Abstract
Background:
Cognitive functions are highly heritable and polygenic, though the source of this genetic influence is unclear. On the neurobiological level, these functions rely on effective neuroplasticity, in which the activity-regulated cytoskeleton associated protein (ARC) plays an essential role.
Objectives:
To examine whether the ARC gene complex may contribute to the genetic components of intellectual function given the crucial role of ARC in brain plasticity and memory formation.
Methods:
The ARC complex was tested for association with intelligence (IQ) in children from the Avon Longitudinal Study of Parents and Children (ALSPAC, N = 5,165). As Alzheimer’s disease (AD) shares genetics with cognitive functioning, the association was followed up in an AD sample (17,008 cases, 37,154 controls).
Results:
The ARC complex revealed association with verbal and total IQ (empirical p = 0.027 and 0.041, respectively) in the ALSPAC. The strongest single variant signal (rs2830077; empirical p = 0.018), within the APP gene, was confirmed in the AD sample (p = 2.76E-03). Functional analyses of this variant showed its preferential binding to the transcription factor CP2.
Discussion:
This study implicates APP in childhood IQ. While follow-up studies are needed, this observation could help elucidate the etiology of disorders associated with cognitive dysfunction, such as AD.
Keywords: Alzheimer’s disease, APP, ARC, Avon Longitudinal Study of Parents and Children, dementia, intelligence, synaptic plasticity
INTRODUCTION
Cognitive function varies between individuals, and is a key predictor of important life outcomes such as mental and physical health as well as longevity [1]. General intellectual function, commonly measured by an aggregated score (intelligence quotient, IQ) across a wide range of cognitive tasks, can be considered a main trait behind this variation [2]. It is well established that intelligence is highly heritable and polygenic [3]. Nonetheless, the extent and nature of this genetic influence is still unknown [4] and warrants further investigation.
Intelligence captures a broad scope of cognitive abilities and can be enumerated by measures of verbal IQ (VIQ) and performance IQ (PIQ), often differentiated into crystallized and fluid types. Crystallized and fluid IQ represent different aspects of intelligence, both behaviorally and biologically [5]. Crystallized intelligence includes knowledge accumulated throughout life, determined by education and experience, while fluid intelligence consists of problem solving and reasoning abilities that has little reliance on stored knowledge [6]. Fluid intelligence is strongly correlated with working memory and functional activity observed by magnetic resonance imaging (MRI) during cognitive tasks [7, 8]. Although these two IQ measures can be combined into a full scale IQ (FSIQ), research on the neural basis of intelligence suggests that it is best represented by at least two dimensions rather than one [5].
The heritability of intelligence has been estimated to increase with age, from ∼30% in childhood up to 80% in adulthood [9]. However, despite such high heritability, intelligence-related phenotypes have not been robustly associated with any single gene or variant [3, 10]. Only recently did a large-scale genome-wide association study (GWAS) on these traits identify 13 variants associated with general cognitive function [11]. This confirms that genetic architecture of intelligence involves many genes of small effect, suggesting that the impact of a single nucleotide polymorphism (SNP) may not be detected without large sample sizes.
The lack of robust association is common in the search for genes behind polygenic traits, such as intelligence [3, 11, 12]. Little progress has been made in finding loci that explain even a small fraction of the phenotypic variance in complex traits [13], posing one of the biggest challenges in present-day behavioral genetics. One promising approach to address this limitation is the examination of functionally related genes as a complex, thereby combining their effects [14].
At the neurobiological level, intellectual and cognitive functions rely on effective neuroplasticity, the brain’s ability to alter neural networks and synapses. Activity-regulated cytoskeleton associated protein (Arc) and its regulators and interactors have been extensively studied due to their essential role in multiple forms of synaptic plasticity [15–17]. Arc appears to play hub-like role in its regulation [18], at least partially by modifying actin cytoskeleton dynamics, receptor endocytosis, and glutamate receptor transcription [15–17]. Thus, ARC (here we refer to the gene as “ARC” and its RNA/protein as “Arc”) and its partners provide a promising set of candidate genes for uncovering genetic components of intellectual function. Here, we collectively refer to these functionally linked genes as the “ARC complex” (Table 1).
Table 1.
Summary of genes included in the curated ARC gene complex. The ARC gene complex was divided into two subgroups: (1) the “ARC expression” subgroup included proteins implicated in the regulation of ARC transcription, mRNA processing, transport, Arc protein translation and degradation and (2) the “ARC function” subgroup included proteins that bind Arc protein or are closely associated with Arc function
| Entrez | Gene | Level of | Gene-Set | References (DOI) |
| Gene ID | Symbol | interaction | subgroup | |
| 23237 | ARC | NA | NA | NA |
| 1385 | CREB1 | DNA | ARC expression | 10.1073/pnas.0806518106 |
| 4205 | MEF2A | DNA | ARC expression | 10.1073/pnas.0806518106 |
| 6722 | SRF | DNA | ARC expression | 10.1073/pnas.0806518106 |
| 3172 | HNF4A | DNA | ARC expression | 10.1073/pnas.0806518106 |
| 3182 | HNRNPAB | mRNA | ARC expression | 10.1091/mbc.E10-11-0904 |
| 3181 | HNRNPA2B1 | mRNA | ARC expression | 10.1091/mbc.E10-11-0904 |
| 5976 | UPF1 | mRNA | ARC expression | 10.1016/j.cell.2007.05.028 |
| 9775 | EIF4A3 | mRNA | ARC expression | 10.1016/j.cell.2007.05.028 |
| 407040 | MIR34A | mRNA | ARC expression | 10.1371/journal.pone.0041688 |
| 406968 | MIR193A | mRNA | ARC expression | 10.1371/journal.pone.0041688 |
| 442900 | MIR326 | mRNA | ARC expression | 10.1371/journal.pone.0041688 |
| 1938 | EEF2 | mRNA | ARC expression | 10.1371/journal.pone.0041688 |
| 10.1016/S0092-8674(03)00079-5 | ||||
| 2332 | FMR1 | mRNA | ARC expression | 10.1016/j.cell.2008.07.031 |
| 8569 | MKNK1 | mRNA | ARC expression | 10.1074/jbc.M109.056077 |
| 2872 | MKNK2 | mRNA | ARC expression | 10.1074/jbc.M109.056077 |
| 23191 | CYFIP1 | mRNA | ARC expression | 10.1016/j.cell.2008.07.031 |
| 6455 | SH3GL1 | protein | Arc function | 10.1016/j.neuron.2006.08.033 |
| 6457 | SH3GL3 | protein | Arc function | 10.1016/j.neuron.2006.08.033 |
| 6456 | SH3GL2 | protein | Arc function | 10.1016/j.neuron.2006.08.033 |
| 1785 | DNM2 | protein | Arc function | 10.1016/j.neuron.2006.08.033 |
| 816 | CAMK2B | protein | Arc function | 10.1016/j.cell.2012.02.062 |
| 1072 | CFL1 | protein | Arc function | 10.1523/JNEUROSCI.2883-07.2007 |
| 4133 | MAP2 | protein | Arc function | 10.1002/jnr.20056 |
| 57731 | SPTBN4 | protein | Arc function | 10.1016/j.brainres.2007.03.079 |
| 1742 | DLG4 | protein | Arc function | 10.1371/journal.pbio.1001478 |
| 4915 | NTRK2 | protein | Arc function | 10.1371/journal.pbio.1001478 |
| 4851 | NOTCH1 | protein | Arc function | 10.1016/j.neuron.2011.01.004 |
| 351 | APP | protein | Arc function | 10.1016/j.cell.2011.09.036 |
| 23621 | BACE1 | protein | Arc function | 10.1016/j.cell.2011.09.036 |
| 5663 | PSEN1 | protein | Arc function | 10.1016/j.cell.2011.09.036 |
| 7329 | UBE2I | protein | Arc function | 10.1074/jbc.M112.356337 |
| 7337 | UBE3A | protein | ARC expression | 10.1016/j.cell.2010.01.026 |
| 54476 | RNF216 | protein | ARC expression | 10.1016/j.neuron.2014.05.016 |
| 7341 | SUMO1 | protein | Arc function | 10.1074/jbc.M112.356337 |
| 5371 | PML | protein | Arc function | 10.1016/j.brainres.2007.03.079 |
| 10524 | KAT5 | protein | Arc function | 10.1523/ENEURO.0019-14.2014 |
The genes in the ARC complex have been implicated in a number of conditions characterized by cognitive disabilities, including schizophrenia [19], Angelman syndrome [20], normal cognitive aging [21], and Alzheimer’s disease (AD) [22]. Notably, the ARC complex contains multiple AD-related genes, including amyloid precursor protein (APP), whose proteolysis generates an AD hallmark, amyloid-β (Aβ).
