Abstract
In multiple myeloma (MM) osteolytic lesions rarely heal because of persistent suppressed osteoblast differentiation resulting in a high fracture risk. Herein, chromatin immunoprecipitation analyses reveal that MM cells induce repressive epigenetic histone changes at the Runx2 locus that prevent osteoblast differentiation. The most pronounced MM-induced changes were at the Runx2-P1 promoter, converting it from a poised bivalent state to a repressed state. Previously it was observed that MM induce the transcription repressor GFI1 in osteoblast precursors, which correlates with decreased Runx2 expression. Thus, prompting detailed characterization of the MM and TNFα-dependent GFI1-response element within the Runx2-P1 promoter. Further analyses reveal that MM-induced GFI1 binding to Runx2 in osteoblast precursors and recruitment of the histone modifiers HDAC1, LSD1, and EZH2 is required to establish and maintain Runx2 repression in osteogenic conditions. These GFI1-mediated repressive chromatin changes persist even after removal of MM. Ectopic GFI1 is sufficient to bind to Runx2, recruit HDAC1 and EZH2, increase H3K27me3 on the gene, and prevent osteogenic induction of endogenous Runx2 expression. Gfi1 knockdown in MC4 cells blocked MM-induced recruitment of HDAC1 and EZH2 to Runx2, acquisition of repressive chromatin architecture, and suppression of OB differentiation. Importantly, inhibition of EZH2 or HDAC1 activity in pre-osteoblasts after MM exposure in vitro or in osteoblast precursors from MM patients reversed the repressive chromatin architecture at Runx2 and rescued osteoblast differentiation.
Implications
This study suggests that therapeutically targeting EZH2 or HDAC1 activity may reverse the profound MM-induced osteoblast suppression and allow repair of the lytic lesions.
Keywords: Multiple myeloma, osteoblast precursors, chromatin, epigenetics, Gfi1, Runx2
Introduction
Multiple myeloma (MM), a malignant plasma cell disorder, is the most frequent cancer to involve bone (1). Over 80% of MM patients develop bone lesions that can result in severe pain and frequent pathological fractures (2), a major contributor to patient morbidity and mortality (3). MM bone disease is characterized by increased osteolytic bone destruction with little or no new bone formation due to persistent MM-induced suppression of bone marrow stromal cell (BMSC) differentiation into bone-forming osteoblasts (OB) (4, 5). This results in lesions that rarely heal, even when patients are in long-term remission. Further, BMSC from MM patients (MM-BMSC) or mouse MM models, and healthy donor BMSC (HD-BMSC) and pre-OB cell lines exposed to MM cells in culture, demonstrate decreased OB differentiation even after removal of the MM cells and extended culture (6). This protracted selective suppression of OB differentiation suggests that MM cells induce a persistent, cell-autonomous change in MM-BMSC. MM-derived TNFα, CCL3, IL3/activin A, Dickkopf1, sclerostin, TGFβ, HGF, and IL7, as well as direct contact, contribute to OB suppression (4, 7), but the mechanisms responsible for the sustained cell-autonomous blockade of OB differentiation in the MM-BMSC are not well understood. MM altered BMSC also support MM cell adhesion, growth, and chemoresistance via increased levels of adhesion molecules, chemokines, and cytokines, and express an altered RANKL(TNFSF11)/osteoprotegerin ratio to favor osteoclastogenesis (8–12).
OB differentiation requires upregulation and activation of the critical transcription factor RUNX2/CBFA1/AML3 (RUNX2) (13). We (6), and others (14), have shown that RUNX2 activity in OB precursors is inhibited in MM, but the mechanism is unclear. Our previous studies of MM-exposed BMSC revealed that Runx2 gene repression was correlated with elevated expression of growth factor independence 1 (GFI1), a transcription repressor (6). We found that BMSC isolated from Gfi1−/− mice were significantly resistant to MM-induced suppression of Runx2. Further, siRNA Gfi1 knockdown in MM-BMSCs restored expression of RUNX2 and OB differentiation markers osteocalcin (OCN, BGLAP) and bone sialoprotein (BSP, IBSP). These studies suggested that GFI1 could be a novel therapeutic target for MM bone disease. However, therapeutic targeting of transcription factors is difficult and GFI1 is a large multi-functional protein with multiple modes of action.
GFI1, a 55 kD zinc-finger containing member of the Snail/Gfi1 transcription repressor family that includes GFI1b, SNAIL (SNAI1), SLUG (SNAI2), IA-1 (INSM1), and MLT1 (ISMN2) (15, 16), has diverse biological functions and mechanisms of action, and regulates various aspects of normal and malignant hematopoiesis as well as inner ear development (17, 18). The 422 aa human (423 aa murine) GFI1 contains an N-terminal SNAG domain, an unstructured intermediate domain, and 6 C-terminal C2-H2 Zn finger domains, of which Zn fingers 3–5 are required for sequence-specific DNA binding to a recognition sequence containing the “AA(T/G)C” core motif (15, 19). GFI1 interacts with various chromatin modifiers to mediate epigenetic repression of target genes. The GFI1 SNAG domain is critical in recruiting lysine specific demethylase 1 (LSD1, KDM1A) with the REST corepressor (CoREST, RCOR1) to target genes regulating hematopoiesis (20). GFI1 recruitment of histone methyltransferase G9a (EHMT2) and histone deacetylase 1 (HDAC1) through the intermediate domain represses the promoter of cell cycle regulator CDKN1A (21). GFI1 can also repress gene expression independently of its DNA binding capability, as shown by its binding to and cooperation with the POZ-ZF transcription factor MIZ-1 (ZBTB17) at the CDKN1A and CDKN2B gene promoters (11, 22). Additionally, GFI1 binding to other transcription factors can interfere with their DNA binding or transactivation properties, thereby repressing their targets without GFI1 DNA binding. For instance, GFI1 can antagonize binding of RELA to its target genes in LPS-stimulated macrophages (23), as well as inhibit PU.1 (SPI1)-dependent gene transcription during granulocyte development (24). Conversely, GFI1 enhances STAT3-mediated gene transactivation by interacting with and sequestering a STAT3-negative regulator PIAS3 (25). GFI1 also regulates gene expression of the T-cell receptor CD45 (PTPRC) at the level of alternative splicing by interacting with the splicing factor U2AF26 (U2AFIL4) (26). Thus, further study was necessary to understand how GFI1 influenced Runx2 expression.
In the current study, we determined if MM cells induce GFI1-mediated epigenetic changes in the chromatin architecture of the Runx2 locus in OB precursors. We identified the chromatin modifiers recruited by GFI1 and explored if inhibition of these enzymatic activities could induce reversal of the persistent suppression of BMSC to osteogenic differentiation, making them potential actionable therapeutic targets to improve bone health in MM patients.
Materials And Methods
Reagents
Reagents used in this study can be found in Supplementary Methods.
Cells and co-culture
All cultures described below contained 10% FCS-1% pen/strep. The pre-OB murine cell line MC3T3-E1 subclone-4 (MC4) was obtained from Dr. Guozhi Xiao (27, 28) in 2009 and subclone-14 (MC14) was obtained from ATCC (CRL-2594) in 2014. Both were maintained in ascorbic acid-free αMEM proliferation media. Murine 5TGM1-GFP-TK (5TGM1) MM cells (6) and human MM1.S-GFP cells (11) were maintained in RPMI1640. Cell lines were authenticated by morphology, gene expression profile, and tumorigenic capacity (MM cells). MC4 cells were grown to 90% confluency prior to co-culture. Direct 5TGM1-MC4 (10:1) co-cultures and indirect co-cultures of MM1.S cells in transwells (10:1) with MC14 cells were carried out in 50:50 RPMI1640/αMEM proliferation media. MM1.S in transwells (Corning Inc., 3450) or 5TGM1 cells were carefully removed (FACS analysis demonstrated that ≤ 1% 5TGM1 cells remained). The MC4 and MC14 cells were isolated immediately or subjected to OB differentiation first. Scrambled control (SHC002, Sigma) and mouse Gfi1 shRNA (Sigma, TRCN0000096706, 5’-CCTCATCACTCATAGCAGAAA-3’) in pLKO.1-puro lentiviruses were generated by the UPCI lentivirus core facility and used to stably transduce (with polybrene) MC4 cells, which were selected and maintained using puromycin (2.5 μg/mL).
Human samples and primary BMSC cultures
BM aspirates and MM bone resections were collected in heparin from 15 healthy donors and 29 MM patients. Human studies were approved by the University of Pittsburgh and Indiana University IRBs. Samples were collected from participants after obtaining written informed consent in accordance with the Declaration of Helsinki. BM mononuclear cells were separated by Ficoll-Hypaque density sedimentation and the nonadherent cells removed after overnight incubation in IMDM-10%FCS. The adherent cultures were then continued for 21 d with media changes every 4 d to obtain BMSC. Subconfluent cells were detached with trypsin and replated (105 cells/10-cm dish) for use at passage 2 and 3.
OB differentiation, and alkaline phosphatase and alizarin red assays
OB differentiation media (αMEM supplemented with 50 μg/ml ascorbic acid and 10 mM β-glycerophosphate; for human cells 10 nM Dex was also added) was added to primary BMSC or MC4 cells with or without prior MM exposure; media was changed every 3 d. Mineralization at times indicated was assessed using alizarin red staining (6). The staining density quantitation was carried out using a ProteinSimple FluorChem™ M imaging system.
