Key Points
Bone marrow progenitors, monocytes, and myeloid DCs contain BRAFV600E alleles in adults with LCH and ECD.
Mutant allele distribution is not disease specific, but precursors have distinct LCH-like and macrophage differentiation capacities.
Abstract
Langerhans cell histiocytosis (LCH) and Erdheim-Chester disease (ECD) are rare histiocytic disorders induced by somatic mutation of MAPK pathway genes. BRAFV600E mutation is the most common mutation in both conditions and also occurs in the hematopoietic neoplasm hairy cell leukemia (HCL). It is not known if adult LCH or ECD arises from hematopoietic stem cells (HSCs), nor which potential blood borne precursors lead to the formation of histiocytic lesions. In this study, BRAFV600E allele–specific polymerase chain reaction was used to map the neoplastic clone in 20 adults with LCH, ECD, and HCL. BRAFV600E was tracked to classical monocytes, nonclassical monocytes, and CD1c+ myeloid dendritic cells (DCs) in the blood, and mutations were observed in HSCs and myeloid progenitors in the bone marrow of 4 patients. The pattern of involvement of peripheral blood myeloid cells was indistinguishable between LCH and ECD, although the histiocytic disorders were distinct to HCL. As reported in children, detection of BRAFV600E in peripheral blood of adults was a marker of active multisystem LCH. The healthy counterparts of myeloid cells affected by BRAF mutation had a range of differentiation potentials depending on exogenous signals. CD1c+ DCs acquired high langerin and CD1a with granulocyte-macrophage colony-stimulating factor and transforming growth factor β alone, whereas CD14+ classical monocytes required additional notch ligation. Both classical and nonclassical monocytes, but not CD1c+ DCs, made foamy macrophages easily in vitro with macrophage colony-stimulating factor and human serum. These studies are consistent with a hematopoietic origin and >1 immediate cellular precursor in both LCH and ECD.
Introduction
Langerhans cell histiocytosis (LCH) and Erdheim-Chester disease (ECD) are rare inflammatory myeloid neoplasms principally caused by mutations in the MEK–extracellular signal-regulated kinase (ERK) signaling pathway, most commonly involving BRAF.1-5 LCH has a peak of incidence in childhood, but an increasing number of adults are now also diagnosed, including some with a poor outcome because of multisystem LCH (MS-LCH).6-8 ECD, once considered a form of “non-Langerhans” histiocytosis, appears to be a related condition by virtue of a common genetic landscape and the histological evidence of coexistent LCH and ECD in a proportion of adult patients.9,10 In contrast with LCH, ECD is rarely seen in children and predominantly affects older males.11,12
In children with MS-LCH caused by BRAFV600E allele–specific polymerase chain reaction (PCR) has been used to demonstrate mutant alleles in CD14+ and CD11c+ peripheral blood populations.5 A hematopoietic origin was corroborated in 2 cases of MS-LCH through expansion of hematopoietic clones in vitro.5 These studies support a model in which MS-LCH arises in the hematopoietic stem cells (HSCs) and is disseminated to the periphery by myeloid cells carrying somatic mutation.13 However, the identity of blood-borne cells that give rise to peripheral tissue histiocytes remains open to further investigation, because >1 cell type carries BRAFV600E and myeloid lineages may have overlapping differentiation potentials.
Human peripheral blood contains several subsets of myeloid mononuclear cells. Monocytes comprise ∼10% of mononuclear cells and may be divided into classical, nonclassical, and intermediate on the basis of CD14 and CD16 expression.14 All human monocytes express CD11c. CD14+ classical monocytes are the most numerous and can be differentiated into macrophages and dendritic cells (DCs) in vitro. Blood DCs comprise 1% to 2% of mononuclear cells and are subdivided into CD123+ plasmacytoid DCs and 2 subsets of CD11c+ myeloid DCs expressing CD1c+ and CD141+.15 CD1c+ DCs are the major population of myeloid DCs in blood and tissues. They may express langerin in vivo16 and readily acquire a Langerhans cell–like phenotype in vitro.17,18
The differentiation potentials of monocytes and blood DCs overlap, and monocyte-derived DCs bear many markers in common with CD1c+ myeloid DCs.19 Monocytes have plasticity in many directions including DC and macrophage axes20,21 and may also express langerin in vitro under certain conditions.22 Thus multiple blood-derived populations have the potential to contribute to histiocytic lesions.
The precursor populations of adults with LCH, ECD, or LCH/ECD crossover disease remain undefined. It is not known whether MS-LCH in adults correlates with the presence of mutant alleles in peripheral blood or if it is possible to discern a similar risk stratification in patients with ECD. NRAS mutation was detected in the CD14+ fraction of peripheral blood mononuclear cells (PBMCs) in 1 patient,23 and ERK activation was reported in 1% to 3% of monocytes of an ECD cohort.24 However, the involvement of other myeloid lineages has not been evaluated in ECD.
Recent RNA sequencing suggests that LCH lesions are enriched for DC gene transcripts, whereas biopsies of ECD are enriched for myeloid precursor and macrophage gene expression.25 It has been suggested that DC pathways of development may be involved in the generation of LCH, whereas monocyte-macrophage differentiation could give rise to ECD. Finally, BRAFV600E has been detected in HSCs of patients with hairy cell leukemia (HCL) and confirmed by xenografting.26 There does not appear to be a clinical overlap between HCL and histiocytosis, and the pathogenesis of these 2 different conditions from a common somatic mutation is poorly understood. HCL is therefore an interesting disease to evaluate in parallel and provides a useful control for the specificity of allele mapping in histiocytosis.
In this report, we have analyzed the distribution of BRAFV600E in the peripheral blood and bone marrow (BM) progenitor compartments of adults with MS-LCH, LCH/ECD crossover, and ECD. We sought to determine whether mutant alleles could be tracked to the HSCs and whether there was a correlation between the architecture of the neoplastic clone and the clinical phenotype of histiocytosis. We also aimed to intersect these observations with studies to determine the potential of DC and monocyte lineages to generate LCH-like cells and foamy macrophages as in vitro surrogates of pathological histiocytes.