In this study, we hypothesized that the ARC complex may contribute to individual differences in intellectual function. We curated the complex and tested whether its SNPs are associated with IQ scores in children of the Avon Longitudinal Study of Parents and Children (ALSPAC). Since polygenic risk for AD is highly correlated with cognitive function [23], we next determined whether SNPs revealing significant association with intelligence in children were also associated with AD in adults in the International Genomics of Alzheimer’s Project (IGAP). Lastly, we examined whether the most strongly associated SNPs may have functional relevance in gene expression.
METHODS
Curation and Assessment of ARC Functional Complex
The ARC gene set was constructed based on Arc interaction partners and regulators, as evidenced in the literature (Table 1). We followed the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) Statement [24]. PubMed entries linked to human, mouse, and rat ARC genes (NCBI Gene IDs: 23237, 11838, and 54323, respectively) were obtained on December 14, 2014 using an NCBI cross-reference query. The search yielded 185 papers, all of which were screened to identify those papers showing experimental evidence for Arc interaction partners and regulators (n = 13). The remaining papers’ references were then examined further, which led to the identification of six additional relevant papers. From these 19 sources, a total of 37 experimentally-verified interaction partners and regulators were identified (Supplementary Figure 1). The gene complex was evaluated in STRING database, version 10 [25], to confirm its network connectivity and assess its enrichment for the biological and cellular processes based on gene ontology (GO). False discovery rate (FDR) was applied to correct for multiple testing.
Participants and genotyping
Avon Longitudinal Study of Parents and Children (ALSPAC) sample
The ALSPAC sample was derived from a well-characterized population-based study carried out in southwest England [26]. Pregnant women residing in the Bristol area of the United Kingdom, who had an estimated date of delivery between April 1,1991 and December 31, 1992, were recruited to take part. Of the original 14,541 pregnancies, 13,988 children were alive at one year. An additional 713 children were enrolled after age seven, resulting in a total sample of 14,701 children. These mother-child pairs have been followed for over 20 years generating an immense amount of data through biological samples, measurements, and questionnaires. The study website contains details of all the data that is available through a fully searchable data dictionary: http://www.bris.ac.uk/alspac/researchers/data-access/data-dictionary/. Ethical approval for the study was obtained from the ALSPAC Ethics and Law Committee and the Local Research Ethics Committees. Phenotype-matched genotype data were available for up to 6,832 children, depending on the variables.
Assessment of intellectual function was performed at 8.5 years of age using the third edition of the Weschler Intelligence Scale for Children (WISC-III) [27]. Intellectual function was measured by the full-scale IQ score (FSIQ), comprised of both a verbal score (VIQ) generated from the vocabulary subtest and a performance score (PIQ) generated from the matrix reasoning subtest. For more details, refer to the ALSPAC data dictionary (http://www.bris.ac.uk/alspac/researchers/data-access/data-dictionary/). Normal distribution of these variables was assured and all outliers were removed (mean±3 standard deviations (SD)). ALSPAC participants’ DNA was extracted from whole blood or buccal swab samples and prepared for genotyping using standard protocols. A total of 9,912 participants were genotyped using the Illumina HumanHap550 platform. SNPs with a minor allele frequency of <1% and call rate of <95% were removed. Furthermore, only SNPs passing the exact test of Hardy–Weinberg equilibrium (p > 5.00×10–7) were considered for analyses. Detailed quality control measures can be found at http://www.bristol.ac.uk/media-library/sites/alspac/migrated/documents/gwas-data-generation.pdf. Known autosomal variants were imputed with MACH 1.0.16 Markov Chain Haplotyping software [28], using CEPH individuals from phase 2 of the HapMap project (HG18) as a reference (release 22). Only SNPs with imputation quality estimates above 0.3 were examined.
Additional quality control was performed in the subset of individuals with available IQ measures to ensure that no SNPs or individuals had poor genotyping rates (<95%) and that no SNPs were rare (minor allele frequency, MAF <1%) or out of Hardy-Weinberg Equilibrium (p < 1.00E-05).
International Genomics of Alzheimer’s Project (IGAP) sample
ARC complex SNPs were extracted from the summary statistics of the IGAP GWAS (http://www.pasteur-lille.fr/en/recherche/u744/igap/igap_download.php) on individuals of European ancestry. IGAP used genotyped and imputed data on 7,055,881 SNPs to meta-analyze four previously-published GWAS datasets [namely, the European Alzheimer’s Disease Initiative (EADI), the Alzheimer Disease Genetics Consortium (ADGC), the Cohorts for Heart and Aging Research in Genomic Epidemiology consortium (CHARGE), and the Genetic and Environmental Risk in AD consortium (GERAD)], consisting of 17,008 AD cases and 37,154 controls [29].
Statistical analyses
To examine the contribution of ARC complex to cognitive function, we carried out the following analyses: (1) tests for association between ARC complex genes and IQ scores in ALSPAC (gene-set tests); (2) genes found to be significant in the gene-set test were further examined for single SNP-based association in ALSPAC; and (3) given reported shared genetics between cognitive function and AD, significant SNPs in ALSPAC were examined in the IGAP to check for association with AD. These analyses are described in more detail below.
Gene-set tests of ARC complex
Here, we tested the entire ARC complex, followed by “ARC expression” and “Arc function” subgroups individually (Table 1) in ALSPAC. The “ARC expression” subgroup included proteins implicated in the regulation of ARC transcription, mRNA processing, transport, Arc protein translation and degradation, while the Arc function subgroup included proteins that bind Arc or are closely associated with Arc function. ARC itself was not considered as a member of “ARC expression” or “Arc function” subgroups as by examining those subgroups we aimed at elucidating the role of ARC regulators and interactors only. SNPs within each gene in the complex (including those within 5,000 base pairs upstream and 1,500 base pairs downstream of each gene) were analyzed in MAGMA, version 1.05 [30], where first the linkage disequilibrium (LD) adjusted gene-based p-values were calculated and then converted to Z-values to regress on gene-set membership, as a predictor. Gene size and gene-sets’ gene density were added as covariates to account for possible confounding effects. Only genes on autosomal chromosomes were examined. Bonferroni correction was applied to correct for multiple testing, with the significance threshold set to 4.17E-03 (12 tests).
Single SNP-based tests
Genes revealing significant association in the gene-set tests were further examined on a single SNP level. Linear regressions were performed to test for association between SNPs and ALSPAC IQ scores in PLINK, version 1.9 [31]. Sex was set as a covariate, and only SNPs on autosomal chromosomes were examined. Correction for multiple testing was achieved by one million permutations, producing a family-wise empirical p-value. SNPs reaching family-wise p-value below 0.05 were considered significant.
Variants found to be significantly associated with IQ after correction for multiple testing were then looked up in the IGAP to check for association with AD.