DNAs
Construction of the -974/+111 mRunx2 P1 promoter-pGL4.10[luc2] reporters containing either wildtype, Δ-37/-7, or the GFI1 site mutations (L mutant GGGCTT, R mutant AAGCCC, and LR mutant GGGCCC) and generation of the expression vectors encoding Myc-tagged mGFI1-1-423 aa, -1-380 aa or -239-423 aa (in pCS2-MT) from mGFI1-wt-pCDNA3.1 are detailed in the Supplementary Methods. All constructs were verified by DNA sequencing.
Transfection of Runx2 P1 promoter-Luc reporters and GFI1 constructs
The mRunx2 P1 promoter-reporters and pRL-TK plasmids (Promega) were transfected into MC4 cells with Lipofectamine2000, along with empty (EV) or wt mGFI1 expression vectors, or treated with TNFα as indicated in Figure legends. Luc and renilla activities were measured in supernatants from lysed cells (48 h) using the Dual-Luciferase® Reporter Assay System (Promega). The normalized (to renilla) relative Luc activities for each reporter construct were calculated as a percent of the activity of the -974/+111 mRunx2-pGL4.10[luc2]-wt co-transfected with EV. Transfections of Myc-mGFI1-wt and Myc-mGFI1-deletions into MC4 for endogenous Runx2 mRNA and ChIP analyses were carried using FugeneHD (E2311, Promega). See Supplementary methods for more details.
Protein lysates and Western blotting
Transfected MC4 cell cultures were treated with 1X lysis buffer (Cell Signaling) to make whole cell lysates, which were examined by western blotting with primary antibodies as indicated. The membranes were then incubated with secondary chemiluminescent antibodies and imaged using a ProteinSimple FluorChem™ M imaging system. Quantitation of protein band densities was performed using the alpha view analysis software package.
Real-time quantitative PCR (qPCR) RNA expression analyses
MC4 RNA was isolated using TRIzol reagent and converted to cDNA using First-Strand cDNA Synthesis System (Life Technologies, 11904-018). qPCR was carried out using 2x Maxima SYBR Green/ROX qPCR Master Mix (K0223, Thermo Fisher) in Fast 96-Well Reaction Plates (Applied Biosystems) using a StepOnePlus (Applied Biosystems). Relative mRNA levels were calculated using the ΔΔCt method using 18SrRNA for normalization. The qPCR primers are listed in Table S1.
ChIP assay
Chromatin from MC4 cells, MM-BMSC, and HD-BMSC was analyzed using a modification of the ChIP Millipore/Upstate protocol (MCPROTO407) as described (29) using Magna ChIP Protein A+G Beads (16-663, Millipore). In brief, a total of 2x107 cells were fixed in 1% formaldehyde (F79-500, Fisher) for 10 min at room temperature. Samples were sonicated (to generate DNA fragments of 250 base pairs (bp) average length) on ice using a Fisher Scientific Sonic Dismembrator (Model 100) and centrifuged at 12000 RPM for 10 min. Chromatin from 4x106 cells was diluted 7-fold in ChIP Dilution Buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH8.1, 167 mM NaCl) and incubated at 4°C overnight with respective antibodies. Aliquots for input and non-specific IgG control samples were included with each experiment. IgG ChIP was run on untreated MC4 samples. ChIP-qPCR primers are listed in Table S2. Fold enrichment was calculated based on Ct as 2(ΔCt), where ΔCt = (CtInput –CtIP). The IgG ΔCt Ct was subtracted from the specific Ab ΔCt to generate ΔΔCt = (ΔCtspecific Ab − ΔCtIgG).
Statistical Analysis
All experiments were repeated at least two independent times. Most data is presented as biological triplicates and results reported as means±SD unless otherwise stated. Statistical significance was evaluated by either the Student’s t test or one-way ANOVA with Tukey’s multiple comparison post-test using Graphpad Prism 6 as indicated. Degree of significance is represented using ρ values: *= ρ≤0.05, **= ρ≤0.01, ***= ρ≤0.001, ****= ρ≤0.0001 (Different symbols may be used to reflect multiple two-way comparisons).
Results
MM induces sustained transcriptional and epigenetic suppression of the mRunx2 promoter in murine pre-OB cells
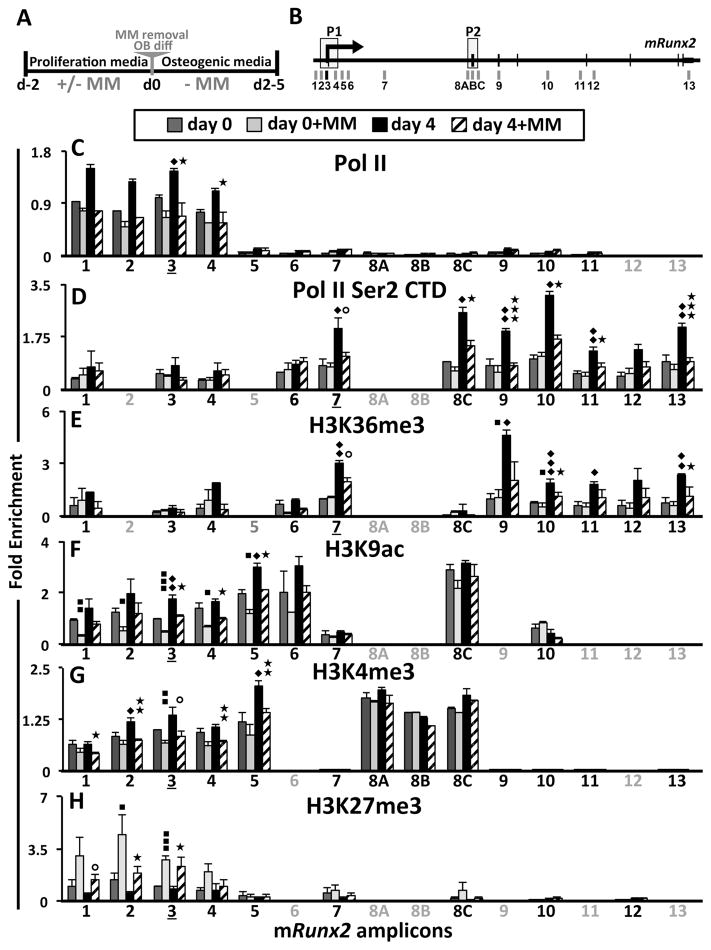
We (Supplementary Fig. S1), and others (30) demonstrated that MM cells and TNFα cause a very rapid decrease of Runx2 mRNA mediated by decreasing Runx2 mRNA half-life. However, since maintenance and propagation of gene silencing is often controlled at the chromatin level, we hypothesized that the long-term suppression of OB differentiation in the MM microenvironment results from epigenetic repression of Runx2 transcription in BMSC. Therefore, we analyzed the effect of 5TGM1-MM cell exposure on RNA Polymerase II (Pol II) occupancy and the histone H3 methylation and acetylation profiles along the murine (m)Runx2 locus during MC3T3-E1 subclone-4 (MC4) cell proliferation and OB differentiation (Fig. 1A) using ChIP-qPCR (Fig. 1B) amplicons as indicated. We found that MC4 exposure to 5TGM1 inhibited the OB-induced recruitment of Pol II to the mRunx2-P1 promoter (Fig. 1C, amplicons 3 and 4), as well as decreased elongating Pol II (marked by Ser2 phosphorylation of the C-terminal domain; Ser2P CTD) downstream of the mRunx2-P1 promoter (Fig. 1D), thus demonstrating that MM exposure down-regulates mRunx2 transcription in MC4 cells. Paused Pol II was not evident at the mRunx2-P2 promoter (Fig. 1C, amplicons 8ABC), but transiting Pol II there was elevated by OB differentiation and decreased by MM exposure (Fig. 1D). Further evidence of prior MM exposure leading to inhibition of mRunx2 transcription during OB induction is revealed by decreased enrichment of tri-methylated H3K36 (H3K36me3) towards the 3’ end of the mRunx2 gene (Fig. 1E), which marks the Pol II elongation footprint (31). However, MM-exposure did not affect presence of Pol II, Ser2P CTD, and H3K36me3 at mRunx2 in proliferating MC4 (Fig. 1C–E). The permissive chromatin marks, acetylation at H3K9 (H3K9ac) (Fig. 1F) and methylation at H3K4 (H3K4me3) (Fig. 1G), were abundant at both mRunx2 promoters prior to OB stimulus, reflecting the poised and basal/constitutive transcription levels in MC4 cells. These marks increased following differentiation (more so at P1 than at P2), consistent with increased mRunx2 activation. MM-exposure significantly reduced the H3K9ac and H3K4me3 levels at mRunx2-P1 in proliferating MC4 (d0) and they were refractory to elevation by OB differentiation induction (d4). In contrast, MM cells upregulated mIl6 mRNA in proliferating MC4, with increased Pol II occupancy and H3K9ac, H3K4me3, and H3K36me3 levels at the mIl6 gene (Supplementary Fig. S2). There is more of the repressive H3K27 tri-methylation (H3K27me3) (32) mark on the mRunx2-P1 promoter than mRunx2-P2 in proliferating cells (Fig. 1H), reflecting the bivalent nature of the poised P1 promoter. Furthermore, MM increased H3K27me3 only at the mRunx2-P1 promoter in MC4 (Fig. 1H), which remained elevated four days after MM cell removal. These data indicate that MM exposure reduced the transcriptionally permissive bivalent chromatin architecture of the mRunx2-P1 promoter in MC4 cells, marked by high H3K9ac and H3K4me3 levels along with H3K27me3, and induced a more repressive H3K27me3-prevalent signature.
