shorter fragments that are subsequently amplified and run on a gel. Since allelic differences affect recognition dramatically, the number and size of the products can be used to determine the genotype in RFLP. With this new method employing a nanopore, it is not necessary to cleave the strand to sort one allele from the other—the difference in threshold voltage could be used to discriminate mutations instead.
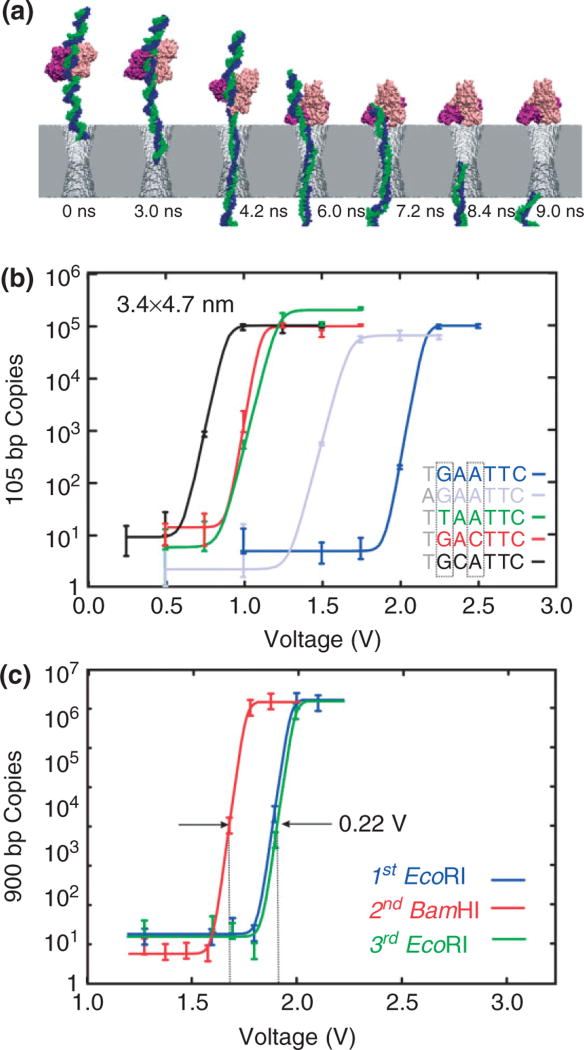
FIGURE 3.
Voltage threshold for permeation through a solid-state nanopore depends on the sequence. (a) Molecular dynamics simulations of EcoRI—DNA complex encountering synthetic nanopore. Snapshots illustrating simulated dissociation of an EcoRI–DNA complex due to the electric field in a nanopore. The Si3N4 membrane is shown in gray, two strands of DNA in blue and green, a protein dimer in pink and purple, water and ions are not shown. Time elapsed from the moment a 4-V transmembrane bias was applied as indicated. (b) Quantitative PCR (qPCR) results indicating the number of 105 base pair (bp) copies from EcoRI–DNA that permeates a 3.4 × 4.7 nm pore with a threshold voltage that depends on the DNA sequence. Superimposed on the data are fits to the curve used to determine the threshold. The cognate sequence GAATTC (blue) has a threshold voltage of about 2.1 V, but in contrast, with a substitution for the first base, TAATTC (green) has a threshold of only 1.1 V. (c) qPCR results indicating the number of 900 bp DNA copies from either EcoRI–DNA or BamHI–DNA that permeates a pore depends on the enzyme. The inset shows a transmission electron micrograph of the 3.4 × 4.5 nm pore. The threshold for the EcoRI and BamHI rupture scales with the bulk dissociation energies.
EcoRI is a type II restriction endonuclease with a palindromic target site on DNA—GAATTC—that it binds to as a dimer. It cleaves foreign DNA in an Escherichia coli host in the presence of a Mg+2 ion cofactor. Similar to other enzymes that specifically recognize DNA, EcoRI finds the cognate site through a three-step process: nonspecific binding to the DNA, linear diffusion along the strand till it encounters the target site, then binding to the target site occurs, accompanied by a large conformation change.55,56 To study the binding of enzymes like EcoRI, we examined the permeability of short strands (1 kb) of dsDNA containing the cognate sequence through pores in nitride membranes as a function of diameter, enzyme, and sequence.53,54
To establish unambiguously if a short strand permeates through the pore, the DNA reaching the anode was analyzed using quantitative PCR (qPCR). We used qPCR to count the number of DNA molecules at the positive electrode that permeate through the pore as a function of the voltage drop across a Si3N4 membrane. We observe a voltage threshold for permeation of dsDNA and the threshold depends on the DNA sequence. Generally, the amount of DNA that permeates the pore rises abruptly over a range of ~250 mV near a threshold that is sensitive to the DNA variant.
The threshold voltage, U, varies widely with base substitutions in the recognition sequence and the flanking nucleotides. Apparently, with EcoRI bound to the cognate sequence (GAATTC), dsDNA (dark blue in Figure 3(b)) does not permeate through the pore unless the voltage exceeds U = 2.15 V. This is in contrast with the corresponding threshold, U = 1.15 V [shown in green in Figure 3(b)], for disrupting the bond between the EcoRI and a DNA variant with a single-base mutation from G to T on the first cognate site from the 5′ end. These thresholds correspond to bulk measurements of the free energy of formation for—GAATTC—and the first-base substitution with- —TAATTC—, which is about 15.2 and 8.6 kcal/mol respectively.57 The threshold even depends on the flanking sequence. Note that for a mutation from T to A on the first element of the flanking sequence near the 5′ end (shown in light blue), the threshold U = 1.62 V, which corresponds to a bulk dissociation energy of 13.2 kcal/mol. The same trends are observed for all of the mutations tested—even using different pores.