Functional analyses
In silico assessment
SNPs surviving correction for multiple testing in association analyses were examined in silico. We used RegulomeDB [32] and HaploReg (version 4.1; CEU population code; http://www.broadinstitute.org/mammals/haploreg/) [33] to explore whether any of the SNPs may affect gene expression. Given the central role of hippocampus in ARC complex processes and memory formation, we focused on expression in hippocampal tissue. To investigate whether any of the SNPs are located within transcription factor binding sites, we utilized MatInspector (Matrix Library 9.3; http://www.genomatix.de/solutions/genomatix-genome-analyzer.html) and MATCH (version 1.0; http://www.gene-regulation.com/pub/programs.html) engines. Variants with predicted functional significance were then assessed further, using electrophoretic mobility shift assays (EMSA) and luciferase reporter assays.
Electrophoretic mobility shift assay (EMSA)
Synthetic 3′ biotin-labeled double-stranded oligonucleotides corresponding to the rs2830077[A] and rs2830077[C] sequences (20 fmol) and recombinant pure TFCP2 (100 ng, Active Motif) were incubated for 20 min using the GelshiftTM Chemiluminescent EMSA kit (Active Motif) in a 1×binding buffer supplemented with 2.5% glycerol, 10 ng/μl Poly d(I-C), 0.1% NP-40, 50 mM KCl, 0.5 mM MgCl2, and 0.12 mM EDTA. Reaction mixtures were separated by 6% PAAG, and products were detected by streptavidin-HRP conjugate. For competition assays, unlabeled oligonucleotides at 100-fold molar excess were added to the reaction mixture 5 min before adding the biotin-labeled probe. Sequences of the double-stranded probes for rs2830077[A] and rs2830077[C] were 5′-GACACGCTGACTTCCAGGCAaAAGCCAGGCACAAGAGAAGC-3′ and 5′-GACACGCTGACTTCCAGGCAcAAGCCAGGCACAAGAGAAGC-3′, respectively.
Luciferase reporter assay
The 300 base pair region of APP intron 1, bearing rs2830077 (NC_000021.9 : 26130583-26130284, GRCh38.p2 reference assembly) was amplified using Phusion® High-Fidelity DNA Polymerase (NEB) and specific primers tagged with XhoI restriction sites (forward: 5′-ATCCTCGAGTAGTTTCTTAAAACATGG-3′ and reverse: 5′-ATCCTCGAGTTATTTAGCTACAAGTTTTAAGA-3′). The amplicon was then cloned into the pGEM-T Easy vector (Promega), and the A allele of rs2830077 was created by site-directed mutagenesis using QuikChange Lightning Multi Site-Directed Mutagenesis Kit(Agilent). The fragment was then excised from the pGEM-T Easy vector using XhoI restriction enzyme (NEB) and ligated into the same site within the pGL3-Basic vector (Promega) to ensure that the vector backbone used for all the promoter constructs remained the same. The sequences and orientation of inserts of all plasmids were verified by direct sequencing.
HEK293FT cells (Thermo Fisher Scientific) were cultured in DMEM (Sigma-Aldrich) supplemented with 10% FBS (Sigma-Aldrich), sodium pyruvate (1 mM), glutamine (2 mM), penicillin (100 U/ml) and streptomycin (100 μg/ml). Cells were plated into 96-well plates (1×104 cells/well) and co-transfected after 24 h using Lipofectamine 2000 (Invitrogen) with 80 ng pGL3 vector constructs and 20 ng pRL-TK control plasmid (Promega) to allow normalization of transfection efficiency. Transfections were carried out in at least triplicates and repeated six times in independent experiments. Cells were assayed for firefly and Renilla luciferase activities 48 h after transfection using the Dual-Luciferase® Reporter Assay System (Promega). Measurements were performed on a VICTOR3 Multilabel Plate Reader (Perkin Elmer). The ratio of firefly to Renilla luciferase activity was calculated.
Image quantification and statistical analyses
Densitometric analysis was done using NIH ImageJ (http://imagej.nih.gov/ij/) and its differences were assessed by one-way analyses of variance (ANOVA), using SigmaPlot 13.0 (Systat Software).
RESULTS
Curation and Assessment of ARC Functional Complex
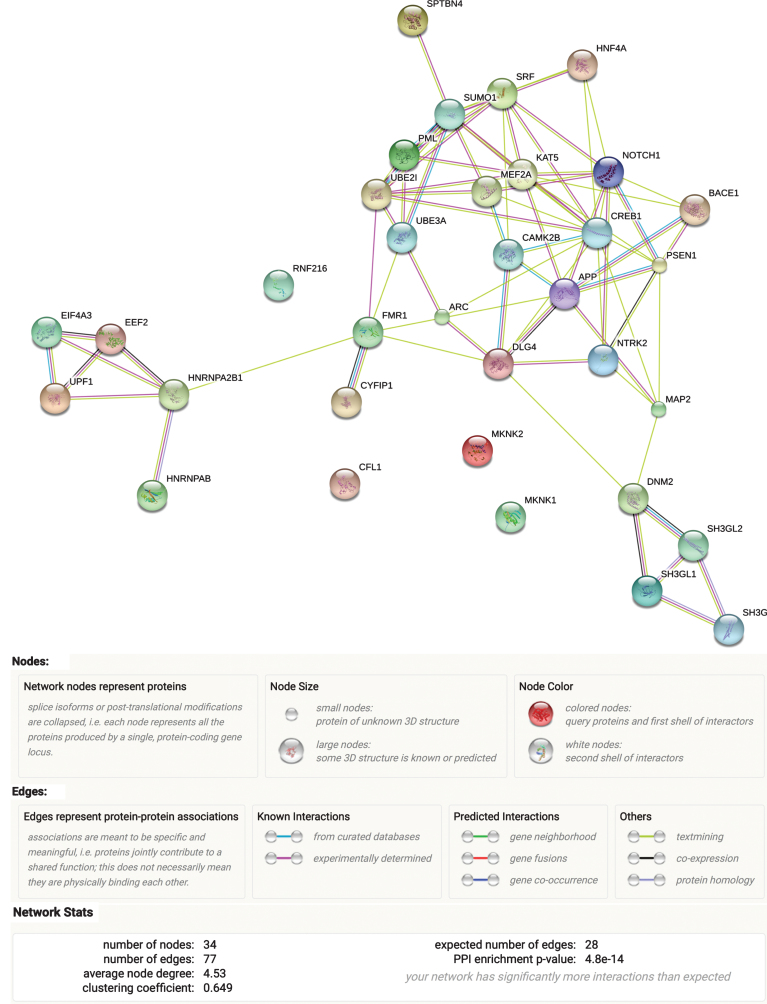
After assembling our ARC complex (Table 1), we evaluated its functional network in STRING database, which uses experimental and predicted protein-protein interaction information to assess connectivity (Fig. 1). Protein-protein interactions were significantly enriched (p = 4.8E-14) in the full ARC complex, indicating that the proteins in the curated gene set are biologically connected as a group. The most significant enrichment of the ARC complex as a whole was noted for cellular components (neuron projection, GO:0043005, FDR = 2.59E-07 and neuron part, GO:0097458, FDR = 2.59E-07). Supplementary Table 1 reflects the main findings from these assessments.
Fig.1.
ARC complex network. Results from the STRING analysis.
Similar to ARC complex, STRING examination of its subgroups (“ARC expression” and “Arc function,” Table 1) revealed that both of them show indication of biological protein-protein interactions, with the “Arc function” subgroup displaying stronger evidence (p = 1.52E-09) than the “ARC expression” subgroup (p = 3.19E-04). Correspondingly, the latter also showed less implication in GO pathways (Supplementary Figure 2) compared to the “Arc function” subgroup, where the strongest enrichment was observed for neuron projection (GO: 0043005, FDR = 1.47E-07), followed by protein binding (GO:0005515, FDR = 1.52E-07). Supplementary Figure 2 depicts the results of ARC complex subgroups STRING examination.