Figure 1. Transcriptional and epigenetic changes at mRunx2 in MM-exposed MC4.
(A) Experimental design schematic of 5TGM1 MM-MC4 co-cultures and induction of OB differentiation. After 48 h co-culture in proliferation media, the MM cells were removed, and the MC4 were either harvested immediately (d0±MM) or first placed in OB differentiation media for 4 days (d4±MM). (B) Schematic of mRunx2 qPCR amplicons with promoters P1 and P2 indicated (see Table S2 for positional numbering and the primer sequences). Amplicon-3 encompasses the Gfi1 binding site. (C–H) ChIP-qPCR analyses of RNA Pol II occupancy and several H3 modifications along mRunx2 in MC4 cells treated as described in A using qPCR amplicons denoted in B (amplicons not done for a particular pull-down are in gray). Enrichment values are plotted relative to amplicons 3 or 7 as indicated by underlining, depending upon whether the focus was on the promoter (C, F–H) or the body of the gene (D, E): (C) total RNA Pol II; (D) phosphorylated Pol II CTD Ser 2P; (E) elongation mark H3K36me3; (F) activation mark H3K9ac; (G) activation mark H3K4me3; and (H) repressive mark H3K27me3. Error bars represent SEM of 3–4 biological replicates (2 replicates for H3K9ac d4±MM). Statistically significant comparisons of: ◆;d4-MM to d0-MM, ■; d0+MM to d0-MM; ★; d4+MM to d4-MM. ⭘ – represents values of p<0.08.
Myeloma induces recruitment of GFI1 to the mRunx2 promoter in pre-OB
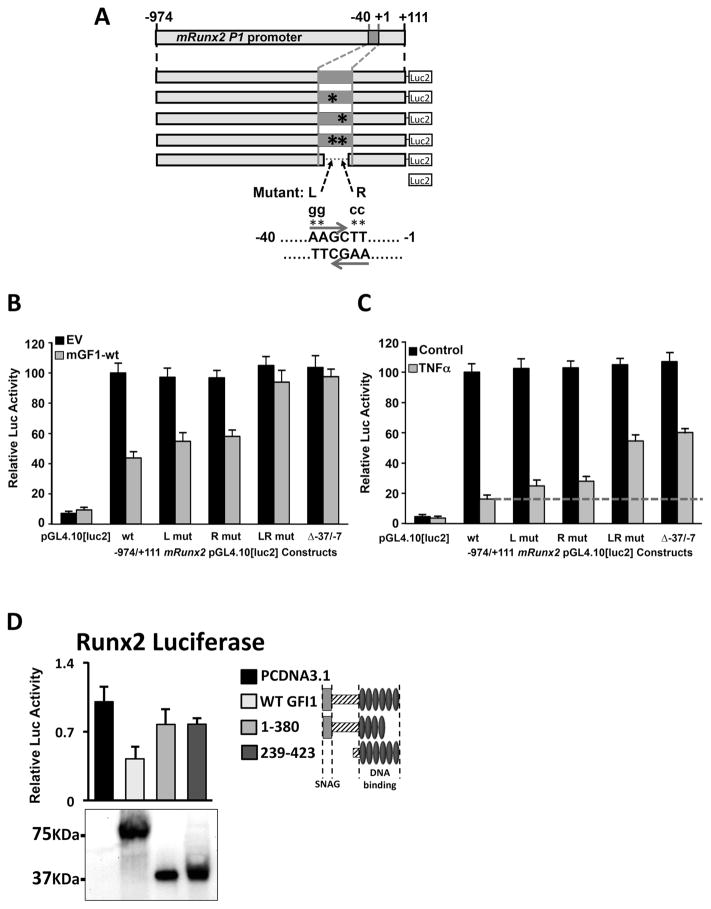
Since we had shown an inverse correlation with GFI1 levels and Runx2 expression (6), we postulated that GFI1 is directly responsible for the MM-induced epigenetic changes by binding at the Runx2 gene and recruiting various co-repressors to establish epigenetic silencing. Therefore, we first needed to establish if GFI1 binds the Runx2 gene. Using Gfi1-WT cotransfections with a set of 5’ and 3’ deletions, as well as internal deletions, of mRunx2-pGL4.10[luc2] reporters, we localized the GFI1-responsivity to the -37/-7 region (Supplementary Fig. S3A,B). There is no consensus GFI1 binding site (15, 19) in the -108/-1 mRunx2 promoter, but the region contains 6 GFI1 binding site cores (AA(T/G)C). Therefore, we used a combination of biotin-oligo (B-oligo) streptavidin agarose bead pull-down assays (Supplementary Fig. S3C–G) and EMSA (Supplementary Fig. S3H,I) to establish that GFI1 binds at an overlapped palindromic pair of GFI1 cores at -21/-18 (L) and -19/-16 (R). Mutation of either core decreased GFI1 binding, but mutation of both (LR) ablated binding (Supplementary Fig. S3G,H). GFI1 cotransfected into MC4 with -974/+111 mRunx2-pGL4.10[luc2] reporters containing site-specific mutations (L, R, and LR) of the -21/-16 double core GFI1 binding site (Fig. 2A) showed that the two single site mRunx2 mutants (L, R) were partially resistant to GFI1, and the double LR mutant and the Δ-37/-7 mRunx2 deletion were entirely resistant (Fig. 2B). Similar results with this set of mRunx2 reporters were obtained using TNFα treatment to repress mRunx2 (Fig. 2C), although the rescue from TNFα repression is only about 60% with LR or Δ-37/-7. This may indicate that a weaker GFI1 binding site at -67/-64 may also play a role in TNFα repression of Runx2 or that another factor is involved. Western blots analysis of the expression of transfected GFI1 protein deletions in HEK293 cells established that the mutant myc-mGFI1 proteins were all expressed as well or better than mGFI1-WT (Input), and -40/-1 B-oligo pulldowns using these extracts demonstrated that only mGFI1-WT and mGFI1;239-423 bound DNA (Supplementary Fig. S4A). Co-transfection of mGFI1-WT, mGFI1;239-423 (lacking recruitment domains for many corepressors) and mGFI1;1-380 (lacking the C-terminal 43aa and doesn’t bind DNA) expression plasmids with the -974/+111 mRunx2-pGL4.10[luc2] reporter revealed that neither mutant mGFI1 could repress reporter expression although they were expressed at similar levels as mGFI1-WT (Fig. 2D).
Figure 2. Mutation of the GFI1 cores at -21/-16 mRunx2 relieves ectopic GFI1 and TNFα repression of the Runx2 promoter.
Reporters -974/+111 mRunx2 promoter-pGL4.10[luc2] WT or containing mutations L, R, or LR or the internal deletion Δ-37/-7 (depicted in A) were transfected into MC4 cells either (B) with pcDNA3.1 (EV) and pcDNA3.1-mGFI1-WT plasmids or (C) that were treated with nothing (Control) or TNFα (0.5 ng/ml) 6 h after transfection. (B, C) Reported luciferase activities in harvested (48 h) cell lysates were evaluated with respect to WT reporter either (B) cotransfected with EV or (C) the untreated control. (D) Myc-mGFI1-WT, deletion constructs which encode mGfi1 aa 1-380 or 239-423, and EV were cotransfected into MC4 cells with mRunx2-Luc-WT reporter depicted in A, and harvested lysates were analyzed for luciferase activities as compared to cells transfected with EV and myc-GFI1 expression by Western Blot (shown below graph). Each experiment above was repeated at least three independent times.