Restriction enzymes specific to hundreds of distinct sequences have already been identified, and the corresponding cognate sites can be found in almost any gene.58 Since the threshold depends on the sequence, one may expect to detect and discriminate mutations similarly. However, the efficacy of using different enzymes depends on the ability to discriminate between the threshold voltages associated with different enzymes. As a test, we measured the threshold for permeation through a pore with a strand of dsDNA (900 bp) that incorporates cognate sites for both EcoRI (GAATTC) as well as BamHI (GGATCC). Since the respective cognate sites differ by 2 bp, only nonspecific binding of one enzyme to the other site is expected. Three qPCR analyses are shown in Figure 3(c): the first measures the threshold associated with the EcoRI–DNA, the second measures the threshold for BamHI–DNA, and the third measures the threshold for EcoRI–DNA again to establish reproducibility. We found that the threshold for rupturing BamHI–DNA complex is 0.22 V lower in correspondence with the dissociation energies (13.2 kcal/mol vs.15.2 kcal/mol). This difference is easily resolved since a second measurement of EcoRI–DNA, after measuring the BamHI–DNA threshold, shows the original threshold.
Detecting mutations with this implementation, which uses a nanopore to disrupt the binding in a DNA–protein complex, suffers from the same weakness as all the rest—PCR is used to amplify the DNA. We employ PCR because it is not possible to identify a unique reliable signature in the blockade current that is associated with the rupture of the enzyme–DNA complex. The main problem with electrically detecting short strands of DNA in these experiments stems from the bandwidth limitation or transient response time of the nanopore, and the corresponding deterioration of the signal-to-noise that accompanies increased bandwidth. A linear extrapolation to an electric field comparable to the thresholds for dissociation of a protein–DNA complex yields a velocity of 48.5 kb/8.3 µs = 1 bp/172 ps (at 2 V/30 nm). At this rate, 100-bp dsDNA would translocate through the pore in about 20 ns.
STRETCHING GENES USING A NANOPORE
What is needed for resequencing DNA is control of translocation kinetics that does not compromise the SNR. A smaller pore diameter helps. The smaller the pore diameter, the larger the blockade in the pore current and the larger the signal. When pore diameter is large (~10 nm), dsDNA may translocate through the pore in folded configuration making single base-pair analysis impracticable.43,44 Therefore, pores with smaller diameters (<3 nm) are of special interest.
The electromechanics involved in a translocation is different in pores with smaller diameters (<3 nm) because of the viscosity of water, the screening, and size of the DNA.38,39,59–61 While only a small voltage is required to force dsDNA through a pore >3 nm in diameter, when the pore diameter is smaller than the double helix, the leading edge of the dsDNA penetrates the membrane into a constriction about ~2.5 nm in diameter and stalls there. However, applying a voltage bias above a threshold provides a differential force exceeding that required to stretch dsDNA (~60 pN),62,63 and the molecule is pulled toward the center of and eventually through the membrane. The two strands comprising the double helix do not pass through pores with diameters 1.6 < d < 2.5 nm in the same way as they do through larger pores.45,46 The confinement of the smaller pores causes the base-pair to tilt. Due to the activation energy required to begin this stretching transition, no DNA will be able to translocate below the threshold. On the other hand, the threshold depends on the pH, the composition of the strand, and the methylation profile.39,64
As illustrated in Figure 4(a), we find that a 2.0-nm pore in a nominally 10-nm thick silicon nitride membrane exhibits a threshold U = 2.9 V for permeation of dsDNA, while a 2.0-nm pore in the 20-nm membrane shows a threshold voltage U = 5.5 V because of the change in the field profile associated with the thickness of the membrane. The estimated differential tensile force on the leading nucleotides in the strand is F = qE > 60 pN, where E is the electric field in the pore, which is large enough to stretch dsDNA. It must be that the origin of the sharp field threshold for permeation is due to the stretching transition.
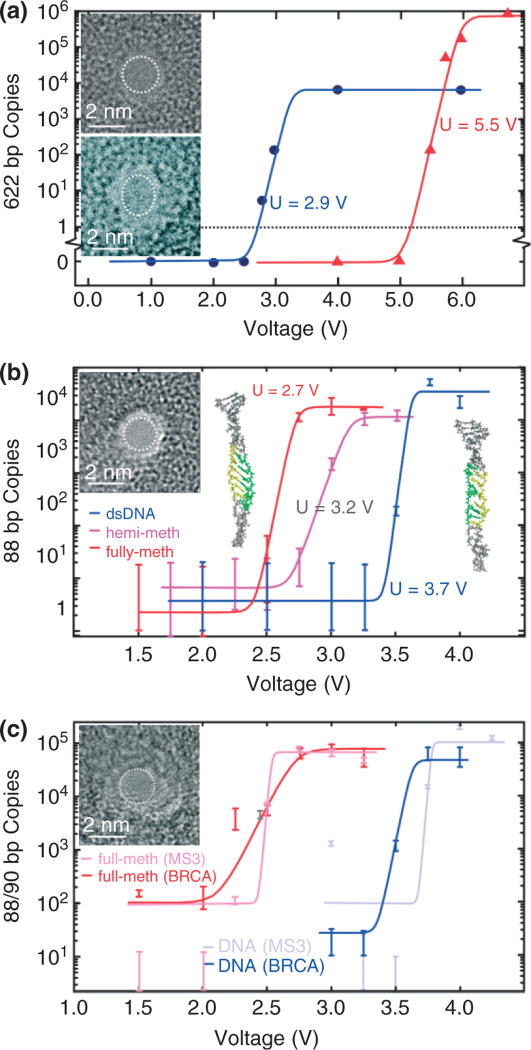
FIGURE 4.