Participants and genotyping
The features of ALSPAC and IGAP samples are summarized in Table 2. The characteristics of IGAP sample were derived from its publication [29], combining 18 datasets. The distribution of the three IQ scores in ALSPAC is presented in Supplementary Figure 3.
Table 2.
ALSPAC and IGAP participant characteristics and ALSPAC IQ scores
| Sample | Number of | Males (%) | Age (years) | WISC-III (mean±standard deviation) | ||
| Participants | Verbal | Performance | Total | |||
| ALSPAC | 5,165 | 49.8 | 8.5 | 108.12 (16.56) | 100.23 (16.86) | 105.12 (16.17) |
| IGAP Cases | 17,008 | 35.1–40.6 | 68.5–82.7* | NA | NA | NA |
| IGAP Controls | 37,154 | 39.3–48.2 | 51.0–76.3** | NA | NA | NA |
NA, non-applicable. *Age range of time of onset across datasets. **Age range at time of examination across datasets.
Statistical analyses
Gene-set tests of ARC complex
Tentative association was observed between the functional subgroup (Table 1) of ARC complex and VIQ as well as FSIQ in the ALSPAC (p = 0.027 and 0.041, respectively). No association signal survived correction for multiple testing. Supplementary Table 2 shows the association results of all gene sets, and individual genes comprising those, for all IQ measures.
Single SNP-based tests
Since tentative association was observed for “ARC function” subgroup only, we tested individual SNPs in those genes for association with IQ measures in ALSPAC. The strongest signals were observed between 13 SNPs in the APP gene and PIQ, with 10 variants surviving the correction for multiple testing (Table 3, Supplementary Figure 4). Another strong signal was noted between rs1491579 in the SH3-domain GRB2-like 3 (SH3GL3) gene (Table 3) and FIQ, though it did not survive multiple test correction. Supplementary Table 3 shows results of association for all the variants reaching point-wise p-values below 1.00E-03.
Table 3.
Properties of the SNPs (and those in high LD with them) revealing significant association between ARC complex variants and IQ measures in the ALSPAC sample
| SNP | Chr. | Base pair | Gene | Beta | Point-wise | Family-wise | OR | p-value | r2 | Chromatin | Active | eQTL |
| (reference allele) | position | (95% CI) | empirical | empirical | (95% CI) | in AD | state in | chromatin marks | ||||
| (GRCh38) | p-value | p-value | hippocampus | in hippocampus | ||||||||
| Performance IQ | ||||||||||||
| rs2830077 (C) | 21 | 26130406 | APP | 1.40 (0.75–2.04) | 2.00E-05 | 0.018 | 0.95 (0.93–0.98) | 2.76E-03 | 1.00 | Transcription Enhancer | H3K4me1_Enh | APP |
| H3K27ac_Enh | ||||||||||||
| rs432766 (C) | 21 | 26135742 | APP | 1.40 (0.75–2.04) | 2.00E-05 | 0.018 | 0.97 (0.94–0.99) | 0.047 | 0.97 | Transcription Enhancer | H3K4me1_Enh | APP |
| H3K27ac_Enh | ||||||||||||
| rs375369 (A) | 21 | 26142055 | APP | 1.40 (0.75–2.04) | 2.00E-05 | 0.018 | 0.96 (0.93–0.99) | 0.015 | 0.94 | none | H3K4me1_Enh | none |
| H3K27ac_Enh | ||||||||||||
| rs467021 (C) | 21 | 26144759 | APP | 1.40 (0.75–2.04) | 2.00E-05 | 0.018 | 0.95 (0.93–0.98) | 3.06E-03 | 0.94 | none | H3K4me1_Enh | none |
| H3K27ac_Enh | ||||||||||||
| rs12483003 (C) | 21 | 26150590 | APP | 1.39 (0.74–2.03) | 2.50E-05 | 0.020 | 0.95 (0.93–0.99) | 3.53E-03 | 0.86 | Transcription Enhancer | H3K4me1_Enh | none |
| H3K27ac_Enh | ||||||||||||
| H3K4me3_Pro | ||||||||||||
| rs368196 (C) | 21 | 26158949 | APP | 1.39 (0.74–2.03) | 2.50E-05 | 0.020 | 0.95 (0.92–0.98) | 2.15E-03 | 0.91 | Transcription Enhancer | H3K4me1_Enh | none |
| H3K27ac_Enh | ||||||||||||
| rs465235 (G) | 21 | 26160704 | APP | 1.39 (0.74–2.03) | 2.50E-05 | 0.020 | 0.95 (0.92–0.98) | 1.27E-03 | 0.89 | none | H3K4me1_Enh | none |
| rs2830072 (T) | 21 | 26128343 | APP | 1.37 (0.72–2.02) | 3.20E-05 | 0.029 | 0.96 (0.93–0.99) | 0.020 | 0.98 | Transcription Enhancer | H3K4me1_Enh | APP |
| H3K27ac_Enh | ||||||||||||
| H3K4me3_Pro | ||||||||||||
| rs2830073 (T) | 21 | 26128825 | APP | 1.37 (0.72–2.02) | 3.20E-05 | 0.029 | 0.95 (0.93–0.98) | 2.47E-03 | 0.98 | Transcription 5 Enhancer | H3K4me1_Enh | APP |
| H3K27ac_Enh | ||||||||||||
| rs2154481 (C) | 21 | 26101558 | APP | 1.36 (0.71–2.01) | 3.50E-05 | 0.032 | 0.95 (0.92–0.98) | 6.18E-04 | 0.84 | Transcription Enhancer | H3K4me1_Enh | APP |
| H3K4me3_Pro | ||||||||||||
| rs2830066 (C) | 21 | 26121885 | APP | 1.27 (0.62–1.92) | 1.10E-04 | 0.096 | NA | NA | 0.80 | Transcription Enhancer | H3K4me1_Enh | APP |
| H3K27ac_Enh | ||||||||||||
| H3K4me3_Pro | ||||||||||||
| rs2830088 (C) | 21 | 26142422 | APP | 1.27 (0.62–1.92) | 1.12E-04 | 0.098 | NA | NA | 0.83 | none | H3K4me1_Enh | none |
| H3K27ac_Enh | ||||||||||||
| rs2830089 (G) | 21 | 26142921 | APP | 1.27 (0.62–1.92) | 1.12E-04 | 0.098 | NA | NA | 0.82 | none | H3K4me1_Enh | none |
| H3K27ac_Enh | ||||||||||||
| Full IQ | ||||||||||||
| rs1491579 (T) | 15 | 83509927 | SH3GL3 | –1.26 (–1.89––0.64) | 9.70E-05 | 0.060 | NA | NA | NA | Active Enhancer | H3K4me1_Enh | SH3GL3 |
| H3K27ac_Enh | ||||||||||||
| H3K4me3_Pro | ||||||||||||
CI, confidence interval; Chr, chromosome; OR, odds ratio; NA, non-applicable; LD, linkage disequilibrium; r2, measure of linkage disequilibrium.
The 10 APP SNPs surviving the correction for multiple testing in association analyses with the PIQ score were then examined for association with AD in IGAP. All 10 SNPs also revealed association signals with AD. Seven of these survived the Bonferroni correction (p < 0.005; Table 3 and Supplementary Figure 4).
Functional analyses
In silico assessment
Among the top APP SNPs significantly associated with PIQ, several were predicted to be located within active chromatin compartments, with transcription enhancer properties in hippocampus as well as expression quantitative trait loci (eQTL) activity in whole blood (Table 3).
H3K27Ac modification and DNAseI hypersensitivity data at rs2830077 also suggested that this region may be an enhancer (Supplementary Figure 5). The potential functional importance of this SNP was supported in HaploReg, which implicated rs2830077 in APP expression [34]. Further, RegulomeDB, MatInspector, and MATCH all indicated the presence of a putative transcription factor CP2 (TFCP2; also known as LBP-1c/CP2/LSF) binding site at rs2830077. We also found that the matrix score of rs2830077[A] is lower than that of the reference allele C, suggesting that the TFCP2 may have higher binding affinity to the C allele than to the A.