Methods
Patient samples, lesional BRAFV600E testing, and clinical data
Blood, skin, BM, and surplus biopsy material were obtained from LCH and ECD patients referred to a local clinic with ethical approval from the Newcastle and North Tyneside Research Ethics Committee. Genomic DNA from formalin-fixed paraffin-embedded LCH lesions was extracted using Promega Maxwell Tissue DNA Purification Kits (Promega; http://www.promega.co.uk), and the region flanking codon 600 amplified by PCR. Amplicons were purified and genotyped by primer extension for the c.1799T>A, p.Val600Glu (V600E) using the Sequenom iPlex protocol (Sequenom; http://www.sequenom.com). The extension products were detected by matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) mass spectrometry using a Sequenom Typer 4.0. Disease activity was assessed by radiological examination including magnetic resonance imaging and positron emission tomography.
Flow cytometry
Absolute quantitation was performed using TruCount tubes on whole blood with a single-step lyse no-wash protocol. For more detailed analysis, whole blood was separated by density centrifugation into mononuclear and red cell/neutrophil fractions. The red cell pellet was subsequently used for extraction of neutrophil DNA. Flow cytometry was performed on an LSRII or LSR Fortessa X20 cytometer from Becton Dickinson (BD; http://www.bd.com), and data were analyzed with FlowJo (Treestar). Fluorescence-activated cell sorting was performed using a BD FACS Aria. Sorted cells were collected into microfuge tubes containing RPMI with 10% fetal calf serum.
Antibodies were from BD unless stated otherwise and are denoted as follows: antigen fluorochrome (clone) CD1a BV421 (HI149; Biolegend; http://www.biolegend.com); CD1c APC (AD5-8E7; Miltenyi Biotec; http://www.miltenyibiotec.com); CD3 FITC (SK7); CD3 V500 (UCHT1); CD7 FITC (4H9); CD10 PE-TR (B-Ly6; Beckman Coulter; http://www.beckmancoulter.com); CD11c A700 (B-ly6); CD14 APCCy7 (MΦP9); CD14 FITC (M5E2); CD16 FITC (NKP15/Leu-11a); CD16 PeCy7 (B73.1); CD19 FITC (4G7); CD19 PECy7 (SJ25C1); CD20 FITC (L27); CD34 PE (8G12); CD34 FITC (8G12); CD34 APC (581); CD38 PECy7 (HB7); CD45 V450 (2D1); CD45 APCCy7 (2D1); CD45RA BV510 (HI100; Biolegend); CD56 FITC (NCAM16.2); CD90 PerCPCY5.5 (5E10; Biolegend); CD123 PerCPCY5.5 (7G3); CD123 PE (9F5); CD207 PE (DCGM4; Beckman Coulter); HLA-DR V500 (L243/G46-6); and HLA-DR A700 (L243/G46-6).
BRAFV600E allele–specific PCR
Genomic DNA from sorted blood cells, BM cells, control cell lines, or cell-free sources was extracted using Qiagen micro-kits (Qiagen; http://www.qiagen.com). BRAF mutation and reference quantitative PCR was performed with competitive allele-specific Taqman mutation detection assays: Mutation Allele Assay, BRAF_476_mu 4465804 Hs00000111_mu; Gene Reference Assay, BRAF_rf - 4465807 Hs00000172_rf, according to the manufacturer’s instructions (Life Technologies; http://www.lifetechnologies.com). A standard curve was derived from 6 serial dilutions of genomic DNA from BRAFV600E melanoma cell line A375 and Epstein-Barr virus–transformed lymphoblastoid cell lines; 0%, 0.1%, and 10% V600E mutation controls were performed with every run. The percentage of BRAFV600E mutation was determined from the dCT (CT reference − CT mutant) using the standard curve. Graphs were plotted with Prism Version 5.0 (GraphPad Software Inc.).
In vitro differentiation
Aliquots of 10 000 sorted cells from healthy controls were cultured in RPMI 1640 with 10% fetal bovine serum or 5% human serum in 100 μL in 96-well plates. Supplements were added: granulocyte-macrophage colony-stimulating factor (GM-CSF), 50 ng/μL; transforming growth factor β (TGF-β), 10 ng/μL; macrophage colony-stimulating factor (M-CSF), 50 ng/μL. Cultures were maintained for 3 days before analysis by flow cytometry. For phase-contrast microscopy, cells were cultured in Thermo Scientific Nunc chamber slides system for 7 days, washed gently, and fixed with 1% paraformaldehyde in phosphate-buffered saline, and images were acquired with Carl Zeiss Axioplan 2 ×40 objective and Carl Zeiss Axiovision software release 4.8.
Statistical methods
Mann-Whitney U tests, Student t tests, and Fisher’s exact test were performed using GraphPad Prism 5.0 software.
Results
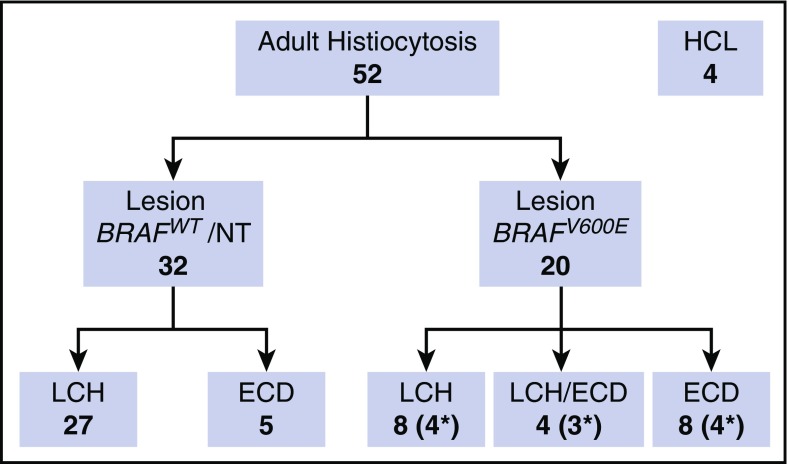
Fifty-two adults with histiocytosis and 4 with HCL were recruited from a local clinic and collaborating centers (Figure 1). Twenty with histiocytosis and all with HCL had confirmed BRAFV600E lesions. Three additional children with BRAFV600E-mutated LCH were included in some analyses for comparison. Further clinical details are given in supplemental Table 1 (available on the Blood Web site).
Figure 1.
Patients. Summary of adult patients included in the study. Asterisks indicate the number of patients with BRAFV600E detectable in PBMCs. The BRAF wild-type (WT) ECD group includes 1 patient with RAS mutation. NT, not tested.