Stretching DNA in a nanopore. (a) Quantitative PCR (qPCR) results obtained for 2-nm pores showing the copy number versus voltage. 622 base pair dsDNA permeates the 2-nm pore in 10-nm thick Si3N4 membrane (top inset) for V > 2.5 V and the 1.8 × 2.2 nm in 20-nm thick Si3N4 membrane (bottom inset) pore for V > 5 V. (b) qPCR results indicating the number of MS3 DNA copies that permeates through the 1.8 ± 0.2 nm pore shown in the inset as a function of the membrane voltage. Insets on the left and right are snapshots of methylated and unmethylated MS3 translocating through the 1.8-nm pore. Both DNA exhibit an ordered B-DNA form, but there is a significant degree of disorder for unmethylated DNA. The highlighted region of the strand shows the portion of the DNA where methylated cytosines are located. The same region is also highlighted in the unmethylated strand for comparison. (c) qPCR results indicating the number of MS3 and BRCA1 DNA copies that permeates through the 1.7 ± 0.2 nm pore shown in the inset as a function of the membrane voltage. Unmethylated MS3 and BRCA1 permeate the U > 3.8 V and U > 3.6 V, respectively while the threshold for fully methylated MS3 and BRCA1 is U > 2.5 V and U > 2.7 V, respectively.
Recently, we established that the voltage threshold for permeation of dsDNA through a nanopore with a diameter smaller than the double helix depends on the methylation level and pattern too, and it can be substantially smaller than for an unmethylated variant of the same DNA.64 We studied two fragments of genomic DNA that are known to control gene expression based on methylation status: MS3 and BRCA1. We investigated the permeability of MS3 and BRCA1 with different methylation levels and profiles through two pores with similar (~1.8 nm) diameters as shown in Figure 4(b) and (c). Figure 4(b) represents the results of three qPCR analyses—one for unmethylated, one for hemi- and another for fully methylated DNA—showing the number of DNA copies permeating the pore as a function of the applied potential. The threshold voltages, U, for fully and hemimethylated MS3 are easily resolved and fall below that observed for the unmethylated variant. For example, the threshold for unmethylated MS3 in Figure 4(b) is U = 3.6 V while hemimethylated and fully methylated MS3 show U = 3.2 and 2.7 V, respectively. A comparison of Figure 4(b) and (c) reveals that similar diameter pores produce similar thresholds too. Both pores show essentially the same threshold for fully methylated and unmethylated MS3 variants, i.e., U = 2.7 V and U = 3.6 V, respectively.
While the threshold is apparently related to the methylation level, it is relatively insensitive to the sequence, as evident from a comparison between the permeation of MS3 and BRCA1 through the pore shown in Figure 4(c). Unmethylated BRCA1 and MS3 sequences are different, but the thresholds for stretching are similar: U = 3.6 and 3.8 V respectively. Likewise, fully methylated BRCA1 that has 12 methylated CpG sites, and fully methylated MS3 that has 10, both show similar shifts in threshold to 2.7 and 2.5 V.
The large shifts in the thresholds with methylation are surprising because the leading nucleotides in the strand, which are important to stretching in a pore, are separated by more than 18 bp (~6 nm) from a methylation site. The structure of methylated DNA, inferred from X-ray diffraction and nuclear magnetic resonance, indicates that the effect of methylation on the conformation of DNA is very subtle and localized near the methylation site.65 On the other hand, MD simulations indicate that the methyl groups reduce the DNA flexibility because of the steric hindrance by bulky methyl groups and because the DNA folds around hydrophobic methyl groups.66 Snapshots of the DNA in the pore shown in the insets of Figure 4(b) reveal the molecular structure with atomic detail, indicating that both methylated and unmethylated DNA exhibit a B-form, but methylated DNA is more ordered and stiffer. The preservation of the B-form in methylated DNA is also evident in the rms deviation in the helix diameter. At 4 V, the interior segments of methylated and unmethylated DNA [shown in yellow in the insets to Figure 4(b)] have an rms deviation 0.29 and 0.49 nm, respectively.
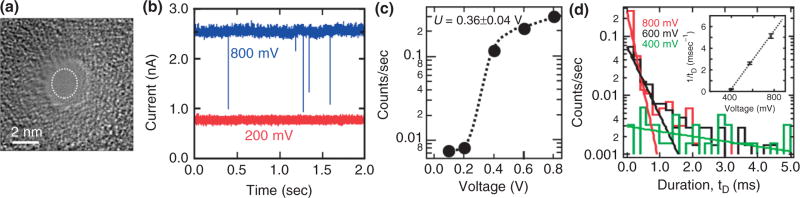
The threshold voltage drops with increasing pore diameter. The permeability of dsDNA through a nanopore with a 2.3 × 2.0 nm cross-section—just smaller than the DNA double helix—in a 15-nm thick membrane is shown in Figure 5(a). When λ-DNA is injected into the electrolyte at the negative electrode and 800 mV applied across the membrane, current transients like those shown in Figure 5(b) are observed. Figure 5(b) also illustrates threshold behavior. Comparing the data of Figure 4(b) obtained using a larger pore with the data taken at 800 mV for this pore, Figure 5(b) shows a dearth of transients in a current trace measured at 200 mV. Figure 5(c) summarizes the frequency of blockade events observed as a function of the voltage applied across the membrane over the range from 100 mV to 1 V, indicating a threshold of U = 0.36 ± 0.04 V.
FIGURE 5.
(a) A transmission electron micrograph of a 2.3 × 2.0 nm nanopore in a 15-nm thick silicon nitride membrane. (b) Electrolytic current measured in 100 mM KCl at 800 mV (blue) and 200 mV through the pore shown in (a) as a function of time. The frequency of blockades decreases dramatically with voltage; at 200 mV (red) no transients are observed. (c) The frequency of blockades observed with the same pore as a function of membrane voltage illustrating the frequency drop as voltage decreases below 0.5 V. The dotted line represents a fit to the data. (d) Distributions illustrating the frequency as a function of the duration of a current blockade, tD, above threshold at 800 mV (red), 600 mV (black), and 400 mV (green). The distribution depends on the voltage. Inset: 1/tD as a function of the applied voltage indicating a threshold.