Electrophoretic mobility shift assay (EMSA)
To validate in silico predictions, we performed EMSA using pure recombinant TFCP2 and synthetic double-stranded oligonucleotides of the two alleles of rs2830077. EMSA demonstrated a shifted band of a DNA-protein complex, with a stronger intensity corresponding to its C allele (46% greater intensity of retarded band; Supplementary Figure 6), confirming its higher binding affinity of TFCP2.
Luciferase reporter assay
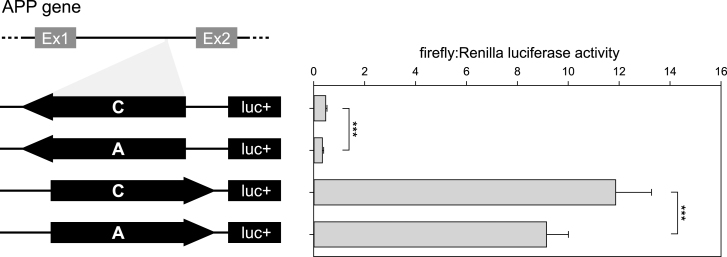
To validate the ability of rs2830077 to modulate transcription, we performed a luciferase assay with the first APP intron harboring this SNP. The luciferase activities for both orientations of the A allele were lower than corresponding activities of the C allele (28% and 23% lower luciferase activity for antisense and sense orientation, respectively; Fig. 2), confirming that rs2830077 may alter enhancer-like activity of this region.
Fig.2.
Results of luciferase reporter assays of APP intronic fragment bearing rs2830077 SNP in HEK293FT cell line. On the left: Schematic drawing of the constructs used in the luciferase assay system. On the right: Corresponding transcription efficiency measurements, which were done in triplicates from 6 independent experiments. Results are expressed as a ratio of firefly to Renilla luciferase activity (means±SD) ***p < 0.001.
DISCUSSION
In this study, we utilized a pathway approach to study genetics of cognition in children (ALSPAC) and followed up the observed association signals in a sample of adults with and without AD (IGAP). Further, we performed a functional examination of associated variants in cell culture. Our experimental approach was prompted by evidence that: (1) cognitive ability may be influenced by genetic variation in synaptic plasticity-related genes [35], (2) AD shares polygenic etiology with cognitive functioning [23], and (3) pathway analyses of functionally related genes can be advantageous as they combine the effect of multiple genes that may be biologically meaningful [14]. Our analyses led us to uncover a common APP variant association with PIQ in children, which was replicated in AD in adults. We further found that this SNP could influence APP expression by affecting TFCP2 binding affinity.
APP encodes the amyloid-β protein precursor (AβPP) that forms Aβ-containing neuritic plaques, the accumulation of which is one of the key histopathological hallmarks in AD. While the role of APP in AD has long been known, its involvement in childhood intelligence has not been previously reported. The association signal observed in this study is between the APP and PIQ (Table 3), believed to reflect fluid intelligence. Examining the data from a recent large-scale GWAS on cognitive functions (N = 112,151) [36], we noted that APP gene revealed signs of association with reaction time (efficiency of information processing) and memory (p = 0.048 and 0.095, respectively; Supplementary Figure 7), further suggesting possible involvement of APP in cognition.
General fluid cognitive functioning in childhood has been proposed to be linked to late-onset dementia [37, 38], with genetic influences possibly being the driving force behind stability of cognitive functioning [39]. In a recent study, carriers of the APOE ɛ4 variant (the best replicated known genetic factor for AD) revealed correlation with working memory and attention in children [40]. Thus, we hypothesize that the genetics of PIQ may be an important determinant of cognitive abilities throughout the lifespan and of age-related dementia.
Genetic overlap between IQ and dementia has previously been reported in other studies, including a large genome-wide analysis in over 100,000 individuals [23]. Another GWAS on general fluid cognitive ability in adults (N = 53,949) identified four genes (namely TOMM40, APOE, ABCG1, and MEF2C) previously associated with AD or neuropathological features of AD and related dementias [11]. Such overlap has also been noted for the APP gene, where a coding variant was shown to be protective against both normal age-related cognitive decline and AD [41]. While the APOE has shown robust association with AD and normal cognitive aging, more in-depth functional studies are needed to more thoroughly understand the functional significance of all genetic contributions of cognitive ability.
In this study, the most prominent significant signal was noted in APP, between the intronic rs2830077 and PIQ in children, also showing association with AD in adults (Table 3). In silico analyses of this SNP prompted its functional evaluation after noting that rs2830077: (1) is located within a region of active chromatin; (2) may have transcriptional enhancer activity in hippocampal tissue; (3) possesses eQTL activity; and (4) is located at a putative TFCP2 binding site. We confirmed via EMSA that TFCP2 binds to this putative enhancer region, with rs2830077 allele-specific binding affinity (Supplementary Figure 5). Further, luciferase assays indicate that the C allele of this SNP confers enhanced expression of APP (Fig. 2). The TFCP2 transcription factor has been implicated in erythroid gene expression [42], repression of HIV transcription [43], and in different cancer types [44, 45]. No direct connections between TFCP2 and AβPP expression have been reported, though a number of studies show association between the TFCP2 gene and AD [46, 47]. The activity of TFCP2 itself is regulated by AβPP-interacting protein Fe65 [48] and the intracellular domain of APP-like protein 2 [49]. Another APP SNP (rs467021), in linkage disequilibrium with rs2830077 (r2 = 0.99952), has been reported to be associated with cognitive decline in AD [50].
The dosage of AβPP has been implicated in altered neuronal endocytosis associated with increased Aβ production and age-related brain atrophy and degeneration, observed in patients with AD and Down syndrome (DS) [51]. Furthermore, duplication of the APP locus is thought to lead to early-onset AD [52] and the trisomy of this locus is likely to contribute to dementia in DS [53]. Indeed, the triplication of the Hsa21 segment including APP in people without DS has been associated with AD [53]. As both AD and DS display dementia, the observed association between PIQ and a SNP potentially altering the expression of APP, and thus its dosage, may help to elucidate the link between variation in intelligence and development of dementia.
We should note that to better estimate the contribution of genetics to IQ through life, examination of longitudinal samples where IQ is measured at early and late ages is desirable. However, since such samples were not available, we took the alternative approach of looking up significant child IQ association signals (ALSPAC) in late adulthood (IGAP). While our findings are intriguing, a direct replication in an independent childhood sample is warranted.
In summary, this study implicates APP in general cognitive abilities. We also show that evidence-based pathway analyses can be useful in identifying genetic factors underlying cognitive function. Follow-up studies are needed to more precisely determine how variants in APP may exert their effects on cognitive function over a lifespan. Such studies may have valuable implications for our understanding of etiology and, eventually, treatment of disorders associated with cognitive dysfunction, such as AD.
Supplementary Material
PRISMA flow diagram of literature search for ARC complex curation.
Summary of pathway analyses of ARC complex subgroups.
Histograms of IQ scores.
Regional plots of association in the APP locus.
H3K27Ac modification and DNAseI hypersensitivity data at rs2830077 locus.
EMSA results using pure recombinant TFCP2 and synthetic double-stranded oligonucleotides of the two alleles of rs2830077.
Regional plots of association between reaction time and memory in the APP locus.