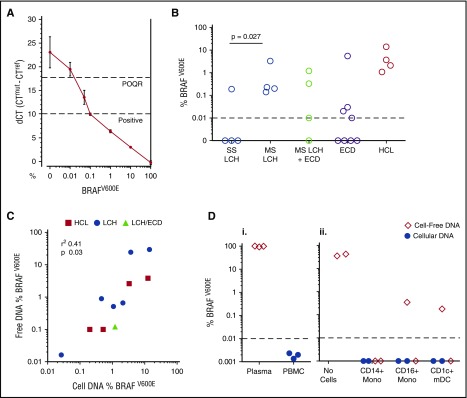
Detection of BRAFV600E was performed using an allele-specific PCR and was sensitive to 0.01% with a quantitative range to 0.1% (Figure 2A). BRAFV600E alleles were detectable in DNA isolated from PBMCs of patients with active MS-LCH with or without coexistent ECD (LCH/ECD crossover) but were only present in 1 patient with single-system (SS) LCH (Figure 2B). ECD is not conventionally classified into SS or MS. Nearly all cases involved multiple systems. There was no clear correlation between clinical features and BRAFV600E detection in the PBMCs (see supplemental Table 1 for details).
Figure 2.
Detection of BRAFV600E alleles. (A) A standard curve constructed using DNA purified from a dilution series of BRAFV600E-positive melanoma cell line A375 into WT Epstein-Barr virus–transformed lymphoblastoid cell line. The quantitative limit of detection was 0.1%; the absolute limit of detection was 0.01% (positive outside of quantitative range [POQR]). (B) BRAFV600E allele frequency in bulk PBMCs in cases of lesion BRAFV600E + LCH, LCH/ECD, and ECD. Contingency of positive PBMCs upon MS-LCH tested by Fisher’s exact test. (C) Correlation between mutated allele burden in cell-free plasma and PBMC DNA in LCH, ECD, and HCL. (Di) Test of exogenous free DNA uptake by PBMCs. BRAF-mutated DNA (derived from melanoma cell line A375) was spiked into whole blood for 24 hours at room temperature. Plasma and PBMCs were isolated by density centrifugation. DNA was extracted from both fractions and subjected to allele-specific PCR. (Dii) Test of exogenous free DNA uptake by sorted cells incubated at 37°C for 24 hours in medium supplemented with 20% human serum from a patient with HCL. DNA from supernatants and cell pellets, as indicated, was subjected to allele-specific PCR. mDC, myeloid dendritic cell.
A simple linear correlation was observed between the frequency of mutant allele detection in PBMCs vs cell-free plasma DNA, with approximately half-log greater sensitivity for detection in cellular DNA (Figure 2C). In order to exclude the uptake of free DNA into the cell-associated pool by phagocytosis or macropinocytosis, purified BRAFV600E-mutated DNA from A375 cells was incubated with whole blood of healthy controls (Figure 2Di), and sorted cell fractions were cultured with medium containing 20% serum from a patient with HCL (Figure 2Dii). Cellular and cell-free DNA was then recovered and subjected to PCR. Mutated BRAF alleles were only recovered from the cell-free fraction, but not the cellular DNA, indicating that uptake of exogenous DNA did not account for cell-associated BRAFV600E. With patient serum, cell cultures not only remained negative but diluted or degraded the cell-free signal.
The pattern of mononuclear cells observed by flow was analyzable by conventional gating, and no atypical populations were detected (supplemental Figure 1). Neither langerin+CD1a+ LCH-like cells nor CD103+CD11c+ hairy cells were detectable in the peripheral blood of any of the histiocytosis patients (supplemental Figure 1). Absolute counting of PBMCs from patients with histiocytosis revealed monocytosis and increased numbers of total CD11c+ and CD11c+CD1c− myeloid cells (supplemental Figures 1 and 2). There were no detectable differences between BRAF-mutated and WT disease, although power to detect subgroup differences was limited. No cytological atypia was observed among monocytes and DC subsets (supplemental Figure 3).
In order to map the distribution of mutant alleles, PBMCs from 9/11 patients with circulating BRAFV600E and 2 children with MS-LCH were fractionated into 4 quadrants according to the expression of HLA-DR and lineage (CD3, CD19, CD20, and CD56). The gates were completely apposed in order to account for all mutant alleles present in PBMCs and to exclude the inadvertent loss of signal to an uncharacterized disease-related population.
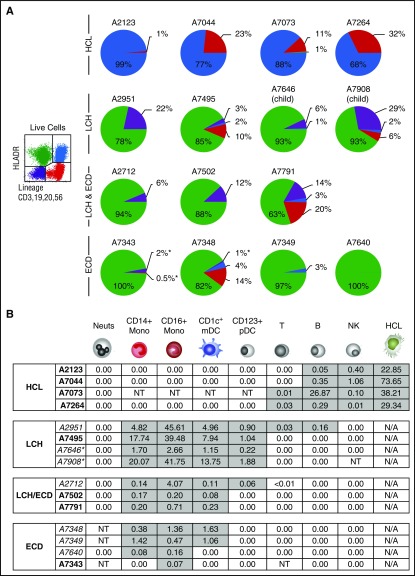
In contrast to patients with HCL, in which BRAFV600E was largely confined to the HLA-DR+lineage+ quadrant containing hairy cells, mutation in patients with histiocytosis was highly enriched in the HLA-DR+lineage− quadrant containing monocytes and DCs (Figure 3A). Some overspill into adjacent quadrants was observed, owing to the close gating. For example, in histiocytosis patients, positivity appeared in the HLA-DR−lineage− quadrant. This was further studied in case A2951 and found to be associated with CD14+ monocytes and CD11c+ myeloid precursors expressing low HLA-DR at the top of the double-negative gate (data not shown).
Figure 3.
Allele-specific PCR of BRAFV600E in PBMCs. (A) The distribution of BRAFV600E in PBMCs sorted into quadrants as shown according to expression of HLA-DR and lineage. Pie charts summarize the distribution of mutant alleles in HCL, LCH, and ECD. The area of each quadrant in the pie is proportional to the total number of mutant alleles in that quadrant (ie, the percentage positivity of the quadrant multiplied by the number of cells in the quadrant). All patients had active disease at the time of sampling. Two children with LCH included for comparison are indicated. Asterisk indicates the threshold of detection for a negative result where cell numbers were limiting. (B) The distribution BRAFV600E among peripheral blood cells showing the percentage of mutated alleles detected. Gray shading indicates positive fractions. Asterisk indicates child with MS-LCH shown for comparison. All patients had active disease at the time of sampling. Italics indicate those who had received prior treatment. B, B cell; Mono, monocyte; Neuts, neutrophil; NK, natural killer; NT, not tested owing to lack of or insufficient material; pDC, plasmacytoid dendritic cell; T, T cell.