Figure 5(d) indicates that it is conceivable that dsDNA can be trapped in a nanopore that is smaller in diameter than a double helix. It shows the frequency of current transients associated with λ-DNA as a function of duration with the voltage as a parameter. If the blockade duration corresponds with the interval that DNA blocks the pore, then the average transient width tD signifies the time required for 48.502 kb λ-DNA to translocate through the pore. The inset of Figure 5(d) shows a plot of the voltage dependence of the reciprocal of the average transient width, i.e., 1/tD, measured above threshold. The line in the inset represents a least-squares fit to the data, which shows a voltage-intercept that is comparable to the threshold voltage inferred from Figure 5(c). It seems that 1/tD falls abruptly near the threshold value, which is consistent with the idea that the molecule may become trapped in the pore near the threshold voltage.
CONTROLLING THE TRANSLOCATION OF DNA AND THE PROSPECTS FOR SEQUENCING IT USING A NANOPORE
Controlling the translocation velocity is one of the key elements in resolving the signal associated with each nucleotide or base-pair. Several schemes have been advanced to slow the DNA in the nanopore.67,68 Trepagnier et al. sought to slow DNA by increasing the opposing force in the pore. When DNA is captured by the nanopore, the electric field on the leading end immediately accelerates it, quickly reaching a terminal velocity due to fluid-based drag forces on the rest of the molecule. By decreasing this terminal velocity either by increasing the resisting force or by decreasing the driving force, electrical measurements can be accomplished within the bandwidth restrictions of the instrument. In addition, there are several other approaches for slowing the DNA molecule in the pore such as increasing the viscosity of the solution to increase the drag force on the DNA molecule41 and selective transport where pore DNA interaction slows the molecule down,69 but these schemes do not offer the control over the translocation dynamics of DNA molecule needed to read out an individual base-pair.
Recent examples have been reported where a λ-DNA has been trapped between the electric field in the pore and a bead held by optical tweezers or a magnet.59,60,70 Using optical tweezers to trap a microsphere with DNA attached, the researchers pulled on the molecule to slow the translocation speed ~1 bp/6 ms through the ~5 nm pore and were even been able to ‘floss’ a short DNA strand back and forth in the pore allowing for repeated measurements. However, this technique was not leveraged to improve the signal-to-noise of the blockade current measurement of the DNA translocating through the pore. A similar setup using magnetic tweezers allows for stronger forces, but less precision is under development by Ling at Brown.70
The stretching transition can also be used to trap DNA and to control the translocation kinetics.68 Figure 5(d) indicates that it is conceivable that dsDNA can be trapped in a nanopore if the membrane voltage is switched to a value below stretching threshold while the molecule is translocating through the pore. We can easily achieve that by forcing dsDNA into a 2.5-nm pore and then, once a blockade in the current is detected, reducing the transmembrane voltage at high speed while the molecule is still in the pore. Once the dsDNA is in the pore, if the bias is reduced below the stretching threshold, the pore acts as a harmonic trap resisting the motion of the molecule.
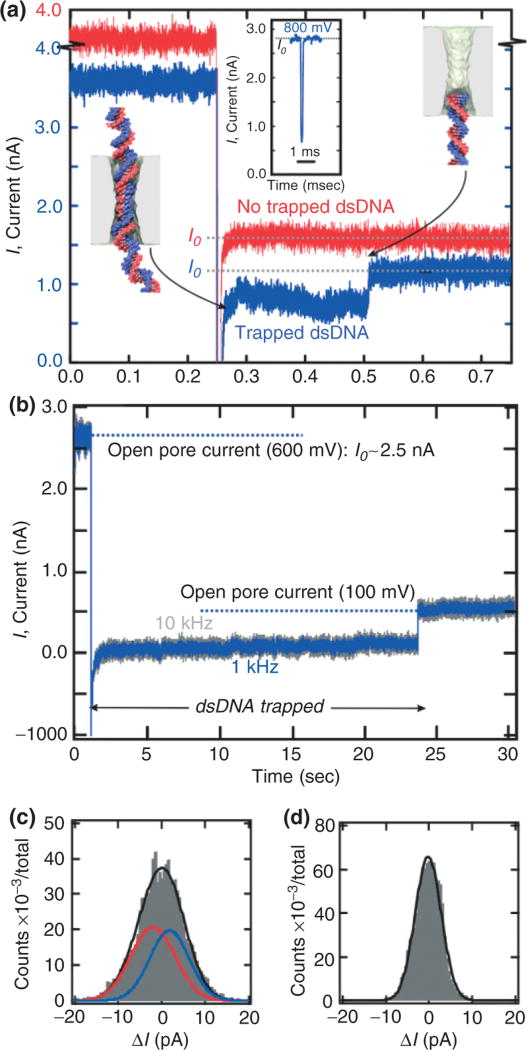
Figure 6(a) and (b) shows data demonstrating that it is possible to weakly trap a single λ-DNA molecule once it is translocating through the pore by switching the electric field at high speed. During normal operation, a transmembrane bias of 800 mV, which is above threshold is applied across the membrane. Once the onset of a blockade is detected a latch switches the voltage from 800 to 200 mV—a value well below the threshold. All the while, the pore current is monitored. Eventually, the current returns to the open pore value consistent with a 200-mV transmembrane bias, but not before we observe a sharp transient like that shown in the blue trace near t = 0.505 s in Figure 6(a). [The red trace, which is offset for clarity in Figure 6(a), shows the pore current without a DNA molecule, for comparison.] While the peak in the distribution obtained at a constant bias of 800 mV occurs near tD = 200 µs, the distribution found when the voltage is switched from 800 to 200 mV occurs near tD ~ 500–600 ms. We reasoned that the long duration current blockades found in a 2.5-nm pore must be evidence of a weakly trapped λ-DNA molecule. If we assume that the molecule is weakly trapped for the duration of the measurement, then the translocation velocity must have slowed substantially to a value of about 1 bp/17 ms, which is more than 1500× times slower than the velocity estimate for tD obtained at 800 mV.