ACKNOWLEDGMENTS
We are grateful to all the families who took part in this study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists, and nurses. The UK Medical Research Council and the Wellcome Trust (Grant ref: 102215/2/13/2) and the University of Bristol provide core support for ALSPAC. This publication is the work of the authors and will serve as guarantors for the contents of this paper. This research was specifically funded by Stiftelsen Kristian Gerhard Jebsen, the University of Bergen, the Western Norwegian Health Authorities (Helse Vest), the Bergen Medical Research Foundation, and the European Community’s Seventh Framework Programme (FP7/2007–2013; Grant no 602805). GWAS data was generated by Sample Logistics and Genotyping Facilities at the Wellcome Trust Sanger Institute and LabCorp (Laboratory Corporation of America) using support from 23andMe.
We thank the International Genomics of Alzheimer’s Project (IGAP) for providing summary results data for these analyses. The investigators within IGAP contributed to the design and implementation of IGAP and/or provided data but did not participate in analysis or writing of this report. IGAP was made possible by the generous participation of the control participants, the patients, and their families. The iSelect chips were funded by the French National Foundation on Alzheimer’s disease and related disorders. EADI was supported by the LABEX (laboratory of excellence program investment for the future) DISTALZ grant, Inserm, Institut Pasteur de Lille, Université de Lille 2 and the Lille University Hospital. GERAD was supported by the Medical Research Council (Grant n° 503480), Alzheimer’s Research UK (Grant n° 503176), the Wellcome Trust (Grant n° 082604/2/07/Z) and German Federal Ministry of Education and Research (BMBF): Competence Network Dementia (CND) grant n° 01GI0102, 01GI0711, 01GI0420. CHARGE was partly supported by the NIH/NIA grant R01 AG033193 and the NIA AG081220 and AGES contract N01–AG–12100, the NHLBI grant R01 HL105756, the Icelandic Heart Association, and the Erasmus Medical Center and Erasmus University. ADGC was supported by the NIH/NIA grants: U01 AG032984, U24 AG021886, U01 AG016976, and the Alzheimer’s Association grant ADGC–10–196728.
Finally, we thank professor Astri J. Lundervold for her invaluable insight on intelligence and help in reviewing this manuscript.
Authors’ disclosures available online (http://j-alz.com/manuscript-disclosures/17-0049r1).
Appendix
The supplementary material is available in the electronic version of this article: http://dx.doi.org/10.3233/JAD-170049.
REFERENCES
- [1]. Deary IJ, Weiss A, Batty GD (2010) Intelligence and personality as predictors of illness and death: How researchers in differential psychology and chronic disease epidemiology are collaborating to understand and address health inequalities. Psychol Sci Public Interest 11, 53–79. [DOI] [PubMed] [Google Scholar]
- [2]. Deary AJ, Strand S, Smith P, Fernandes C (2007) Intelligence and educational achievement. Intelligence 35, 13–21. [Google Scholar]
- [3]. Davies G, Tenesa A, Payton A, Yang J, Harris SE, Liewald D, Ke X, Le Hellard S, Christoforou A, Luciano M, McGhee K, Lopez L, Gow AJ, Corley J, Redmond P, Fox HC, Haggarty P, Whalley LJ, McNeill G, Goddard ME, Espeseth T, Lundervold AJ, Reinvang I, Pickles A, Steen VM, Ollier W, Porteous DJ, Horan M, Starr JM, Pendleton N, Visscher PM, Deary IJ (2011) Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Mol Psychiatry 16, 996–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4]. Visscher PM, Hill WG, Wray NR (2008) Heritability in the genomics era–concepts and misconceptions. Nat Rev Genet 9, 255–266. [DOI] [PubMed] [Google Scholar]
- [5]. Nisbett RE, Aronson J, Blair C, Dickens W, Flynn J, Halpern DF, Turkheimer E (2012) Intelligence: New findings and theoretical developments. Am Psychol 67, 130–159. [DOI] [PubMed] [Google Scholar]
- [6]. Toga AW, Thompson PM (2005) Genetics of brain structure and intelligence. Annu Rev Neurosci 28, 1–23. [DOI] [PubMed] [Google Scholar]
- [7]. Prabhakaran V, Rypma B, Gabrieli JD (2001) Neural substrates of mathematical reasoning: A functional magnetic resonance imaging study of neocortical activation during performance of the necessary arithmetic operations test. Neuropsychology 15, 115–127. [DOI] [PubMed] [Google Scholar]
- [8]. Gray JR, Chabris CF, Braver TS (2003) Neural mechanisms of general fluid intelligence. Nat Neurosci 6, 316–322. [DOI] [PubMed] [Google Scholar]
- [9]. Deary IJ, Penke L, Johnson W (2010) The neuroscience of human intelligence differences. Nat Rev Neurosci 11, 201–211. [DOI] [PubMed] [Google Scholar]
- [10]. Luciano M, Lind PA, Deary IJ, Payton A, Posthuma D, Butcher LM, Bochdanovits Z, Whalley LJ, Visscher PM, Harris SE, Polderman TJ, Davis OS, Wright MJ, Starr JM, de Geus EJ, Bates TC, Montgomery GW, Boomsma DI, Martin NG, Plomin R (2008) Testing replication of a 5-SNP set for general cognitive ability in six population samples. Eur J Hum Genet 16, 1388–1395. [DOI] [PubMed] [Google Scholar]
- [11]. Davies G, Armstrong N, Bis JC, Bressler J, Chouraki V, Giddaluru S, Hofer E, Ibrahim-Verbaas CA, Kirin M, Lahti J, van der Lee SJ, Le Hellard S, Liu T, Marioni RE, Oldmeadow C, Postmus I, Smith AV, Smith JA, Thalamuthu A, Thomson R, Vitart V, Wang J, Yu L, Zgaga L, Zhao W, Boxall R, Harris SE, Hill WD, Liewald DC, Luciano M, Adams H, Ames D, Amin N, Amouyel P, Assareh AA, Au R, Becker JT, Beiser A, Berr C, Bertram L, Boerwinkle E, Buckley BM, Campbell H, Corley J, De Jager PL, Dufouil C, Eriksson JG, Espeseth T, Faul JD, Ford I, Generation S, Gottesman RF, Griswold ME, Gudnason V, Harris TB, Heiss G, Hofman A, Holliday EG, Huffman J, Kardia SL, Kochan N, Knopman DS, Kwok JB, Lambert JC, Lee T, Li G, Li SC, Loitfelder M, Lopez OL, Lundervold AJ, Lundqvist A, Mather KA, Mirza SS, Nyberg L, Oostra BA, Palotie A, Papenberg G, Pattie A, Petrovic K, Polasek O, Psaty BM, Redmond P, Reppermund S, Rotter JI, Schmidt H, Schuur M, Schofield PW, Scott RJ, Steen VM, Stott DJ, van Swieten JC, Taylor KD, Trollor J, Trompet S, Uitterlinden AG, Weinstein G, Widen E, Windham BG, Jukema JW, Wright AF, Wright MJ, Yang Q, Amieva H, Attia JR, Bennett DA, Brodaty H, de Craen AJ, Hayward C, Ikram MA, Lindenberger U, Nilsson LG, Porteous DJ, Raikkonen K, Reinvang I, Rudan I, Sachdev PS, Schmidt R, Schofield PR, Srikanth V, Starr JM, Turner ST, Weir DR, Wilson JF, van Duijn C, Launer L, Fitzpatrick AL, Seshadri S, Mosley TH Jr, Deary IJ (2015) Genetic contributions to variation in general cognitive function: A meta-analysis of genome-wide association studies in the CHARGE consortium (N=53949). Mol Psychiatry 20, 183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12]. Myrum C, Giddaluru S, Jacobsen K, Espeseth T, Nyberg L, Lundervold AJ, Haavik J, Nilsson LG, Reinvang I, Steen VM, Johansson S, Wibrand K, Le Hellard S, Bramham CR (2015) Common variants in the ARC gene are not associated with cognitive abilities. Brain Behav 5, e00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13]. Goldstein DB (2009) Common genetic variation and human traits. N Engl J Med 360, 1696–1698. [DOI] [PubMed] [Google Scholar]