The mononuclear compartment was further fractionated to explore the distribution of mutant alleles at higher resolution (supplemental Figures 1 and 2; Figure 3B). As expected, BRAFV600E was localized to hairy cells with a low level of alleles detectable in phenotypically normal B and NK cells but none in the remaining monocytes and myeloid cells. In contrast, the majority of alleles in patients with histiocytosis were found in CD14+ classical monocytes, CD16+ nonclassical monocytes, and CD11c+CD1c+ myeloid DCs. Plasmacytoid DCs harbored some alleles in LCH patients, and in 1 case, BRAFV600E was also detectable at a low level in B and T cells. The distribution of alleles did not vary significantly between LCH, ECD, and patients with LCH/ECD. In addition to these compartments, we studied CD14+CD16+ intermediate monocytes and CD11c+CD1c− myeloid cells in a subset of patients (see supplemental Figures 1, 2, and 4 for details). These did not differ significantly from the allele level in CD14+ classical monocytes and CD1c+ myeloid DCs, respectively.
BRAFV600E mutation was previously reported in children with MS-LCH, using a more simple gating strategy to obtain CD14+ and CD11c+ myeloid cells.5 CD11c+ myeloid cells comprise multiple fractions including CD1c+ myeloid DCs, CD16+ nonclassical monocytes, and CD11c+CD1c− cells, which were analyzed separately here. The relationship between these populations is summarized in supplemental Figure 4.
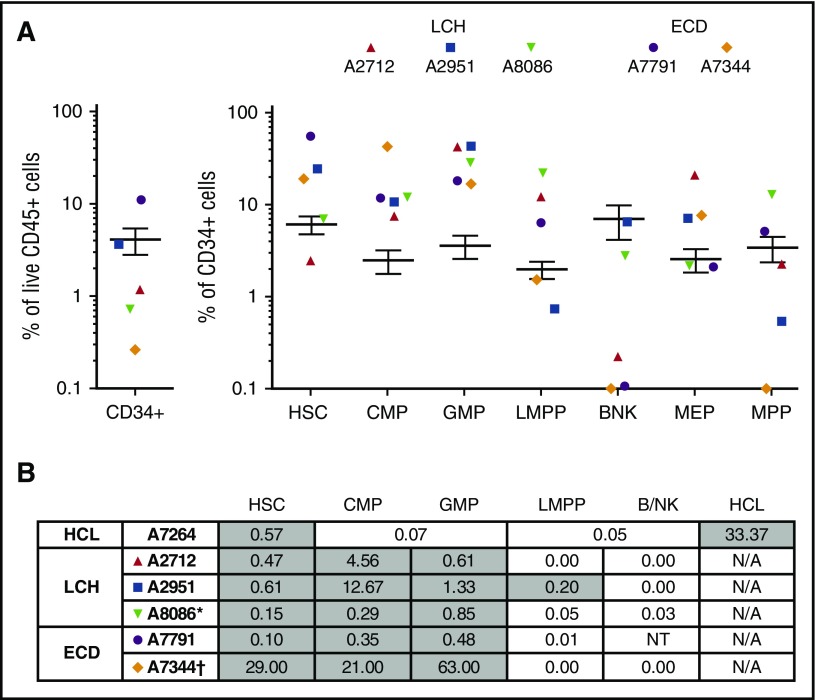
In order to probe the hematopoietic origin of LCH and ECD, BM was analyzed from 2 patients with MS-LCH and 2 with ECD. One ECD patient (A7344) had mutated NRAS rather than BRAF and was previously reported to have a high level of NRAS mutation in CD14+ peripheral blood monocytes (A7344).23 A commonly used schema for immunophenotyping (supplemental Figure 5) revealed that the common myeloid progenitor (CMP) and granulocyte-macrophage progenitor (GMP) compartments were relatively expanded (Figure 4A). BRAFV600E was detectable in CD90+CD45RA−CD38low HSCs, CD38highCD45RA− CMP, and CD45RA+ GMP (Figure 4B). The mutant allele was present at <1% in HSCs and was most enriched in the CMP fraction. Mature histiocytes arising from the mutated clone were rigorously excluded in this analysis (supplemental Figure 5). A pediatric patient included for comparison showed a similar distribution of alleles. The NRAS-mutated patient was quantified by peak height on Sanger sequencing at 21% to 63% in myeloid precursor fractions. Progenitor cells from this patient were demonstrated to engraft in immunodeficient mice, in the accompanying paper.27
Figure 4.
Mononuclear profile and BRAF allele frequency in CD34+ BM progenitors. (A) Relative proportions of progenitor fractions among CD34+ BM mononuclear cells compared with healthy controls (n = 21), expressed as percentage of total live cells. Error bars depict 95% confidence intervals of healthy controls. Populations: B/NK, B/NK cell progenitors; LMPP, lymphoid-primed multipotent progenitors; MEP, megakaryocyte–erythroid progenitor; MPP, multipotent progenitor. (B) The distribution of BRAFV600E among CD34+ BM progenitors. Gray shading indicates positive fractions. BM aspirate was obtained in parallel with the peripheral blood shown previously. *Child; †NRASQ61R Sanger sequencing. NT, not tested owing to insufficient number of cells.
These results suggest that LCH and ECD arise in the HSCs and are transmitted to peripheral tissues by blood-borne myeloid cells. However, there was no obvious disease-specific bias in the distribution of mutant alleles among myeloid cells that might correlate phenotypic differences between LCH and ECD. We therefore evaluated the potential of each myeloid fraction to give rise to the characteristic cells of LCH and ECD lesions through in vitro differentiation of sorted fractions from healthy controls.