FIGURE 6.
Trapping a single γ-DNA molecule in a nanopore.(a) Triggered by the onset of a current blockade indicating that dsDNA is translocating through the 2.5-nm pore, the voltage across the pore is switched from 800 mV (above the threshold for the stretching transition) to 200 mV (below threshold). As a result, the mean width of the current transient (blue) increases from about 200 µs to about 0.5 s. The red trace, which is offset by 0.5 nA shows the current after the switch is manually triggered—in the absence of a blockade—without DNA translocating through the nanopore. Inset: an example of a current blockade observed in the same pore as a function of time at a constant V = 800 mV bias. (b) Triggered by the onset of a current blockade indicating that dsDNA is translocating through a 2.6-nm diameter pore, the voltage is switched from 600 mV (above the stretching threshold) to 100 mV (below threshold). As a result, the molecule is trapped in the pore till t = 23.4 s. (c) Histogram showing the distribution of the current during the blockade in an interval t = 0.5 s when λ-DNA is trapped for 53.2 s and (d) the open pore after the molecule exits the trap. The distribution for the trapped molecule must be fit to at least two Gaussians: one (solid blue) offset from the median (ΔI = 0) by +1.8 pA with a width of 14.4 pA and another (solid red line) offset by −2.1 pA with a width of 12.9 pA. The black line represents the sum. In contrast, the data in (d) representing the open pore can be fit by a single Gaussian with a width 8.6 pA.
Figure 6(b) represents another, extreme example of a trapped molecule in a 2.5-nm pore. During normal operation, a transmembrane bias of 600 mV, which is above threshold, is applied across the membrane resulting in an open pore current >2 nA. Once the onset of a blockade is detected, a latch switches the voltage from 600 to 100 mV—a value well below the threshold. Subsequently, dsDNA translocates through the pore at a greatly reduced speed. In Figure 6(b), the molecule exits the pore near t = 23.4 s as evident by the return of the current to the open pore value, I0. The corresponding translocation speed for this 22.2-s long event is 1 bp/0.8 ms, which is about 50,000× slower than a typical translocation occurring for a voltage above threshold. With the molecule trapped under the conditions, we filtered the current data as shown in Figure 6(b) using a 10–600 Hz bandpass filter and formed histograms of the current fluctuations using 0.5-s windows. When the molecule is trapped for about 60 s, each window shows a histogram similar to that shown in Figure 6(c), which can be represented by the superposition of two Gaussian distributions: one (solid blue) offset from the median (ΔI = 0) by +1.8 pA with a width of 14.4 pA and another (solid red line) offset by −2.1 pA with a width of 12.9 pA. The black line represents the sum. In contrast, the data in (d) representing the open pore can be fit by a single Gaussian with a width 8.6 pA.
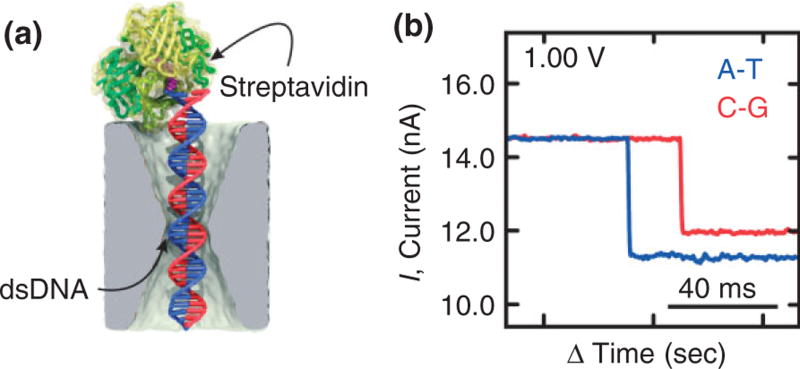
We attribute these separate peaks to resolved C–G/G–C and A–T/T–A bp, respectively. This assertion is corroborated by even longer duration measurements of blockade currents associated with streptavidin bound, 100-bp long, C–G and A–T biotinylated duplexes trapped by the electric field in a pore in a configuration represented schematically in Figure 7(a). Streptavidin has an extraordinary affinity for biotin, which we leveraged to measure the blockade current for a trapped biotin–DNA duplex in a 2.6 × 2.1 nm pore immersed in 1 M KCl. At low voltage, the duration of a blockade is interminable—the blockade ends only if the voltage is manually reversed and the dsDNA is impelled out of the pore, streptavidin, and all. However, the dwell time is an exponentially decreasing function of the applied voltage. In our analysis of the blockades due to the two variants, C–G and A–T, we focused on dwell times at the peak of the distribution or longer and on traces with essentially the same open pore current measured after intervening flushes and cleans. Figure 7(b) shows a typical current blockade observed at 1 V for dwell times >10 s, respectively. Clearly, it is easy to discriminate C–G base-pair stretched in the pore from the smaller blockade current associated with either A–T—the difference is 533 ± 98 pA at 1 V. It seems that C–G can easily be discriminated from A–T under these conditions over a range of voltage. Apparently, the increase in molarity (10 ×) and larger voltage (10 ×) exaggerates the effect of stretching on the blockade current.
FIGURE 7.
Streptavidin bound biotin DNA duplex trapped by the electric field in a nanopore. (a) Model of biotinylated dsDNA bound to streptavidin in a 2.6 nm × 2.1 nm cross-section pore in a 23-nm thick membrane. (b) A current blockade measured at 1.0 V transmembrane bias in 1 M KCl in a 2.6 × 2.1 nm pore associated with C–G and A–T duplexes biotinylated to streptavidin. The difference in blockade current can be used to discriminate C–G from A–T.