- [14]. Sun YV (2012) Integration of biological networks and pathways with genetic association studies. Hum Genet 131, 1677–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15]. Bramham CR, Alme MN, Bittins M, Kuipers SD, Nair RR, Pai B, Panja D, Schubert M, Soule J, Tiron A, Wibrand K (2010) The Arc of synaptic memory. Exp Brain Res 200, 125–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16]. Korb E, Finkbeiner S (2011) Arc in synaptic plasticity: From gene to behavior. Trends Neurosci 34, 591–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17]. Shepherd JD, Bear MF (2011) New views of Arc, a master regulator of synaptic plasticity. Nat Neurosci 14, 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18]. Myrum C, Baumann A, Bustad HJ, Flydal MI, Mariaule V, Alvira S, Cuellar J, Haavik J, Soule J, Valpuesta JM, Marquez JA, Martinez A, Bramham CR (2015) Arc is a flexible modular protein capable of reversible self-oligomerization. Biochem J 468, 145–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19]. Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, Ruderfer D, Moran J, Chambert K, Toncheva D, Georgieva L, Grozeva D, Fjodorova M, Wollerton R, Rees E, Nikolov I, van de Lagemaat LN, Bayes A, Fernandez E, Olason PI, Bottcher Y, Komiyama NH, Collins MO, Choudhary J, Stefansson K, Stefansson H, Grant SG, Purcell S, Sklar P, O’Donovan MC, Owen MJ (2012) De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry 17, 142–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20]. Greer PL, Hanayama R, Bloodgood BL, Mardinly AR, Lipton DM, Flavell SW, Kim TK, Griffith EC, Waldon Z, Maehr R, Ploegh HL, Chowdhury S, Worley PF, Steen J, Greenberg ME (2010) The Angelman Syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell 140, 704–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21]. Fletcher BR, Hill GS, Long JM, Gallagher M, Shapiro ML, Rapp PR (2014) A fine balance: Regulation of hippocampal Arc/Arg3.1 transcription, translation and degradation in a rat model of normal cognitive aging. Neurobiol Learn Mem 115, 58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22]. Wu J, Petralia RS, Kurushima H, Patel H, Jung MY, Volk L, Chowdhury S, Shepherd JD, Dehoff M, Li Y, Kuhl D, Huganir RL, Price DL, Scannevin R, Troncoso JC, Wong PC, Worley PF (2011) Arc/Arg3.1 regulates an endosomal pathway essential for activity-dependent beta-amyloid generation. Cell 147, 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23]. Hagenaars SP, Harris SE, Davies G, Hill WD, Liewald DC, Ritchie SJ, Marioni RE, Fawns-Ritchie C, Cullen B, Malik R, METASTROKE Consortium, International Consortium for Blood Pressure GWAS, SpiroMeta Consortium, CHARGE Consortium Pulmonary Group, CHARGE Consortium Aging and Longevity Group, Worrall BB, Sudlow CL, Wardlaw JM, Gallacher J, Pell J, McIntosh AM, Smith DJ, Gale CR, Deary IJ (2016) Shared genetic aetiology between cognitive functions and physical and mental health in UK Biobank (N=112 151) and 24 GWAS consortia. Mol Psychiatry 21, 1624–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24]. Moher D, Liberati A, Tetzlaff J, Altman DG, Group P (2009) Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med 6, e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25]. Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C (2015) STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43, D447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26]. Boyd A, Golding J, Macleod J, Lawlor DA, Fraser A, Henderson J, Molloy L, Ness A, Ring S, Davey Smith G (2013) Cohort Profile: The ‘children of the 90s’–the index offspring of the Avon Longitudinal Study of Parents and Children. Int J Epidemiol 42, 111–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27]. Wechsler D, Golombok S, Rust J (1992) WISC-III UK Wechsler Intelligence Scale for Children: UK Manual, Psychol. Corp, Sidcup, UK. [Google Scholar]
- [28]. Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR (2010) MaCH: Using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol 34, 816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29]. Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier-Boley B, Russo G, Thorton-Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, Vardarajan BN, Kamatani Y, Lin CF, Gerrish A, Schmidt H, Kunkle B, Dunstan ML, Ruiz A, Bihoreau MT, Choi SH, Reitz C, Pasquier F, Cruchaga C, Craig D, Amin N, Berr C, Lopez OL, De Jager PL, Deramecourt V, Johnston JA, Evans D, Lovestone S, Letenneur L, Morón FJ, Rubinsztein DC, Eiriksdottir G, Sleegers K, Goate AM, Fiévet N, Huentelman MW, Gill M, Brown K, Kamboh MI, Keller L, Barberger-Gateau P, McGuiness B, Larson EB, Green R, Myers AJ, Dufouil C, Todd S, Wallon D, Love S, Rogaeva E, Gallacher J, St George-Hyslop P, Clarimon J, Lleo A, Bayer A, Tsuang DW, Yu L, Tsolaki M, Bossú P, Spalletta G, Proitsi P, Collinge J, Sorbi S, Sanchez-Garcia F, Fox NC, Hardy J, Deniz Naranjo MC, Bosco P, Clarke R, Brayne C, Galimberti D, Mancuso M, Matthews F; European Alzheimer’s Disease Initiative (EADI), Genetic and Environmental Risk in Alzheimer’s Disease, Alzheimer’s Disease Genetic Consortium, Cohorts for Heart and Aging Research in Genomic Epidemiology, Moebus S, Mecocci P, Del Zompo M, Maier W, Hampel H, Pilotto A, Bullido M, Panza F, Caffarra P, Nacmias B, Gilbert JR, Mayhaus M, Lannefelt L, Hakonarson H, Pichler S, Carrasquillo MM, Ingelsson M, Beekly D, Alvarez V, Zou F, Valladares O, Younkin SG, Coto E, Hamilton-Nelson KL, Gu W, Razquin C, Pastor P, Mateo I, Owen MJ, Faber KM, Jonsson PV, Combarros O, O’Donovan MC, Cantwell LB, Soininen H, Blacker D, Mead S, Mosley TH Jr, Bennett DA, Harris TB, Fratiglioni L, Holmes C, de Bruijn RF, Passmore P, Montine TJ, Bettens K, Rotter JI, Brice A, Morgan K, Foroud TM, Kukull WA, Hannequin D, Powell JF, Nalls MA, Ritchie K, Lunetta KL, Kauwe JS, Boerwinkle E, Riemenschneider M, Boada M, Hiltuenen M, Martin ER, Schmidt R, Rujescu D, Wang LS, Dartigues JF, Mayeux R, Tzourio C, Hofman A, Nöthen MM, Graff C, Psaty BM, Jones L, Haines JL, Holmans PA, Lathrop M, Pericak-Vance MA, Launer LJ, Farrer LA, van Duijn CM, Van Broeckhoven C, Moskvina V, Seshadri S, Williams J, Schellenberg GD, Amouyel P (2013) Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 45, 1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30]. de Leeuw CA, Mooij JM, Heskes T, Posthuma D (2015) MAGMA: Generalized gene-set analysis of GWAS data. PLoS Comput Biol 11, e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31]. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC (2007) PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32]. Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, Cherry JM, Snyder M (2012) Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 22, 1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33]. Ward LD, Kellis M (2016) HaploReg v4: Systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease.D. Nucleic Acids Res 44, 877–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34]. Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, Christiansen MW, Fairfax BP, Schramm K, Powell JE, Zhernakova A, Zhernakova DV, Veldink JH, Van den Berg LH, Karjalainen J, Withoff S, Uitterlinden AG, Hofman A, Rivadeneira F, t Hoen PA, Reinmaa E, Fischer K, Nelis M, Milani L, Melzer D, Ferrucci L, Singleton AB, Hernandez DG, Nalls MA, Homuth G, Nauck M, Radke D, Volker U, Perola M, Salomaa V, Brody J, Suchy-Dicey A, Gharib SA, Enquobahrie DA, Lumley T, Montgomery GW, Makino S, Prokisch H, Herder C, Roden M, Grallert H, Meitinger T, Strauch K, Li Y, Jansen RC, Visscher PM, Knight JC, Psaty BM, Ripatti S, Teumer A, Frayling TM, Metspalu A, van Meurs JB, Franke L (2013) Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet 45, 1238–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35]. Hill WD, Davies G, van de Lagemaat LN, Christoforou A, Marioni RE, Fernandes CP, Liewald DC, Croning MD, Payton A, Craig LC, Whalley LJ, Horan M, Ollier W, Hansell NK, Wright MJ, Martin NG, Montgomery GW, Steen VM, Le Hellard S, Espeseth T, Lundervold AJ, Reinvang I, Starr JM, Pendleton N, Grant SG, Bates TC, Deary IJ (2014) Human cognitive ability is influenced by genetic variation in components of postsynaptic signalling complexes assembled by NMDA receptors and MAGUK proteins. Transl Psychiatry 4, e341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36]. Davies G, Marioni RE, Liewald DC, Hill WD, Hagenaars SP, Harris SE, Ritchie SJ, Luciano M, Fawns-Ritchie C, Lyall D, Cullen B, Cox SR, Hayward C, Porteous DJ, Evans J, McIntosh AM, Gallacher J, Craddock N, Pell JP, Smith DJ, Gale CR, Deary IJ (2016) Genome-wide association study of cognitive functions and educational attainment in UK Biobank (N=112 151). Mol Psychiatry 21, 758–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37]. Whalley LJ, Starr JM, Athawes R, Hunter D, Pattie A, Deary IJ (2000) Childhood mental ability and dementia. Neurology 55, 1455–1459. [DOI] [PubMed] [Google Scholar]
- [38]. Starr JM, Lonie J (2008) Estimated pre-morbid IQ effects on cognitive and functional outcomes in Alzheimer disease: A longitudinal study in a treated cohort. BMC Psychiatry 8, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39]. Hoekstra RA, Bartels M, Boomsma DI (2007) Longitudinal genetic study of verbal and nonverbal IQ from early childhood to young adulthood. Learn Individ Differ 17, 97–114. [Google Scholar]
- [40]. Chang L, Douet V, Bloss C, Lee K, Pritchett A, Jernigan TL, Akshoomoff N, Murray SS, Frazier J, Kennedy DN, Amaral DG, Gruen J, Kaufmann WE, Casey BJ, Sowell E, Ernst T, Pediatric Imaging, Neurocognition, and Genetics (PING) Study Consortium (2016) Gray matter maturation and cognition in children with different APOE epsilon genotypes. Neurology 87, 585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41]. Jonsson T, Atwal JK, Steinberg S, Snaedal J, Jonsson PV, Bjornsson S, Stefansson H, Sulem P, Gudbjartsson D, Maloney J, Hoyte K, Gustafson A, Liu Y, Lu Y, Bhangale T, Graham RR, Huttenlocher J, Bjornsdottir G, Andreassen OA, Jonsson EG, Palotie A, Behrens TW, Magnusson OT, Kong A, Thorsteinsdottir U, Watts RJ, Stefansson K (2012) A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. Nature 488, 96–99. [DOI] [PubMed] [Google Scholar]
- [42]. Chae JH, Kim CG (2003) CP2 binding to the promoter is essential for the enhanced transcription of globin genes in erythroid cells. Mol Cells 15, 40–47. [PubMed] [Google Scholar]
- [43]. Yoon JB, Li G, Roeder RG (1994) Characterization of a family of related cellular transcription factors which can modulate human immunodeficiency virus type 1 transcription in vitro. Mol Cell Biol 14, 1776–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44]. Jiang H, Du J, Jin J, Qi X, Pu Y, Fei B (2014) LSF expression and its prognostic implication in colorectal cancer. Int J Clin Exp Pathol 7, 6024–6031. [PMC free article] [PubMed] [Google Scholar]
- [45]. Yoo BK, Emdad L, Gredler R, Fuller C, Dumur CI, Jones KH, Jackson-Cook C, Su ZZ, Chen D, Saxena UH, Hansen U, Fisher PB, Sarkar D (2010) Transcription factor Late SV40 Factor (LSF) functions as an oncogene in hepatocellular carcinoma. Proc Natl Acad Sci U S A 107, 8357–8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46]. Bertram L, Parkinson M, McQueen MB, Mullin K, Hsiao M, Menon R, Moscarillo TJ, Blacker D, Tanzi RE (2005) Further evidence for LBP-1c/CP2/LSF association in Alzheimer’s disease families. J Med Genet 42, 857–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47]. Lambert JC, Goumidi L, Vrieze FW, Frigard B, Harris JM, Cummings A, Coates J, Pasquier F, Cottel D, Gaillac M, St Clair D, Mann DM, Hardy J, Lendon CL, Amouyel P, Chartier-Harlin MC (2000) The transcriptional factor LBP-1c/CP2/LSF gene on chromosome 12 is a genetic determinant of Alzheimer’s disease. Hum Mol Genet 9, 2275–2280. [DOI] [PubMed] [Google Scholar]
- [48]. Bruni P, Minopoli G, Brancaccio T, Napolitano M, Faraonio R, Zambrano N, Hansen U, Russo T (2002) Fe65, a ligand of the Alzheimer’s beta-amyloid precursor protein, blocks cell cycle progression by down-regulating thymidylate synthase expression. J Biol Chem 277, 35481–35488. [DOI] [PubMed] [Google Scholar]
- [49]. Xu Y, Kim HS, Joo Y, Choi Y, Chang KA, Park CH, Shin KY, Kim S, Cheon YH, Baik TK, Kim JH, Suh YH (2007) Intracellular domains of amyloid precursor-like protein 2 interact with CP2 transcription factor in the nucleus and induce glycogen synthase kinase-3beta expression. Cell Death Differ 14, 79–91. [DOI] [PubMed] [Google Scholar]
- [50]. Belbin O, Beaumont H, Warden D, Smith AD, Kalsheker N, Morgan K (2009) PSEN1 polymorphisms alter the rate of cognitive decline in sporadic Alzheimer’s disease patients. Neurobiol Aging 30, 1992–1999. [DOI] [PubMed] [Google Scholar]
- [51]. Prandini P, Deutsch S, Lyle R, Gagnebin M, Delucinge Vivier C, Delorenzi M, Gehrig C, Descombes P, Sherman S, Dagna Bricarelli F, Baldo C, Novelli A, Dallapiccola B, Antonarakis SE (2007) Natural gene-expression variation in Down syndrome modulates the outcome of gene-dosage imbalance. Am J Hum Genet 81, 252–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52]. Rovelet-Lecrux A, Hannequin D, Raux G, Le Meur N, Laquerriere A, Vital A, Dumanchin C, Feuillette S, Brice A, Vercelletto M, Dubas F, Frebourg T, Campion D (2006) APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat Genet 38, 24–26. [DOI] [PubMed] [Google Scholar]
- [53]. Wiseman FK, Alford KA, Tybulewicz VL, Fisher EM (2009) Down syndrome–recent progress and future prospects. Hum Mol Genet 18, R75–R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PRISMA flow diagram of literature search for ARC complex curation.
Summary of pathway analyses of ARC complex subgroups.
Histograms of IQ scores.
Regional plots of association in the APP locus.
H3K27Ac modification and DNAseI hypersensitivity data at rs2830077 locus.
EMSA results using pure recombinant TFCP2 and synthetic double-stranded oligonucleotides of the two alleles of rs2830077.
Regional plots of association between reaction time and memory in the APP locus.
The supplementary material is available in the electronic version of this article: http://dx.doi.org/10.3233/JAD-170049.