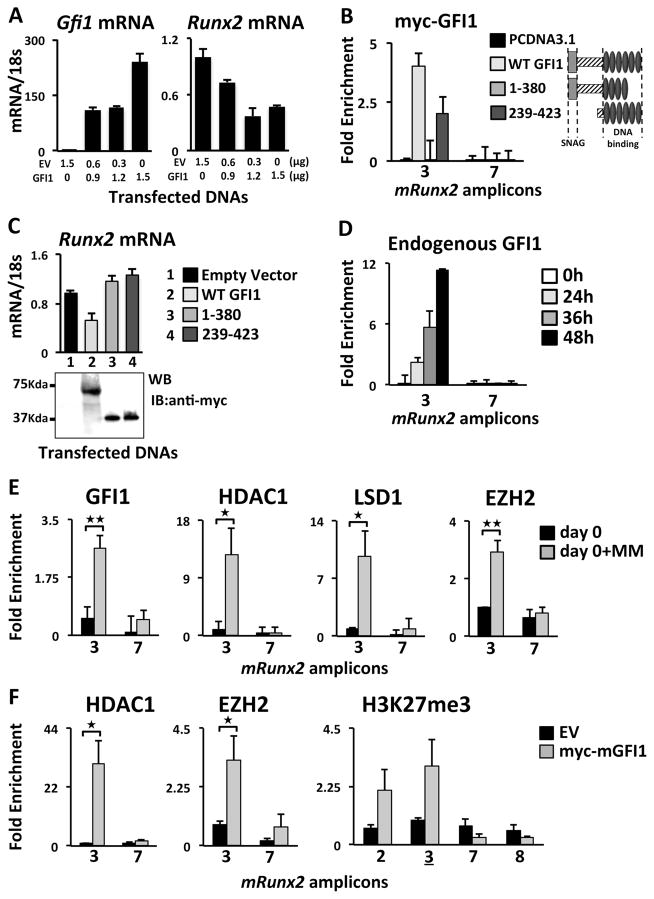
Consistent with the reporter experiments, we observed that ectopic mGFI1 dose-dependently decreased endogenous mRunx2 mRNA in proliferating undifferentiated MC4 cells (Fig. 3A), indicating that increased GFI1 was sufficient for endogenous mRunx2 repression. The increased GFI1 did not alter expression of Sp1, Il6 or the RUNX2 targets Osx (Sp7), Ocn and Bsp (Supplementary Fig. 4B); the latter because these genes were not yet stimulated. We analyzed the capacity of ectopic mGFI1-WT and mGFI1-deletions (1-380 and 239-423) to bind (Fig. 3B) and regulate endogenous mRunx2 expression (Fig. 3C). ChIP-qPCR analysis demonstrated ectopic mGFI1-WT and mGFI1;239-423 occupancy on the endogenous mRunx2 promoter in MC4 cells using amplicon-3 (centered on -36) that included the -21/-16 GFI1 sites, whereas mGFI1;1-380 did not bind (Fig. 3B). Further, mGFI1-WT repressed endogenous mRunx2 expression; while neither mGFI1;1-380 nor mGFI1;239-423 was able to repress mRunx2 expression (Fig. 3C). A ChIP-qPCR scan for ectopic GFI1-WT binding along the Runx2 gene showed that it did not bind near the Runx2-P2 promoter (Supplementary Fig. 4C). Kinetic ChIP-qPCR analyses of MM-exposed MC4 cells revealed that endogenous GFI1 recruitment to mRunx2-P1 is not detectable until 36-h of MM treatment with increased occupancy at 48-h (Fig. 3D).
Figure 3. Analysis of recruitment to mRunx2 of histone modifier enzymes in MM-exposed or GFI1-transfected MC4.
(A) Varying amounts of mGFI1-WT and EV plasmids transfected as indicated into MC4 cells and mGfi1 and endogenous mRunx2 mRNA levels were evaluated by qPCR. (B,C) Myc-mGFI1-WT, myc-mGFI1 deletion constructs encoding aa 1-380 or 239-423, or EV were transfected into MC4. Transfected cells were analyzed for (B) myc-mGFI1 binding at the Runx2 promoter amplicon-3 by ChIP-qPCR using anti-myc Ab and (C) the effect on endogenous Runx2 mRNA levels by qPCR with expression of the transfected myc-mGFI1s by Western Blot displayed underneath. (D) ChIP-qPCR analysis of endogenous GFI1 recruitment to the Runx2 promoter amplicon-3 (Fig 1B) in MC4 cells co-cultured with 5TGM1 MM cells for the indicated times. For all, biological triplicates within two separately run experiments were averaged together and the SEM calculated. (E) ChIP-qPCR analyses of MC4 cells after MM-exposure per scheme in Fig. 1A (d0±MM) for GFI1 binding and HDAC1, LSD1, and EZH2 occupancy within the mRunx2 amplicon-3. (F) ChIP-qPCR analyses of ectopic GFI1 recruitment of HDAC1 and EZH2, and consequent enhancement of H3K27me3 at the Runx2 promoter in MC4. Error bars represent SEM for 3 biological replicates except H3K27me3 in F had only two. (B, D–F) Amplicon-7 was used as a negative control for GFI1 binding.
GFI1 recruits chromatin co-repressors to induce epigenetic suppression of the Runx2 promoter in myeloma-exposed pre-OB
The pleiotropic effects of GFI1-targeted epigenetic gene repression are associated with its recruitment of various histone co-repressors (20, 21, 33). Since we demonstrated that MM cells induce recruitment of endogenous GFI1 to the Runx2 gene (Fig. 3D,E) with concomitant epigenetic repression of the Runx2 locus (Fig. 1C–H), we screened for MM-induced occupancy of chromatin modifiers near the GFI1 binding site in mRunx2 in pre-OBs. MM treatment induced HDAC1 and LSD1 (Fig. 3E) binding to the mRunx2 promoter in MC4 cells, which is consistent with the observed decrease in transcription activation marks H3K9ac and H3K4me3 (Fig. 1). Since we detected a significant MM-induced increase in H3K27me3 levels at mRunx2 (Fig. 1H), we used ChIP to confirm MM-induced occupancy of EZH2, the methyltransferase component of the polycomb repressive complex 2 (PRC2) responsible for generating H3K27me3 (34), near the mRunx2 GFI1 binding site (Fig. 3E).
Ectopically expressed Myc-mGFI1-WT in MC4 cells resulted in recruitment of the histone modifiers HDAC1 and EZH2 to the mRunx2 amplicon-3 (Fig. 3F), thus demonstrating that GFI1 is capable of recruiting these co-repressors to mRunx2 in the absence of MM signals. Further, the increased recruitment of endogenous EZH2 resulted in enhanced deposition of the repressive H3K27me3 mark (Fig. 3F).
GFI1 is required for MM-induced recruitment of repressive chromatin modifiers to the Runx2 gene in pre-OB
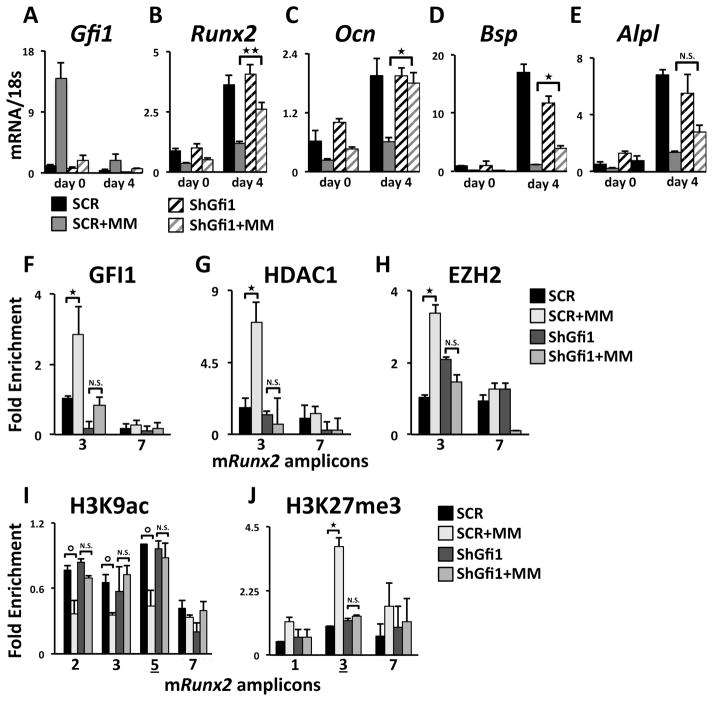
The direct involvement of MM-induced GFI1 recruitment of epigenetic co-repressors was further delineated using a stable Gfi1 knockdown MC4 cell line (shGfi1-MC4) (Fig. 4A), with ~50% reduction in GFI1 protein levels (Supplementary Fig. S4D). 5TGM1-MM co-culture with control shSCR-MC4 resulted in the expected reduction of mRunx2 mRNA expression (Fig. 4B). MM co-culture with shGfi1-MC4 still resulted in a rapid decrease in mRunx2 mRNA (Fig. 4B d0), likely due to mRNA destabilization. However, decreased GFI1 prevented the sustained mRunx2 repression observed following induction of OB differentiation (Fig. 4B d4). Further, RUNX2 target genes mOcn and mBsp also exhibited significant resistance to MM inhibition in shGfi1-MC4 compared to SCR-MC4 (Fig. 4C,D). Alkaline phosphatase (mAlpl) expression trended up, but the change was not significant (Fig. 4E). Consistent with the MM-resistant mRunx2 mRNA expression in shGfi1-MC4, lack of MM-induced GFI1 binding to the mRunx2 promoter (Fig. 4F) results in deficient recruitment of co-repressors HDAC1 (Fig. 4G) and EZH2 (Fig. 4H). Further, lack of GFI1-mediated HDAC1 and EZH2 recruitment rescued levels of H3K9ac at mRunx2 after MM co-culture (Fig. 4I). Concomitantly, we observed significantly reduced enrichment of the repressive mark H3K27me3 (Figure 4J), further arguing for the importance of GFI1-directed EZH2 recruitment to the mRunx2 promoter in pre-OB during MM co-culture conditions. Thus lack of GFI1 recruitment directly correlates with the inability of the MM cells to induce epigenetic suppression of the mRunx2 promoter. These results reveal that destabilization of mRunx2 mRNA is not sufficient to prevent OB differentiation in the absence of GFI1-mediated epigenetic alteration of the mRunx2 gene.