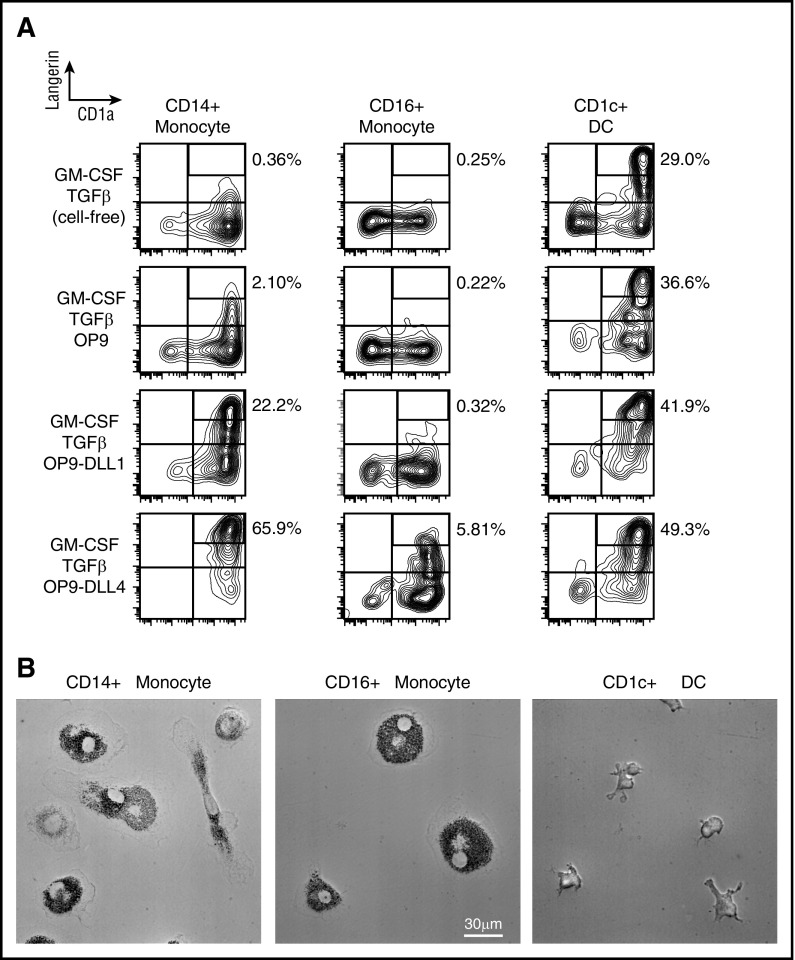
It has previously been demonstrated that monocytes and CD1c+ DCs can express langerin in vitro under different conditions, but only cells with high langerin and CD1a contain Birbeck granules.17,18,22,28,29 Previous reports are discordant and have not examined monocytes and DCs from peripheral blood and compared their differentiation capacity in the same platform. We therefore tested the development of a Langerhans cell (LC)–like phenotype by blood monocytes and CD1c+ DCs using conditions that had been previously described in either monocytes or DCs17,18,22,28,29: soluble factors GM-CSF and TGF-β and the notch ligands δ-like protein 1 (DLL1) and DLL4, expressed on mouse OP9 cells. In order to study the potential to develop foamy macrophage morphology, we used M-CSF with 5% human serum.
As shown previously, CD1c+ DCs attained high langerin expression with GM-CSF and TGF-β alone. Soluble factors GM-CSF and TGF-β were not sufficient to induce high langerin in CD14+ monocytes, but this potential was revealed by coculture with notch ligands DLL1 and DLL4. CD16+ monocytes did not express high langerin under any conditions, but notch ligation also further enhanced the expression of langerin by CD1c+ DCs in response to soluble factors (Figure 5; supplemental Figure 6). In macrophage differentiation conditions, CD1c+ DCs were unable to form foam cells, whereas both monocyte subsets developed this phenotype. A summary of the distribution of BRAF alleles and differentiation potential of myeloid lineage affected is shown in Table 1.
Figure 5.
In vitro differentiation potential of monocytes and myeloid DCs. (A) Sorted CD14+ monocytes, CD16+ monocytes, and CD1c+ myeloid DCs from healthy controls cultured for 3 days in conditions as shown. LangerinhighCD1a+ gates depicted contain LCH-like cells with Birbeck granules. Notch ligands DLL1 And DLL4 were presented on transfected mouse OP9 cells. Representative data of 5 independent experiments. (B) Sorted CD14+ monocytes, CD16+ monocytes, and CD1c+ myeloid DCs from healthy controls cultured for 7 days with M-CSF and 5% human serum (HS) to reveal potential to differentiate into foamy macrophages. Representative phase contrast images of 5 independent experiments.
Table 1.
Table summarizing BRAFV600E mutant allele frequency among blood monocytes and myeloid DCs (data from Figure 3) and the potential of WT cells to form langerinhigh LCH-like cells or foamy macrophages in vitro
| CD14 | CD16 | CD1c | |
|---|---|---|---|
| % BRAFV600E | |||
| Median | 0.38 | 1.36 | 0.2 |
| Range | 0-18 | 0-45 | 0-8 |
| Langerin+ potential | |||
| GM-CSF + TGF-β | − | − | ++ |
| +DLL1 | + | − | +++ |
| +DLL4 | +++ | − | +++ |
| Foamy mac potential | +++ | +++ | − |
Frequency of langerinhigh indicated as follows: −, <10%; +, 10%-25%; ++, 26%-35%; +++, >35%.
Discussion
Prior to the discovery of BRAFV600E, there was accumulating evidence that both LCH and ECD were clonal proliferations of histiocytes, including a remarkable insight by Chetritt et al that ECD was the “macrophage counterpart to LCH.”30,31 Here we have exploited the recurrent BRAFV600E mutation to map the neoplastic clones of LCH and ECD. The results further reinforce the concept that LCH and ECD are closely related hematopoietic neoplasms, regardless of their distinctive pathological features.32
A high-resolution analysis of myeloid cells demonstrated the highest frequency of mutant alleles in CD14+ classical monocytes, CD16+ nonclassical monocytes, and CD1c+ myeloid DCs. Unexpectedly, the nonclassical monocyte is a reservoir of BRAF mutation and accounts for a proportion of the signal previously tracked to CD11c+ myeloid cells.5 We also mapped BRAF mutation to cells defined by phenotype as HSCs in LCH, ECD, and HCL. BRAFV600E has previously been reported in HSCs of children with MS-LCH5 and patients with HCL,26 and it remains unclear how the same mutation present in HSCs generates such unrelated pathologies. Although there is a documented overlap between LCH and ECD,9,10 HCL is unrelated in incidence, and we did not observe HCL leukocytes in any patient with histiocytosis.