MD and Brownian dynamics (BD) simulations of these experiments reveal that, when trapped, dsDNA is stretched in the pore constriction in a way which preserves the natural inclination of the base-pair, the angle they make relative to the helical axis.68 The simulations also show that it should be possible to determine the sequence of dsDNA from the configuration of the stretched dsDNA in the pore by simply measuring the pore current in the trapped configuration without ambiguities between A–T and T–A or G–C and C–G. A smaller pore diameter and higher electrolyte concentration improve the resolving power. As pore diameter is reduced to d = 2.0 nm, the conformation of the trapped DNA molecule will resemble that shown in Figure 1(b) with the stretching exaggerated by the narrower constriction. Thus, in the future we envision using a similar trap as part of a sequencing protocol, whereby the translocation kinetics of dsDNA in a pore is stringently controlled and measurements can then be performed, taking the time necessary to extract information from the pore current about the identities of the nucleotides in the constriction.
CONCLUSION
Sequencing a single molecule of DNA using a nanopore represents the ultimate sequencing technology, which has the potential to extract the maximum amount of information from a minimum of material for applications in genomics, epigenetics, and proteomics. The nanopore sequencing concept is revolutionary because it combines the potential for long read lengths (>5 kb) with high speed (1 bp/10 ns), while obviating the need for costly and error-prone procedures like PCR amplification due to the exquisite single molecule sensitivity. However, high fidelity reads demand stringent control over both the molecular configuration in the pore and the translocation kinetics. The molecular configuration determines how the ions passing through the pore come into contact with the nucleotides, while the translocation kinetics affects the time interval in which the same nucleotides are held in the constriction as the data is acquired.
The bandwidth and noise performance present severe limitations for high-throughput resequencing with a solid-state nanopore, however. The membrane capacitance affects both aspects of the performance and consequently miniaturization of the membrane (likely to submicron dimensions) is a prerequisite for sequence. Finally, if nanopore sequencing proves to be feasible, the long read length and high translocation velocity could make sequencing inexpensive and push the fruits of genomics within the grasp of everyday medicine.
Acknowledgments
We would like to thank our collaborators in these efforts: A. Aksimentev, K. Schulten, J. Comer, A. Feinberg, K. Schulten, S. Sligar, Q. Zhao, and X. Zou. We gratefully acknowledge the use of the Center for Microanalysis of Materials supported by the US DOE grant DEFG02-91-ER45439. This work was funded by grants from NIH (R01 HG003713A) and the National Science Foundation (TH 2008-01040 ANTC).
References
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Michael Zody MC, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Venter JC, Adams MD, Myers E, Li PW, Mural RJ, et al. The sequence of human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.Shendure J, Mitra R, Varma C, Church GM. Advanced sequencing technologies: methods and goals. Nat Rev Genet. 2004;5:335–344. doi: 10.1038/nrg1325. [DOI] [PubMed] [Google Scholar]
- 4.Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Branton D, Deamer D, Marziali A, Bayley H, Benner SA, et al. The potential and challenges of nanopore sequencing. Nat Biotechnol. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dekker C. Solid-state nanopores. Nat Nanotechnol. 2007;2:209–215. doi: 10.1038/nnano.2007.27. [DOI] [PubMed] [Google Scholar]
- 7.Coulter W. Means for Counting Particles Suspended in a Fluid. USA, USPTO: 1953. [Google Scholar]
- 8.Wood LD, Parsons DW, Jones S, Lin J, Sjöblom T, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 9.Botstein D, White RL, Skolnick M, Davis RW. Construction of a genetic linkage map in man using restriction length polymporphisms. Am J Hum Genet. 1980;32:314–331. [PMC free article] [PubMed] [Google Scholar]
- 10.Packer BR, Yeager M, Burdett L, Welch R, Beerman M, et al. PharmGKB: the Pharmacogentics Knowledge Base. Nucleic Acids Res. 2002;30:158–162. [Google Scholar]
- 11.Kim S, Misra A. SNP genotyping: technologies and biomedical applications. Annu Rev Biomed Eng. 2007;9:289–320. doi: 10.1146/annurev.bioeng.9.060906.152037. [DOI] [PubMed] [Google Scholar]
- 12.Mitra RD, Butty VL, Shendure J, Williams BR, Houseman DE, et al. Digital genotyping and haplotyping with polymerase colonies. Proc Natl Acad Sci USA. 2003;100:5926–5931. doi: 10.1073/pnas.0936399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thorisson G, Smith A, Krishnan L, Stein L. The international HapMap project web site. Genome Res. 2005;15:1592–1593. doi: 10.1101/gr.4413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Istrail S, Waterman MS, Clark AG, editors. Dimacs/Recomb Satellite Workshop. New York: Springer-Verlag; 2004. Computational methods for SNPs and haplotype inference. [Google Scholar]
- 15.Brena RM, Huang THM, Plass C. Toward a human epigenome. Nat Genet. 2006;38:1359–1360. doi: 10.1038/ng1206-1359. [DOI] [PubMed] [Google Scholar]