Figure 4. mGfi1 knockdown in MC4 cells prevents MM-induced repression of mRunx2 and OB differentiation markers, the recruitment of HDAC1 and EZH2, and repressed chromatin architecture acquisition.
qPCR analysis of mRNAs from SCR- and shGfi1-MC4 cells treated as described in Fig. 1A for: (A) Gfi1, (B) Runx2, (C) Ocn, (D) Bsp, and (E) Alpl mRNA expression. ChIP-qPCR analyses of MM-induced recruitment to the Runx2 promoter of (F) GFI1, (G) HDAC1, and (H) EZH2 and enrichment profiles for (I) H3K9ac and (J) H3K27me3 in SCR and shGfi1-MC4 at d0±MM. IgG ChIP was run on SCR-MC4 cells. Error bars represent SEM for (A–E) 3–4 or (F–J) 2 biological replicates ⭘ – represents values of p<0.08. Amplicons as indicated in Fig 1B.
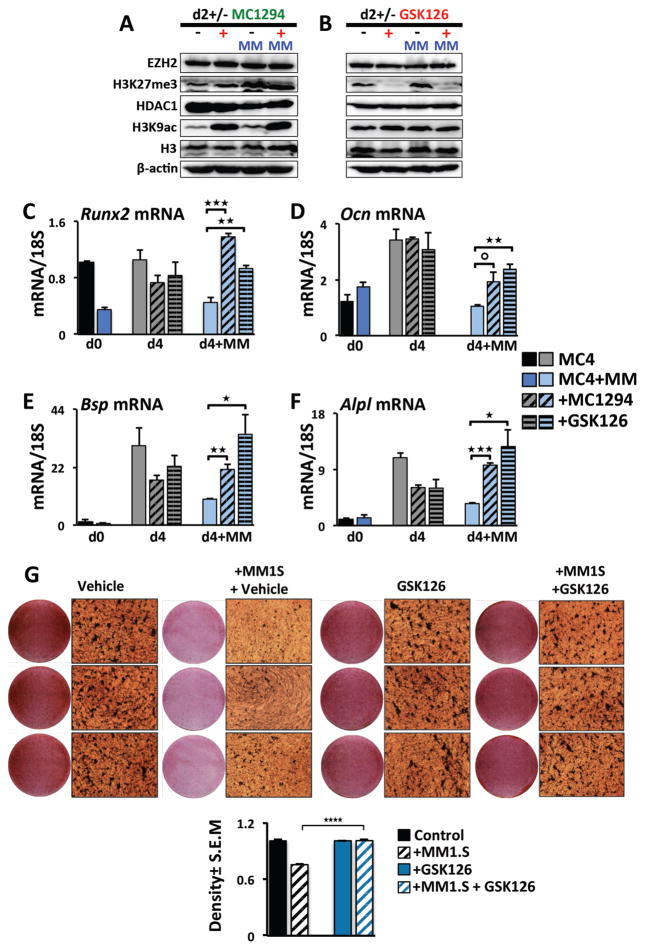
MM suppression of mRunx2 and OB differentiation of MC4 cells is reversed by HDAC1 or EZH2 inhibition
We used small molecule inhibitors of HDAC1 (MC1294) and EZH2 (GSK126) enzymatic activities to investigate if the MM-induced GFI1-mediated epigenetic repression of mRunx2 is reversible. Following 5TGM1-MC4 co-cultures in proliferation media, we removed the MM cells and subjected the MC4 cells to OB differentiation in the presence of either vehicle, MC1294 or GSK126 (Fig. 5). Western analyses demonstrated that the HDAC inhibitor MC1294 increased global H3K9Ac levels in MC4 cells after 2 days regardless of whether or not the cells had been pre-exposed to MM cells, while not affecting the H3, HDAC1, EZH2, or H3K27me3 levels (Fig. 5A). Similarly, the EZH2 inhibitor GSK126 decreased global H3K27me3 levels in MC4 cells after 2 days without affecting H3, EZH2, HDAC1, and H3K9ac levels (Fig. 5B). MC4 treatment with MC1294 or GSK126 did not alter standard OB differentiation-stimulated mRunx2 mRNA expression at d4 (Fig. 5C). However, inhibition of HDAC1 or EZH2 activity significantly rescued mRunx2 mRNA from the MM-mediated sustained repression at d4 (Fig. 5C). HDAC1 and EZH2 inhibition similarly rescued mRNA expression of several downstream RUNX2 targets critical for OB differentiation, including mOcn, mBsp and mAlpl (Fig. 5D–F). Mineralization assays confirmed that EZH2 inhibition reversed the OB differentiation block established by human MM1.S transwell co-culture with MC14 cells (Fig. 5G). Our results argue that GFI1 recruitment of the epigenetic histone modifiers HDAC1 and EZH2 and their actions at the mRunx2 histones facilitate the suppressive MM effects on mRunx2 in pre-OB MC4 cells, and that this effect is reversible after short-term (48–72 h) MM exposure.
Figure 5. Inhibition of histone modifiers HDAC1 and EZH2 rescues OB differentiation of MM-exposed MC4 cultures.
(A–F) MC4 cells were exposed to 5TGM1 MM cells as diagrammed in Fig 1A in the absence of inhibitors. After MM removal at d0, the MC4 were cultured in OB differentiation media for 2 to 4 days with either DMSO vehicle, MC1294 (10 μM), or GSK126 (5 μM) added as indicated. (A,B) The effects of the inhibitors (A) MC1294 (HDACi) and (B) GSK126 (EZH2i) on global levels of H3K9ac, H3K27me3, H3, HDAC1, EZH2 levels in MC4 cells on day 2 were assessed by Western blot using antibodies as indicated. (C–F) Effects of the inhibitors MC1294 and GSK126 on (C) Runx2, (D) Ocn, (E) Bsp, and (F) Alpl mRNA expression during differentiation of control and 5TGM1 MM-exposed MC4 at day 0 (no inhibitor) or after 4 days of differentiation (d0±MM, d4±MM). Error bars represent SEM for 3 biological replicates. (G) Human MM1.S MM cells in transwells (or empty control transwells) were co-cultured with MC14 cells for 72 h. Following transwell removal, the MC14 cells were cultured in osteogenic media +/- GSK126 (2.5 μM) and mineralization was assessed using Alizarin Red staining at d21; the GSK126 was absent d14–21. Shown is density quantitation for the average of 6 wells with SEM and significance indicated.
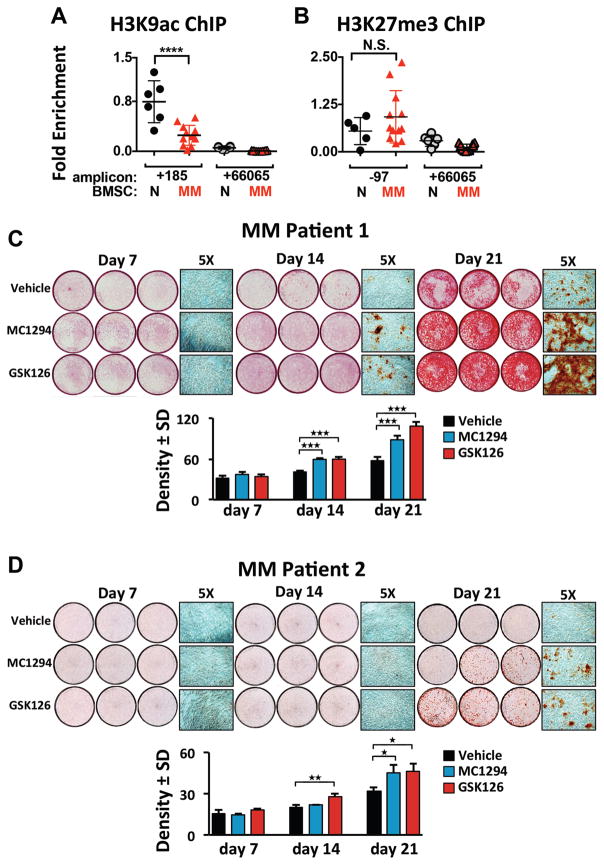
MM induces sustained transcriptional and epigenetic suppression of the hRunx2 promoter in human MM patient BMSC that is reversed by HDAC1 or EZH2 inhibition
To demonstrate involvement of MM-induced hRunx2 epigenetic suppression in preventing OB differentiation in patients, we used ChIP-qPCR to analyze the activation mark H3K9ac at the hRunx2 promoter in BMSC from MM patients (MM-BMSC) and healthy donors (HD-BMSC). Chromatin isolated from MM-BMSC (n=12) revealed significant reduction of H3K9ac at the hRunx2 promoter as compared to HD-BMSC samples (n=6) (Fig. 6A). Analysis of additional samples demonstrated that the repressive mark H3K27me3 at the hRunx2 promoter was higher on average for MM-BMSC (n=12) as compared to HD-BMSC (n=6) (Fig. 6B), though the difference did not reach statistical significance. Therefore, we treated MM-BMSC from two patients with vehicle, MC1294, or GSK126 for 7, 14, and 21 days in osteogenic culture conditions and assayed mineralization/calcium deposition (Fig. 6C,D). Both MC1294 and GSK126 permitted significantly more OB differentiation as compared to vehicle for each MM-BMSC sample. MM-BMSC from three additional patients assayed only at 21 days gave similar results (Supplementary Fig. S5A–D). In contrast, EZH2 inhibition did not change OB differentiation of HD-BMSC (Supplementary Fig. S5E). These data, together with our results from MC4 cells (Fig. 5), demonstrate that MM-induced GFI1 recruitment of EZH2 mediates H3K27me3 epigenetic repression of Runx2, which contributes to the long-term suppression of hBMSC differentiation into functioning OB, and, importantly, that it is reversible even after long-term MM-exposure in vivo.