The initial cell-sorting approach adopted apposed gating of the HLA-DR vs lineage plot of total PBMCs in order to account for all the mutated alleles present in PBMC DNA (neutrophils were consistently negative). Although we localized most of the BRAFV600E alleles in peripheral blood to the HLA-DR+lineage− compartment of both LCH and ECD, some overspill was observed into the HLA-DR−lineage− cells, owing to this strategy. Although a proportion of this signal was contained in HLA-DRloCD14+ monocytes, CD11c+CD1c− myeloid progenitors that may generate macrophages are also found in this region. In particular, ECD patients frequently have expanded myelopoiesis with an element of left shift that mobilizes these cells into the peripheral compartment; thus, we cannot exclude a contribution of more primitive myeloid precursors to the lesions of ECD. Notably, recent gene expression studies described enrichment for myeloid progenitor signatures in ECD lesions.25
There is significant interest in cell-free DNA as a marker of neoplasia, and this is also detectable in histiocytosis.33 In our hands, plasma cell-free BRAFV600E was approximately half-log lower in abundance than cell-associated alleles. We considered the possibility that exogenous DNA might account for the mutant alleles found in PBMCs, especially as the signal was localized to monocytes with phagocytic activity. However, exposure of healthy control blood to BRAF-mutated genomic DNA did not result in a cell-associated signal. Furthermore, the lack of mutated alleles in neutrophils, but the localization of mutation to nonphagocytic CD34+ progenitor cells, argues against phagocytic uptake of exogenous DNA as a significant source of artifact.
Total RNA profiling reveals a strong DC-related and macrophage-related signature in LCH and ECD, respectively.25 However, there was no bias in the distribution of mutated alleles to suggest that the phenotypic differences of LCH and ECD lesions are determined by the myeloid lineages harboring BRAF mutation. This may not be surprising given the relatively high incidence of overlap cases now documented.9,10 Many alternatives may explain the variation between LCH and ECD, such as genetic background, age-related myeloid hematopoietic bias,34 additional somatic mutation,35 or extrinsic inflammatory signals arising independently in the local tissue environment. The difference in median age of onset between LCH and ECD is consistent with an influence of age-related myeloid bias or clonal hematopoiesis. From a pathological perspective, the occurrence of LCH and ECD together in a single lesion suggests a cell-intrinsic mechanism.10 However, in cases where an overlap syndrome occurs with distinct and characteristic anatomical distributions of each disease, the tissue environment may direct either LCH- or ECD-like pathology, through extrinsic signals. In keeping with previous reports of expanded myelopoiesis in MS-LCH,36 we observed an increase in blood myeloid cells and BM myeloid progenitors in adult LCH and ECD.
The differentiation of myeloid cells in response to cytokine and notch-mediated signaling showed that >1 precursor has the potential to form cells with characteristic properties of LCH and ECD histiocytes (Figure 6). The critical role of notch signaling in permitting LC development from monocytes was first reported in response to DLL1.22 Without this signal, CD14+ classical monocytes only express low langerin and are unable to form langerinhighCD1a+ cells containing Birbeck granules in response to GM-CSF and TGF-β. In contrast, CD1c+ DCs spontaneously express langerin in vivo16 and rapidly acquire LC-like properties when exposed to GM-CSF and TGF-β.17,18 In vitro differentiation experiments presented here clearly show that 2 pathways may generate langerinhigh cells and that DLL4 provides a more potent signal for monocytes than DLL1. Recent molecular studies have shown that DLL1 induces LC development in monocytes by downregulating KLF and depressing RUNX3.29 Elevated GM-CSF, M-CSF, and FLT3 ligand have been reported in the serum of patients with LCH, consistent with a pathological role of these cytokines.36,37 Intrinsic notch signaling was previously suggested as a key mechanism promoting LCH pathogenesis, through expression of Jagged 2 by lesional histiocytes.28 Taken together, these observations do not support a simple dichotomous model that LCH arises from the DC pathway and ECD from the monocyte-macrophage pathway of myeloid cell development. Classical monocytes appear able to contribute to histiocytes of both disorders, depending on the signals they receive. CD1c+ DCs, on the other hand, were unable to form foamy macrophages when exposed to strongly macrophage-promoting conditions of M-CSF and lipid-containing human serum. Both classical and nonclassical monocytes rapidly differentiated into typical foam cells under these conditions. CD16+ nonclassical monocytes are likely to be heterogeneous and have also been characterized as DC-like.38,39 It is possible that a subpopulation formed the macrophages observed in culture as the final density was lower than for equivalent numbers of CD14+ monocytes. Given the high proportion of BRAF-mutated alleles in CD16+ monocytes, they may contribute to histiocytes in the lesions of ECD, although they are reported to be unable to form giant cells. The occurrence of mutated alleles in monocytes also presents the possibility that macrophages in LCH lesions arise from the neoplastic clone. Previous studies show that M-CSF, one of the factors used to differentiate foam cells, is significantly increased in MS-LCH.36
Figure 6.
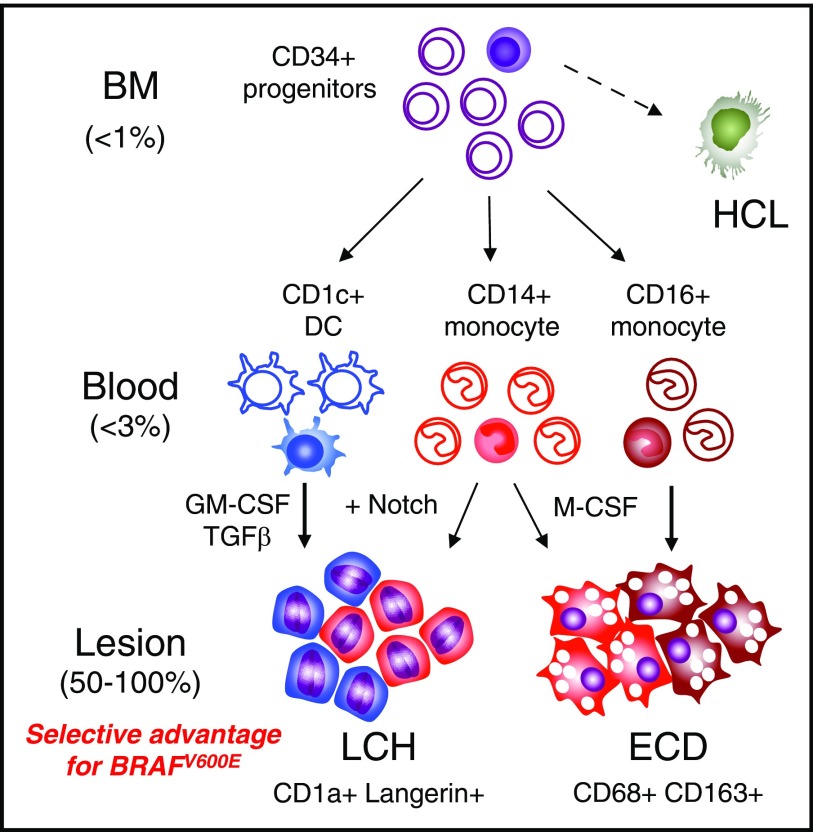
Potential precursor pathways in MS-LCH and ECD. Schema summarizing potential precursor pathways in MS-LCH and ECD based on the distribution of BRAFV600E alleles in peripheral blood and the differentiation potentials of healthy control cells in vitro. A minority of cells (filled) in the BM and blood contain BRAF mutation. The principal observation is that <1% clonal hematopoiesis gives rise to <3% mutated blood precursors that appear to be strongly selected for in peripheral tissue environments, resulting in lesional histiocyte mutation levels of up to 100%. Lesions potentially consist of >1 precursor as indicated by blue for CD1c+ DCs, red for classical monocytes, and brown for nonclassical monocytes. The depiction of lesion composition is conjectural and not based on experimental observation. Furthermore, additional contributions from other, more primitive myeloid progenitors cannot be excluded at present.