- 16.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 17.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci USA. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irizarry RA, Ladd-Acosta C, Chavalho B, Wu H, Brand-burg SA, et al. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Res. 2008;18:780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, et al. MethylLight: a high throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:e32–3. doi: 10.1093/nar/28.8.e32. doi: 10.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasianowicz J, Brandin E, Branton D, Deamer DW. Proc Natl Acad Sci USA. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stoddart D, Heron AJ, Mikhailova E, Magila G, Bayley H. Single nucleotide discrimination in immobilized DNA oligonucleotides with a biological nanopore. Proc Natl Acad Sci USA. 2009;106:7702–7707. doi: 10.1073/pnas.0901054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clarke J, Wu HC, Jayasinghe L, Patel A, Reid S, et al. Continous base identification for single-molecule nanopore DNA sequencing. Nat Nanotechnol. 2009;4:265–270. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- 24.Movileanu L. Squeezing a single polypeptide through a nanopore. Soft Matter. 2008;4:925–931. doi: 10.1039/b719850g. [DOI] [PubMed] [Google Scholar]
- 25.Bayley H, Cremer PS. Stochastic sensors inspired by biology. Nature. 2001;413:226–230. doi: 10.1038/35093038. [DOI] [PubMed] [Google Scholar]
- 26.Movileanu L. Interrogating single proteins through nanopores: challenges and opportunities. Trends Biotechnol. 2009;27:333–341. doi: 10.1016/j.tibtech.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 27.Heng JB, Ho C, Kim T, Timp R, Aksimentiev A, et al. Sizing DNA using a nanometer-diameter pore. Biophys J. 2004;87:2905–2911. doi: 10.1529/biophysj.104.041814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischbein MD, Drndić M. Sub-10 nm device fabrication in a transmission electron microscope. Nano Lett. 2007;7:1329–1337. doi: 10.1021/nl0703626. [DOI] [PubMed] [Google Scholar]
- 29.Martin CR, Nishizawa M, Jiarge K, Kang M, Lee SB. Adv Mater. 2001;13:1351–1362. [Google Scholar]
- 30.Li J, Stein D, McMullan C, Branton D, Aziz MJ, et al. Ion-beam sculpting at nanometer scales. Nature. 2001;412:166–169. doi: 10.1038/35084037. [DOI] [PubMed] [Google Scholar]
- 31.Storm AJ, Chen JH, Ling XS, Zandbergen HW, Dekker C. Fabrication of solid-state nanopores with single nanometer precision. Nat Matter. 2003;2:537–540. doi: 10.1038/nmat941. [DOI] [PubMed] [Google Scholar]
- 32.Smeets RMM, Keyser UF, Dekker NH, Dekker C. Noise in solid-state nanopores. Proc Natl Acad Sci USA. 2008;105:417–421. doi: 10.1073/pnas.0705349105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dimitrov V, Mirsaidov U, Wang D, Sorsch T, Mansfield W, Miner J, Klemens F, Cirelli R, Yemenicioglu S, Timp G. yNanopores in solid-state membranes engineered for single molecule detection. Nanotechnology. 2010;21(6):065502. doi: 10.1088/0957-4484/21/6/065502. [DOI] [PubMed] [Google Scholar]
- 34.Ho C, Qiao R, Heng JB, Chatterjee A, Timp R, et al. Electrolytic transport through a synthetic nanometer- diameter pore. Proc Natl Acad Sci USA. 2005;102:10445–10450. doi: 10.1073/pnas.0500796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim MJ, Wanunu M, Bell DC, Meller A. Rapid fabrication of uniformly sized nanopores and nanopore arrays for parallel DNA analysis. Adv Mater. 2006;18:3149–3153. [Google Scholar]
- 36.Smeets RMM, Keyser UF, Krapf D, Wu M-Y, Dekker NH, et al. Salt dependence of ion transport and DNA translocation through solid-state nanopores. Nano Lett. 2005;6:89–95. doi: 10.1021/nl052107w. [DOI] [PubMed] [Google Scholar]
- 37.Vinkatesan M, Dorvel B, Yemenicioglu S, Watkins N, Petrov I, et al. Highly sensitive, mechanically stable nanopore sensors for DNA analysis. Adv Mater. 2009;21:1–6. doi: 10.1002/adma.200803786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heng JB, Aksimentiev A, Ho C, Marks P, Grinkova Y, et al. Stretching DNA using the electric field in a synthetic nanopore. Nano Lett. 2005;5:1883–1888. doi: 10.1021/nl0510816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heng JB, Aksimentiev A, Ho C, Marks P, Grinkova YV, et al. The electromechanics of DNA in a synthetic nanopore. Biophys J. 2006;90:1098–1106. doi: 10.1529/biophysj.105.070672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang H, Venkatesan BM, Iqbal SM, Andreadakis G, Kosari G. DNA counterion current and saturation examined by a MEMS-based solid state nanopore sensor. Biomed Microdevices. 2006;8:263–269. doi: 10.1007/s10544-006-9144-x. [DOI] [PubMed] [Google Scholar]
- 41.Fologea D, Uplinger J, Thomas B, McNabb DS, Li JL. Slowing DNA translocation in a solid-state nanopore. Nano Lett. 2005;5:1734–1737. doi: 10.1021/nl051063o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aksimentiev A, Heng JB, Timp G, Schulten K. Microscopic kinetics of DNA translocation through synthetic nanopores. Biophys J. 2004;87:2086–2097. doi: 10.1529/biophysj.104.042960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen P, Gu J, Brandin E, Kim YR, Wang Q, et al. Probing single DNA molecule transport using fabricated nanopores. Nano Lett. 2004;4:2293–2298. doi: 10.1021/nl048654j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Storm AJ, Chen JH, Zandbergen HW, Dekker C. Translocation of double stranded DNA through a silicon oxide nanopore. Phys Rev E. 2005;71:051903. doi: 10.1103/PhysRevE.71.051903. [DOI] [PubMed] [Google Scholar]