Figure 6. MM-BMSC samples exhibited decreased H3K9Ac at the hRUNX2 promoter compared to HD-BMSC and inhibition of either HDAC1 or EZH2 rescues MM-BMSC OB differentiation.
(A) Anti-H3K9Ac (and IgG) ChIP-qPCR analysis of HD-BMSC (N) (n=6) and MM-BMSC (MM) (n=12, patient characteristics in Table S3) using amplicons +185 and +66065 relative to the hRUNX2 P1 TSS. One anti-H3K9Ac ChIP amplicon +185 N sample result was used as the reference sample for all other data and ΔΔCt shown. (B) Anti-H3K27me3 (and IgG) ChIP-qPCR analysis of HD-BMSC (n=6), which included two donors used in A, and a unique set of MM-BMSC (n=12, patient characteristics in Table S4), using amplicons -97 and +66065 as described in A. There were no significant differences in the IgG pulldown results across all samples and between the amplicons. The significance of differences between N and MM samples for each amplicon were determined by one-way ANOVA with Tukey’s multiple comparison post-test using Graphpad Prism 6. (C, D) MM-BMSC from two different patients (Table S5) were cultured 7, 14, or 21 days in osteogenic media supplemented with vehicle, MC1294 (10 μM) or GSK126 (2.5 μM); the inhibitors were absent d14–21. Mineralization was assessed using Alizarin Red staining. Three independent wells from each treatment group are shown as well as a representative 5X magnification. Below each set is the density quantitation for the average of 6 wells/condition with SEM and significance indicated.
Discussion
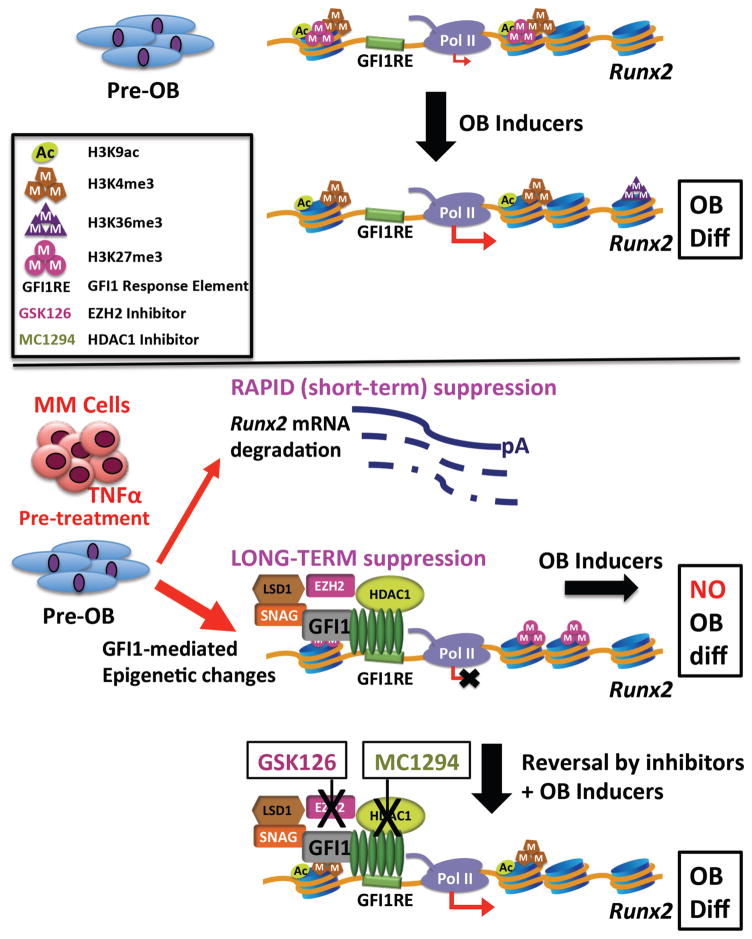
Our studies demonstrate that the key mechanism by which MM cells establish persistent suppression of OB differentiation in MMBD (5) is via induction of direct GFI1 binding to the Runx2-P1 promoter in pre-OB cells resulting in Runx2 repression. While MM cells enhance Runx2 mRNA degradation in proliferating pre-OB, this effect is not sufficient to establish OB suppression. MM cells induce GFI1 binding to a novel GFI1 response element within the Runx2-P1 promoter. GFI1 then acts as a platform molecule for formation of a repressive complex containing histone modifier enzymes HDAC1, LSD1 and EZH2, which decrease H3K9ac and H3K4me3 and increase H3K27me3 modifications, respectively, to establish a repressive chromatin architecture at Runx2 that is refractory to OB inducer activation (Fig. 7). Importantly, we’ve shown that this refractory state requires active maintenance and is reversible by inhibition of HDAC1 or EZH2 activity.
Figure 7. Schematic of the mechanism of GFI1-induced epigenetic repression of the Runx2 locus in MM exposed pre-OB.
In proliferating pre-OB cells, Runx2-P1 is in a poised bivalent configuration with paused Pol II and prominent levels of activation-ready promoter chromatin marks H3K4me3 and H3K9ac, as well as H3K27me3, with low levels of basal transcription. OB differentiation induction stimulates increased accumulation of these active chromatin marks, as well as release of Pol II into the Runx2 structural region as marked by increased Pol II Ser2P-CTD and accumulation of the H3K36me3 mark. MM exposure acts in a dual mode to repress Runx2 expression. The rapid TNFα-induced decrease in Runx2 mRNA is mediated by increased mRNA degradation. However, this is insufficient to block induction of OB differentiation. The sustained suppression of OB differentiation requires modifications of the Runx2 chromatin architecture. GFI1 binds to Runx2 and facilitates recruitment of histone co-repressors HDAC1, LSD1 and EZH2, which results in decreased active H3K9ac and H3K4me3 and increased repressive H3K27me3 chromatin marks, causing an epigenetic block refractory to transcriptional activation in response to OB differentiation signals. Inhibition of either HDAC1 or EZH2 can reverse the inhibition and allow OB differentiation.
We identified a functional GFI1 response element with two overlapped palindromic cores at -21/-16, that each contributes to the strength of GFI1 binding. It’s possible that the presence of both sides of the palindromic recognition sites generates some cooperative binding, although this is unlikely to be through Zn fingers 3–5 of two GFI1 molecules interacting with the DNA at the same time due to steric hindrance (19, 35, 36). GFI1 has not been reported to dimerize, although other C2H2 Zn finger transcription factors, such as Ikaros, TRPS1, and Drosophila Hunchback can homodimerize via the alpha helices of two Zn fingers that are not involved in protein-DNA interactions (37). It remains to be established if these palindromic sites can induce a pair of GFI1 molecules to bind at the same time, perhaps by each only contributing a subset of Zn fingers 3–5.
Gene expression is closely associated with histone exchange and histone post-translational modifications, which regulate the states of chromatin compaction and assembly of transcription machinery at gene promoters (38). In proliferating pre-OB, we found that the Runx2-P1 promoter has a poised bivalent chromatin architecture with moderate levels of active histone marks H3K9ac and H3K4me3, as well as the repressive mark H3K27me3, is pre-loaded with Pol II, and undergoes a low level of basal transcription detectable by qPCR. Many developmental genes have a similar poised promoter architecture that can swiftly respond to external stimuli but lacks the transcription elongation properties associated with active gene expression (39). Similar to a previous report (40), stimulation of OB differentiation induced changes in the Runx2 epigenetic profile (increased H3K36me3, H3K9ac, and H3K4me3, and decreased H3K27me3) that were consistent with the expected activation of the Runx2 gene.
We found that MM cells induced significant chromatin alterations on the Runx2 gene in pre-OB cells in proliferation media that included a profound decrease in the activation mark H3K9ac together with increased levels of the repressive mark H3K27me3. ChIP-qPCR of human BMSC samples from MM patients and healthy donors also revealed significantly decreased H3K9ac and a trend towards higher H3K27me3 in MM-BMSC as compared to HD-BMSC. H3K27me3 has been reported to be elevated in primary undifferentiated BMSC, with removal by the demethylase Jumonji domain-containing protein 3 (JMJD3) required to allow Runx2 activation during OB induction (41). Thus, the difference in H3K27me3 levels between MM-BMSC and HD-BMSC after OB differentiation induction would likely be larger. In summary, the effect of these MM-induced chromatin changes in proliferating pre-OB is to make the Runx2 gene refractory to activation by OB differentiation stimulation, even in the absence of MM cells, by blocking the normal epigenetic changes induced during OB differentiation. Thus, leaving the Runx2 chromatin in a state similar to the undifferentiated, proliferating pre-OB in spite of exposure to activation signals.