An important caveat of the in vitro differentiation experiment is that healthy control cells without BRAF mutation were used. In the absence of a surface antigen that can distinguish mutated and unmutated progeny, this analysis cannot be performed with enriched BRAF-mutated cells isolated from patients. The presence of mutation may not matter because ERK activation is a consequence of many of the stimuli used in vitro. However, it remains hypothetically possible that intrinsic BRAF activation has other unanticipated effects.
The finding that BRAF-mutated PBMCs are detected at higher levels in MS-LCH has potential utility as an indicator of clinical risk and response to therapy, as suggested in children.5 Our conclusion in adults is based on only 8 patients and should be expanded to a larger cohort ideally. The sensitivity of detection using allele-specific PCR is critical as less sensitive techniques failed to find BRAF mutation in peripheral blood or BM fractions previously.2,4 BRAFV600E was detected in PBMCs of a subset of ECD patients, and it remains to be determined, using a large group, whether this segregates with disease risk in ECD.
Acknowledgments
The authors are grateful to Andrew Filby and the Newcastle University Flow Cytometry Core Facility for assistance with the generation of flow cytometry data, and Alex Laude and the Bioimaging core for assistance with microscopy.
This work was supported by a grant from Cancer Research UK (C30484/A21025) and by the Histiocytosis Association and Bright Red. V.B. is supported by a Wellcome Trust Intermediate Clinical Fellowship (101155/Z/13/Z); M.H. is supported by a Wellcome Trust Senior Clinical Fellowship (WT088555MA); S.B. was supported by a National Institute for Health Research Clinical Lectureship; B.H.D. is supported by the American Society of Hematology Senior Research Training Award for Fellows and the New York State Council on Graduate Medical Education Empire Clinical Research Investigator Program Fellowship; and E.L.D. and O.A.-W. are supported by grants from the Erdheim-Chester Disease Global Alliance and the Histiocytosis Association.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: P.M. designed research, performed experiments, analyzed data, and wrote the manuscript: V.B. designed research, analyzed data, and contributed clinical data and patient material; C.M.B. designed research, performed experiments, and analyzed data; A.N. contributed clinical data and patient material; N.M. performed experiments and analyzed data; S.B. contributed clinical data and patient material; M.H. designed research and contributed clinical data and patient material; E.L.D., B.H.D., J.V., D.H., H.G., and M.M. contributed clinical data and patient material; K.L.M. and C.A. designed research; O.A.-W. designed research and contributed clinical data and patient material; and M.C. supervised the study, designed research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Matthew Collin, Human Dendritic Cell Laboratory, Institute of Cellular Medicine, Newcastle University, Framlington Pl, Newcastle upon Tyne NE2 4HH, United Kingdom; e-mail: matthew.collin@ncl.ac.uk.
References
- 1.Badalian-Very G, Vergilio JA, Degar BA, et al. . Recurrent BRAF mutations in Langerhans cell histiocytosis. Blood. 2010;116(11):1919-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sahm F, Capper D, Preusser M, et al. . BRAFV600E mutant protein is expressed in cells of variable maturation in Langerhans cell histiocytosis. Blood. 2012;120(12):e28-e34. [DOI] [PubMed] [Google Scholar]
- 3.Haroche J, Charlotte F, Arnaud L, et al. . High prevalence of BRAF V600E mutations in Erdheim-Chester disease but not in other non-Langerhans cell histiocytoses. Blood. 2012;120(13):2700-2703. [DOI] [PubMed] [Google Scholar]
- 4.Satoh T, Smith A, Sarde A, et al. . B-RAF mutant alleles associated with Langerhans cell histiocytosis, a granulomatous pediatric disease[published correction appears in PLoS One. 2012;7(6). doi:10.1371/annotation/74a67f4e-a536-4b3f-a350-9a4c1e6bebbd]. PLoS One. 2012;7(4):e33891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berres ML, Lim KP, Peters T, et al. . BRAF-V600E expression in precursor versus differentiated dendritic cells defines clinically distinct LCH risk groups[published correction appears in J. Exp. Med 2015;212(2):281. J Exp Med. 2014;211(4):669-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aricò M, Girschikofsky M, Généreau T, et al. . Langerhans cell histiocytosis in adults. Report from the International Registry of the Histiocyte Society. Eur J Cancer. 2003;39(16):2341-2348. [DOI] [PubMed] [Google Scholar]
- 7.Teng CL, Lin TH, Young JH, Chou G, Young CS. Rapidly fatal Langerhans’ cell histiocytosis in an adult. J Formos Med Assoc. 2005;104(12):955-959. [PubMed] [Google Scholar]
- 8.Girschikofsky M, Arico M, Castillo D, et al. . Management of adult patients with Langerhans cell histiocytosis: recommendations from an expert panel on behalf of Euro-Histio-Net. Orphanet J Rare Dis. 2013;8:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin J, Zhang F, Zhang H, et al. . Hand-Schüller-Christian disease and Erdheim-Chester disease: coexistence and discrepancy. Oncologist. 2013;18(1):19-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hervier B, Haroche J, Arnaud L, et al. ; French Histiocytoses Study Group. Association of both Langerhans cell histiocytosis and Erdheim-Chester disease linked to the BRAFV600E mutation. Blood. 2014;124(7):1119-1126. [DOI] [PubMed] [Google Scholar]
- 11.Diamond EL, Dagna L, Hyman DM, et al. . Consensus guidelines for the diagnosis and clinical management of Erdheim-Chester disease. Blood. 2014;124(4):483-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haroche J, Arnaud L, Cohen-Aubart F, et al. . Erdheim-Chester disease. Curr Rheumatol Rep. 2014;16(4):412. [DOI] [PubMed] [Google Scholar]