- 45.Zhao Q, Comer J, Dimitrov V, Yemenicioglu S, Aksimentiev A, et al. Stretching and unzipping nucleic acid hairpins using a synthetic nanopore. Nucleic Acids Res. 2008;36:1532–1541. doi: 10.1093/nar/gkm1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Comer J, Dimitrov V, Zhao Q, Timp G, Aksimentiev A. Microscopic mechanics of hairpin DNA translocation through synthetic nanopores. Biophys J. 2009;96:593–608. doi: 10.1016/j.bpj.2008.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soni GV, Meller A. Progress toward ultrafast DNA sequencing using solid-statenanopores. Clin Chem. 2007;53:1996–2001. doi: 10.1373/clinchem.2007.091231. [DOI] [PubMed] [Google Scholar]
- 48.Gierhart BC, David G, Howitt DG, Chen SJ, Zhu Z, et al. Nanopore with transverse nanoelectrodes for electrical characterization and sequencing of DNA. Sens Actuators B Chem. 2008;132:593–600. doi: 10.1016/j.snb.2007.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zwolak M, di Ventra M. Electronic signature of DNA nucletides via transverse transport. Nano Lett. 2005;5:421–424. doi: 10.1021/nl048289w. [DOI] [PubMed] [Google Scholar]
- 50.Sigalov G, Comer J, Timp G, Aksimentiev A. Detection of DNA sequences using an alternating electric field in a nanopore. Nano Lett. 2008;8:56–63. doi: 10.1021/nl071890k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heng J, Aksimentiev A, Ho C, Dimitrov V, Sorsch TW, et al. Beyond the gene chip. Bell Labs Tech J. 2005;10:5–22. doi: 10.1002/bltj.20102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lederer H, May RP, Kjems JK, Baer G, Heumann H. Solution structure of a short DNA fragment studied by neutron scattering. Eur J Biochem. 1986;161:191–196. doi: 10.1111/j.1432-1033.1986.tb10141.x. [DOI] [PubMed] [Google Scholar]
- 53.Zhao Q, Sigalov G, Dimitrov V, Dorvel B, Mirsaidov U, et al. Detecting SNPs using a synthetic nanopore. Nano Lett. 2007;7:1680–1685. doi: 10.1021/nl070668c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dorvel B, Sigalov G, Zhao Q, Comer J, Dimitrov V, et al. Analyzing the forces binding a restriction endonuclease to DNA using a synthetic nanopore. Nucleic Acids Res. 2009;37:4170–4179. doi: 10.1093/nar/gkp317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sidorova NY, Rau DC. Linkage of EcoRI dissociation from its specific DNA recognition site to water activity, salt concentration, and pH: separating their roles in specific and nonspecific binding. J Mol Biol. 2001;310:801–816. doi: 10.1006/jmbi.2001.4781. [DOI] [PubMed] [Google Scholar]
- 56.Jeltsch A, Alaves J, Wolfes H, Maas G, Pingoud A. Pausing of the restriction endonuclease EcoRI during linear diffusion on DNA. Biochemistry. 1994;33:10215–10219. doi: 10.1021/bi00200a001. [DOI] [PubMed] [Google Scholar]
- 57.Jen-Jacobsen L. Protein-DNA recognition complexes: conservation of structure and binding energy in the transition state. Biopolymers. 1997;44:153–180. doi: 10.1002/(SICI)1097-0282(1997)44:2<153::AID-BIP4>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 58.Lesser DR, Kurpiewski MR, Jen-Jacobson L. The Energetic Basis of Specificity in the Eco-RI endonuclease-DNA Interaction. Science. 1990;250:776–786. doi: 10.1126/science.2237428. [DOI] [PubMed] [Google Scholar]
- 59.Keyser UF, Koeleman BN, Dorp S, Krapf D, Smeets RM, et al. Direct force measurements on DNA in a solid-state nanopore. Nat Phys. 2006;2:473–477. [Google Scholar]
- 60.Dorp S, Keyser UF, Dekker NH, Dekker C, Lemay SG. Origin of the electrophoretic force on DNA in solid-state nanopores. Nat Phys. 2009;5:347–351. [Google Scholar]
- 61.Luan B, Aksimentiev A. Strain softening in stretched DNA. Phys Rev Lett. 2008;101:11–15. doi: 10.1103/PhysRevLett.101.118101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith SB, Cui Y, Bustamante C. Overstretching B-DNA: the elastic response of individual doublestranded and single-stranded DNA molecules. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 63.Clausen-Schaumann H, Rief M, Tolksdorf C, Gaub HE. Mechanical stability of single DNA molecules. Biophys J. 2000;78:1997–2007. doi: 10.1016/S0006-3495(00)76747-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mirsiadov U, Timp W, Zou X, Dimitrov V, Schulten K, et al. Nanoelectromechanics of methylated DNA in a synthetic nanopore. Biophys J. 2009;96:L32–L34. doi: 10.1016/j.bpj.2008.12.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heinemann U, Hahn M. CCAGGC-m5C-TGG. Helical fine structure, hydration, and comparison with CCAGGCCTGG. J Biol Chem. 1992;267:7332–7341. [PubMed] [Google Scholar]
- 66.Derreumaux S, Chaoui M, Tevanian G, Fermandjian S. Impact of CpG methylation on structure, dynamics and solvation of cAMP DNA responsive element. Nucleic Acids Res. 2001;29:2314–2326. doi: 10.1093/nar/29.11.2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trepagnier EH, Radenovic A, Sivak D, Geissler P, Liphardt J. Controlling DNA capture and propagation through artificial nanopores. Nano Lett. 2007;7:2824–2830. doi: 10.1021/nl0714334. [DOI] [PubMed] [Google Scholar]
- 68.Mirsaidov U, Dimitrov V, Comer J, Wang D, Timp W, et al. Trapping a double-stranded DNA molecule in a nanopore and the prospects for sequencing it. Submitted to. Proc Natl Acad Sci USA. 2009 [Google Scholar]
- 69.Iqbal SM, Akin D, Bashir R. Solid-state nanopore channels with DNA selectivity. Nat Nanotechnol. 2007;2:243–248. doi: 10.1038/nnano.2007.78. [DOI] [PubMed] [Google Scholar]
- 70.Peng H, Ling XS. Reverse DNA translocation through a solid-state nanopore by magnetic tweezers. Nanotechnology. 2009;20:185101. doi: 10.1088/0957-4484/20/18/185101. [DOI] [PMC free article] [PubMed] [Google Scholar]