ChIP-qPCR analysis of MM-MC4 co-culture time courses revealed that GFI1 recruitment to the Runx2-P1 promoter is not rapid, taking at least 36 h to become detectable. This result supports our previous report that GFI1 translocates from the cytoplasm to the nucleus following 5TGM1 MM co-culture or TNFα treatment of more than 24 h (6). MM-induced GFI1 recruitment to the Runx2 promoter coincided with an increased presence of LSD1, HDAC1, and EZH2, the enzymes responsible for the histone modifications that established an epigenetic block to osteoblastogenesis. Ectopic expression of GFI1 in MC4 cells in the absence of MM exposure was sufficient to recruit HDAC1 and EZH2, alter the chromatin architecture, and repress the Runx2 gene. GFI1 can repress target genes by recruiting HDAC1 and LSD1 co-repressors to establish epigenetic silencing in other cell systems (20, 21), and their presence at Runx2 is consistent with the MM-induced decrease in activating marks H3K9ac and H3K4me3, respectively. Of note, LSD1 primarily acts on H3K4me1/2 substrates (42), but its presence regulating the H3K4 methylation state is primarily associated with gene repression and decreased levels of H3K4me3 at promoters (43). We made the novel observation that Gfi1 mediates the recruitment of EZH2 to Runx2, facilitating deposition of H3K27me3 at the Runx2 promoter. Snail1, another member of the SNAG family of zinc finger transcription repressors (44), has also been implicated in recruiting components of PRC2 during the repression of the E-cadherin (CDH1) gene in tumor cells (45) via the N-terminal repressor SNAG domain.
Studies with MC4 with a stable Gfi1 knockdown demonstrated that lack of GFI1 binding to the Runx2 promoter in MM-exposed pre-OB caused diminished recruitment of both HDAC1 and EZH2, preventing MM-induced H3K9ac loss and H3K27me3 increase on Runx2. These changes allowed OB differentiation, as evidenced by increased expression of Runx2 and the OB differentiation markers Ocn and Bsp. Interestingly, Gfi1 knockdown did not prevent the early MM-induced decrease of Runx2 mRNA. This indicates that destabilization of the Runx2 mRNA is not sufficient to repress OB differentiation, and that GFI1-mediated chromatin changes are necessary for the MM alteration of pre-OB fate.
Several studies indicate that both HDAC1 and EZH2 are associated with negative regulation of osteoblastogenesis. Human mesenchymal stem cells exhibited increased osteogenic differentiation due to CDK1 dependent phosphorylation of EZH2, thereby causing disruption of PRC2 complex formation on Runx2 and OB-related gene promoters (46). Dudakovic et al. (47) reported that human stromal cells from the vascular fraction of adipose tissue displayed enhanced OB differentiation if treated with EZH2 inhibitor or shRNA. Similarly, down-regulation of HDAC1 activity was shown to promote OB differentiation due to hyperacetylation of osteogenic gene promoters (48). Using the selective inhibitors MC1294 (HDAC1i) and GSK126 (EZH2i) to treat MC4 cells placed into OB differentiation media after 72 h MM exposure, we demonstrated that blockade of either of these epigenetic modifiers rescued expression of Runx2 as well as its downstream target OB genes Ocn, Bsp, and Alpl from MM-triggered repression. HDAC1 and EZH2 have a plethora of roles during OB differentiation and we observed that the universal targeting of these enzymes was slightly repressive on Bsp and Alpl expression in normal OB differentiation samples. Despite this effect, the inhibitors had profound positive effects on the expression of these genes during OB differentiation after MM exposure. Further, we reported that siRNA knockdown of Gfi1 in BMSC isolated from MM patients or after MM exposure of MC4 cells also rescued the expression of these genes during induction of OB differentiation (6). These results suggest that the MM-induced epigenetic suppression of the Runx2 promoter is a very dynamic and reversible process that requires continuous maintenance by GFI1 and its recruited repressive chromatin modifiers to prevent Runx2 activation by stimulators of OB differentiation. How GFI1 remains elevated in MM-BMSC in the absence of MM cells remains to be determined.
Here we provide evidence that suppression of the transition of BMSC to functioning OB in the pro-inflammatory myeloma BM microenvironment is likely due to Gfi1-mediated and maintained epigenetic repression of the key OB differentiation factor Runx2 via recruitment of HDAC1 and EZH2. Interfering either with Gfi1 expression or with HDAC1 or EZH2 activity reverses the epigenetic repression and permits OB differentiation. These results suggest that treatment of MM patients with clinically available HDAC1 or EZH2 inhibitors may block or reverse the profound OB suppression in MM and allow repair of lytic lesions. Understanding the mechanisms associated with the repressive effects of GFI1 in BMSC may also lead to the development of novel therapeutics for MMBD, as well as various inflammatory diseases such as rheumatoid arthritis, that cause homeostatic imbalance in the bone microenvironment.
Supplementary Material
Acknowledgments
Financial Support: This work was supported by the National Institutes of Health Grants (R01AR059679 to DLG & GDR and K08CA177927 to KRW), the Veterans Administration (Merit Review to GDR), and the Sarcoma Foundation of America (KRW). This project used the UPCI Lentiviral and FACS facilities that are supported in part by National Institutes of Health Grant P30CA047904.
The authors gratefully thank the UPCI Lentivirus Facility for the SCR and shGfi1 lentiviruses; and the Veterans Administration Pittsburgh Healthcare System, Research and Development for use of the facilities.
Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIAMS, NCI, NIH, the Department of Veterans Affairs, or the United States Government.
Footnotes
Conflict of Interest: GDR participates on an advisory board for Amgen. The other authors have declared that no conflict of interest exists.
AUTHOR CONTRIBUTIONS
Study design: JA, SJ, GDR, DLG; Conducting experiments: JA, SJ, QS, PZ; Provided samples from human subjects: RS, KRW; Processed human samples: PZ, JA, JLA; Data analysis: JA, SJ, QS, DLG; Data interpretations: JA, SJ, QS, GDR, DLG; Drafting manuscript: JA, SJ, DLG; Revising manuscript content: JA, RS, GDR, DLG; Approving final version of manuscript: all authors. DLG takes responsibility for the integrity of the data analysis.
References
- 1.Roodman GD. Pathogenesis of myeloma bone disease. J Cell Biochem. 2010;109:283–91. doi: 10.1002/jcb.22403. [DOI] [PubMed] [Google Scholar]
- 2.Saad F, Lipton A, Cook R, Chen YM, Smith M, Coleman R. Pathologic fractures correlate with reduced survival in patients with malignant bone disease. Cancer. 2007;110:1860–7. doi: 10.1002/cncr.22991. [DOI] [PubMed] [Google Scholar]
- 3.Sonmez M, Akagun T, Topbas M, Cobanoglu U, Sonmez B, Yilmaz M, et al. Effect of pathologic fractures on survival in multiple myeloma patients: a case control study. Journal of experimental & clinical cancer research : CR. 2008;27:11. doi: 10.1186/1756-9966-27-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galson DL, Silbermann R, Roodman GD. Mechanisms of multiple myeloma bone disease. BoneKEy reports. 2012;1:135. doi: 10.1038/bonekey.2012.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giuliani N, Rizzoli V, Roodman GD. Multiple myeloma bone disease: Pathophysiology of osteoblast inhibition. Blood. 2006;108:3992–6. doi: 10.1182/blood-2006-05-026112. [DOI] [PubMed] [Google Scholar]
- 6.D'Souza S, del Prete D, Jin S, Sun Q, Huston AJ, Kostov FE, et al. Gfi1 expressed in bone marrow stromal cells is a novel osteoblast suppressor in patients with multiple myeloma bone disease. Blood. 2011;118:6871–80. doi: 10.1182/blood-2011-04-346775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Accardi F, Toscani D, Bolzoni M, Dalla Palma B, Aversa F, Giuliani N. Mechanism of Action of Bortezomib and the New Proteasome Inhibitors on Myeloma Cells and the Bone Microenvironment: Impact on Myeloma-Induced Alterations of Bone Remodeling. BioMed research international. 2015;2015:172458. doi: 10.1155/2015/172458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arnulf B, Lecourt S, Soulier J, Ternaux B, Lacassagne MN, Crinquette A, et al. Phenotypic and functional characterization of bone marrow mesenchymal stem cells derived from patients with multiple myeloma. Leukemia. 2007;21:158–63. doi: 10.1038/sj.leu.2404466. [DOI] [PubMed] [Google Scholar]
- 9.Corre J, Mahtouk K, Attal M, Gadelorge M, Huynh A, Fleury-Cappellesso S, et al. Bone marrow mesenchymal stem cells are abnormal in multiple myeloma. Leukemia. 2007;21:1079–88. doi: 10.1038/sj.leu.2404621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garderet L, Mazurier C, Chapel A, Ernou I, Boutin L, Holy X, et al. Mesenchymal stem cell abnormalities in patients with multiple myeloma. Leukemia & lymphoma. 2007;48:2032–41. doi: 10.1080/10428190701593644. [DOI] [PubMed] [Google Scholar]