- 13.Collin M, Bigley V, McClain KL, Allen CE. Cell(s) of origin of Langerhans cell histiocytosis. Hematol Oncol Clin North Am. 2015;29(5):825-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ziegler-Heitbrock L, Ancuta P, Crowe S, et al. . Nomenclature of monocytes and dendritic cells in blood. Blood. 2010;116(16):e74-e80. [DOI] [PubMed] [Google Scholar]
- 15.Collin M, McGovern N, Haniffa M. Human dendritic cell subsets. Immunology. 2013;140(1):22-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bigley V, McGovern N, Milne P, et al. . Langerin-expressing dendritic cells in human tissues are related to CD1c+ dendritic cells and distinct from Langerhans cells and CD141high XCR1+ dendritic cells. J Leukoc Biol. 2015;97(4):627-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martínez-Cingolani C, Grandclaudon M, Jeanmougin M, Jouve M, Zollinger R, Soumelis V. Human blood BDCA-1 dendritic cells differentiate into Langerhans-like cells with thymic stromal lymphopoietin and TGF-β. Blood. 2014;124(15):2411-2420. [DOI] [PubMed] [Google Scholar]
- 18.Milne P, Bigley V, Gunawan M, Haniffa M, Collin M. CD1c+ blood dendritic cells have Langerhans cell potential. Blood. 2015;125(3):470-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manh TP, Alexandre Y, Baranek T, Crozat K, Dalod M. Plasmacytoid, conventional, and monocyte-derived dendritic cells undergo a profound and convergent genetic reprogramming during their maturation. Eur J Immunol. 2013;43(7):1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chomarat P, Banchereau J, Davoust J, Palucka AK. IL-6 switches the differentiation of monocytes from dendritic cells to macrophages. Nat Immunol. 2000;1(6):510-514. [DOI] [PubMed] [Google Scholar]
- 21.Xue J, Schmidt SV, Sander J, et al. . Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 2014;40(2):274-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoshino N, Katayama N, Shibasaki T, et al. . A novel role for Notch ligand Delta-1 as a regulator of human Langerhans cell development from blood monocytes. J Leukoc Biol. 2005;78(4):921-929. [DOI] [PubMed] [Google Scholar]
- 23.Emile JF, Diamond EL, Hélias-Rodzewicz Z, et al. . Recurrent RAS and PIK3CA mutations in Erdheim-Chester disease. Blood. 2014;124(19):3016-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cangi MG, Biavasco R, Cavalli G, et al. . BRAFV600E-mutation is invariably present and associated to oncogene-induced senescence in Erdheim-Chester disease. Ann Rheum Dis. 2015;74(8):1596-1602. [DOI] [PubMed] [Google Scholar]
- 25.Diamond EL, Durham BH, Haroche J, et al. . Diverse and targetable kinase alterations drive histiocytic neoplasms. Cancer Discov. 2016;6(2):154-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chung SS, Kim E, Park JH, et al. . Hematopoietic stem cell origin of BRAFV600E mutations in hairy cell leukemia. Sci Transl Med. 2014;6(238):238ra71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Durham BH, Roos-Weil D, Baillou C, et al. . Functional evidence for derivation of systemic histiocytic neoplasms from hematopoietic stem/progenitor cells. Blood. 2017;130(2):176-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hutter C, Kauer M, Simonitsch-Klupp I, et al. . Notch is active in Langerhans cell histiocytosis and confers pathognomonic features on dendritic cells. Blood. 2012;120(26):5199-5208. [DOI] [PubMed] [Google Scholar]
- 29.Jurkin J, Krump C, Köffel R, et al. . Human skin dendritic cell fate is differentially regulated by the monocyte identity factor KLF4 during steady state and inflammation [published online ahead of print 11 October 2016]. J Allergy Clin Immunol. doi: 10.1016/j.jaci.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Willman CL, Busque L, Griffith BB, et al. . Langerhans’-cell histiocytosis (histiocytosis X)--a clonal proliferative disease. N Engl J Med. 1994;331(3):154-160. [DOI] [PubMed] [Google Scholar]
- 31.Chetritt J, Paradis V, Dargere D, et al. . Chester-Erdheim disease: a neoplastic disorder. Hum Pathol. 1999;30(9):1093-1096. [DOI] [PubMed] [Google Scholar]
- 32.Emile JF, Abla O, Fraitag S, et al. ; Histiocyte Society. Revised classification of histiocytoses and neoplasms of the macrophage-dendritic cell lineages. Blood. 2016;127(22):2672-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janku F, Vibat CR, Kosco K, et al. . BRAF V600E mutations in urine and plasma cell-free DNA from patients with Erdheim-Chester disease. Oncotarget. 2014;5(11):3607-3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geiger H, de Haan G, Florian MC. The ageing haematopoietic stem cell compartment. Nat Rev Immunol. 2013;13(5):376-389. [DOI] [PubMed] [Google Scholar]
- 35.Jaiswal S, Fontanillas P, Flannick J, et al. . Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rolland A, Guyon L, Gill M, et al. . Increased blood myeloid dendritic cells and dendritic cell-poietins in Langerhans cell histiocytosis. J Immunol. 2005;174(5):3067-3071. [DOI] [PubMed] [Google Scholar]
- 37.Emile JF, Tartour E, Brugières L, et al. . Detection of GM-CSF in the sera of children with Langerhans’ cell histiocytosis. Pediatr Allergy Immunol. 1994;5(3):162-163. [DOI] [PubMed] [Google Scholar]
- 38.MacDonald KP, Munster DJ, Clark GJ, Dzionek A, Schmitz J, Hart DN. Characterization of human blood dendritic cell subsets. Blood. 2002;100(13):4512-4520. [DOI] [PubMed] [Google Scholar]
- 39.Piccioli D, Tavarini S, Borgogni E, et al. . Functional specialization of human circulating CD16 and CD1c myeloid dendritic-cell subsets. Blood. 2007;109(12):5371-5379. [DOI] [PubMed] [Google Scholar]