- 11.Hiruma Y, Honjo T, Jelinek DF, Windle JJ, Shin J, Roodman GD, et al. Increased signaling through p62 in the marrow microenvironment increases myeloma cell growth and osteoclast formation. Blood. 2009;113:4894–902. doi: 10.1182/blood-2008-08-173948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu G, Liu K, Anderson J, Patrene K, Lentzsch S, Roodman GD, et al. Expression of XBP1s in bone marrow stromal cells is critical for myeloma cell growth and osteoclast formation. Blood. 2012;119:4205–14. doi: 10.1182/blood-2011-05-353300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi T, Kronenberg H. Minireview: transcriptional regulation in development of bone. Endocrinology. 2005;146:1012–7. doi: 10.1210/en.2004-1343. [DOI] [PubMed] [Google Scholar]
- 14.Giuliani N, Colla S, Morandi F, Lazzaretti M, Sala R, Bonomini S, et al. Myeloma cells block RUNX2/CBFA1 activity in human bone marrow osteoblast progenitors and inhibit osteoblast formation and differentiation. Blood. 2005;106:2472–83. doi: 10.1182/blood-2004-12-4986. [DOI] [PubMed] [Google Scholar]
- 15.Grimes HL, Gilks CB, Chan TO, Porter S, Tsichlis PN. The Gfi-1 protooncoprotein represses Bax expression and inhibits T-cell death. Proc Natl Acad Sci U S A. 1996;93:14569–73. doi: 10.1073/pnas.93.25.14569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tateno M, Fukunishi Y, Komatsu S, Okazaki Y, Kawai J, Shibata K, et al. Identification of a novel member of the snail/Gfi-1 repressor family, mlt 1, which is methylated and silenced in liver tumors of SV40 T antigen transgenic mice. Cancer Res. 2001;61:1144–53. [PubMed] [Google Scholar]
- 17.van der Meer LT, Jansen JH, van der Reijden BA. Gfi1 and Gfi1b: key regulators of hematopoiesis. Leukemia. 2010;24:1834–43. doi: 10.1038/leu.2010.195. [DOI] [PubMed] [Google Scholar]
- 18.Fiolka K, Hertzano R, Vassen L, Zeng H, Hermesh O, Avraham KB, et al. Gfi1 and Gfi1b act equivalently in haematopoiesis, but have distinct, non-overlapping functions in inner ear development. EMBO Rep. 2006;7:326–33. doi: 10.1038/sj.embor.7400618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zweidler-Mckay PA, Grimes HL, Flubacher MM, Tsichlis PN. Gfi-1 encodes a nuclear zinc finger protein that binds DNA and functions as a transcriptional repressor. Mol Cell Biol. 1996;16:4024–34. doi: 10.1128/mcb.16.8.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saleque S, Kim J, Rooke HM, Orkin SH. Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol Cell. 2007;27:562–72. doi: 10.1016/j.molcel.2007.06.039. [DOI] [PubMed] [Google Scholar]
- 21.Duan Z, Zarebski A, Montoya-Durango D, Grimes HL, Horwitz M. Gfi1 coordinates epigenetic repression of p21Cip/WAF1 by recruitment of histone lysine methyltransferase G9a and histone deacetylase 1. Mol Cell Biol. 2005;25:10338–51. doi: 10.1128/MCB.25.23.10338-10351.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Q, Basu S, Qiu Y, Tang F, Dong F. A role of Miz-1 in Gfi-1-mediated transcriptional repression of CDKN1A. Oncogene. 2010;29:2843–52. doi: 10.1038/onc.2010.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharif-Askari E, Vassen L, Kosan C, Khandanpour C, Gaudreau MC, Heyd F, et al. Zinc finger protein Gfi1 controls the endotoxin-mediated Toll-like receptor inflammatory response by antagonizing NF-kappaB p65. Mol Cell Biol. 2010;30:3929–42. doi: 10.1128/MCB.00087-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dahl R, Iyer SR, Owens KS, Cuylear DD, Simon MC. The transcriptional repressor GFI-1 antagonizes PU.1 activity through protein-protein interaction. The Journal of biological chemistry. 2007;282:6473–83. doi: 10.1074/jbc.M607613200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodel B, Tavassoli K, Karsunky H, Schmidt T, Bachmann M, Schaper F, et al. The zinc finger protein Gfi-1 can enhance STAT3 signaling by interacting with the STAT3 inhibitor PIAS3. EMBO J. 2000;19:5845–55. doi: 10.1093/emboj/19.21.5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heyd F, ten Dam G, Moroy T. Auxiliary splice factor U2AF26 and transcription factor Gfi1 cooperate directly in regulating CD45 alternative splicing. Nat Immunol. 2006;7:859–67. doi: 10.1038/ni1361. [DOI] [PubMed] [Google Scholar]
- 27.Xiao G, Cui Y, Ducy P, Karsenty G, Franceschi RT. Ascorbic acid-dependent activation of the osteocalcin promoter in MC3T3-E1 preosteoblasts: requirement for collagen matrix synthesis and the presence of an intact OSE2 sequence. Molecular endocrinology. 1997;11:1103–13. doi: 10.1210/mend.11.8.9955. [DOI] [PubMed] [Google Scholar]
- 28.Wang D, Christensen K, Chawla K, Xiao G, Krebsbach PH, Franceschi RT. Isolation and characterization of MC3T3-E1 preosteoblast subclones with distinct in vitro and in vivo differentiation/mineralization potential. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 1999;14:893–903. doi: 10.1359/jbmr.1999.14.6.893. [DOI] [PubMed] [Google Scholar]
- 29.Adamik J, Wang KZ, Unlu S, Su AJ, Tannahill GM, Galson DL, et al. Distinct mechanisms for induction and tolerance regulate the immediate early genes encoding interleukin 1beta and tumor necrosis factor alpha. PloS one. 2013;8:e70622. doi: 10.1371/journal.pone.0070622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilbert L, He X, Farmer P, Rubin J, Drissi H, van Wijnen AJ, et al. Expression of the osteoblast differentiation factor RUNX2 (Cbfa1/AML3/Pebp2alpha A) is inhibited by tumor necrosis factor-alpha. The Journal of biological chemistry. 2002;277:2695–701. doi: 10.1074/jbc.M106339200. [DOI] [PubMed] [Google Scholar]
- 31.Henikoff S, Shilatifard A. Histone modification: cause or cog? Trends in genetics : TIG. 2011;27:389–96. doi: 10.1016/j.tig.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 32.Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, et al. A model for transmission of the H3K27me3 epigenetic mark. Nature cell biology. 2008;10:1291–300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- 33.Montoya-Durango DE, Velu CS, Kazanjian A, Rojas ME, Jay CM, Longmore GD, et al. Ajuba functions as a histone deacetylase-dependent co-repressor for autoregulation of the growth factor-independent-1 transcription factor. The Journal of biological chemistry. 2008;283:32056–65. doi: 10.1074/jbc.M802320200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–9. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zarebski A, Velu CS, Baktula AM, Bourdeau T, Horman SR, Basu S, et al. Mutations in growth factor independent-1 associated with human neutropenia block murine granulopoiesis through colony stimulating factor-1. Immunity. 2008;28:370–80. doi: 10.1016/j.immuni.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee S, Doddapaneni K, Hogue A, McGhee L, Meyers S, Wu Z. Solution structure of Gfi-1 zinc domain bound to consensus DNA. Journal of molecular biology. 2010;397:1055–66. doi: 10.1016/j.jmb.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 37.McCarty AS, Kleiger G, Eisenberg D, Smale ST. Selective dimerization of a C2H2 zinc finger subfamily. Mol Cell. 2003;11:459–70. doi: 10.1016/s1097-2765(03)00043-1. [DOI] [PubMed] [Google Scholar]
- 38.Venkatesh S, Workman JL. Histone exchange, chromatin structure and the regulation of transcription. Nature reviews Molecular cell biology. 2015;16:178–89. doi: 10.1038/nrm3941. [DOI] [PubMed] [Google Scholar]
- 39.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tai PW, Wu H, Gordon JA, Whitfield TW, Barutcu AR, van Wijnen AJ, et al. Epigenetic landscape during osteoblastogenesis defines a differentiation-dependent Runx2 promoter region. Gene. 2014;550:1–9. doi: 10.1016/j.gene.2014.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang D, Okamura H, Nakashima Y, Haneji T. Histone demethylase Jmjd3 regulates osteoblast differentiation via transcription factors Runx2 and osterix. The Journal of biological chemistry. 2013;288:33530–41. doi: 10.1074/jbc.M113.497040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–53. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 43.Zentner GE, Henikoff S. Regulation of nucleosome dynamics by histone modifications. Nature structural & molecular biology. 2013;20:259–66. doi: 10.1038/nsmb.2470. [DOI] [PubMed] [Google Scholar]
- 44.Chiang C, Ayyanathan K. Snail/Gfi-1 (SNAG) family zinc finger proteins in transcription regulation, chromatin dynamics, cell signaling, development, and disease. Cytokine Growth Factor Rev. 2013;24:123–31. doi: 10.1016/j.cytogfr.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herranz N, Pasini D, Diaz VM, Franci C, Gutierrez A, Dave N, et al. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol Cell Biol. 2008;28:4772–81. doi: 10.1128/MCB.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei Y, Chen YH, Li LY, Lang J, Yeh SP, Shi B, et al. CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of human mesenchymal stem cells. Nature cell biology. 2011;13:87–94. doi: 10.1038/ncb2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dudakovic A, Camilleri ET, Xu F, Riester SM, McGee-Lawrence ME, Bradley EW, et al. Epigenetic Control of Skeletal Development by the Histone Methyltransferase Ezh2. The Journal of biological chemistry. 2015;290:27604–17. doi: 10.1074/jbc.M115.672345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee HW, Suh JH, Kim AY, Lee YS, Park SY, Kim JB. Histone deacetylase 1-mediated histone modification regulates osteoblast differentiation. Molecular endocrinology. 2006;20:2432–43. doi: 10.1210/me.2006-0061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.