Abstract
Antibody derivatives, such as antibody fragments (Fab) and single-chain variable fragments (scFv), are now being used to image traditionally hard-to-see protein subpopulations, including nascent polypeptides being translated and post-translationally modified proteins. This has allowed researchers to directly image and quantify for the first time (1) translation initiation and elongation kinetics with single-transcript resolution and (2) the temporal ordering and kinetics of post-translational histone and RNA polymerase II modifications. Here we review these developments and discuss the strengths and weaknesses of live-cell imaging with antibody-based probes. Further development of these probes will increase their versatility and open up new avenues of research for dissecting complex gene regulatory dynamics.
Imaging the full central dogma with antibody-based probes
Sixty years ago, Francis Crick first stated the central dogma of molecular biology: DNA makes RNA makes protein [1]. At that time, the central dogma could only be imagined, but over the past two decades revolutionary advances in fluorescence microscopy have now made it possible to directly image the dogma in living cells and organisms, as it plays out in real-time, one molecule at a time [2–5]. A key breakthrough was the discovery and development of the green fluorescent protein [6–8], which can be genetically fused to other proteins to selectively light them up and track their expression in vivo (see glossary). While this powerful technology can illuminate a good portion of the central dogma, key processes remain in the dark. For one, the translation of a nascent peptide chain from mRNA cannot be imaged with fluorescent fusion tags because they take too long to mature and light up [9,10]. By the time the fluorescence becomes visible, translation is over and the protein has long separated from its parental mRNA strand. Second, once tags do light up, they cannot discriminate post-translational protein modifications [11–13] – such as acetylation, methylation, and phosphorylation – even though these modifications can dramatically alter the protein's behavior [14–16]. These two fundamental challenges have made it difficult to image, quantify, and distinguish translational and post-translational gene regulatory mechanisms in living cells and organisms.
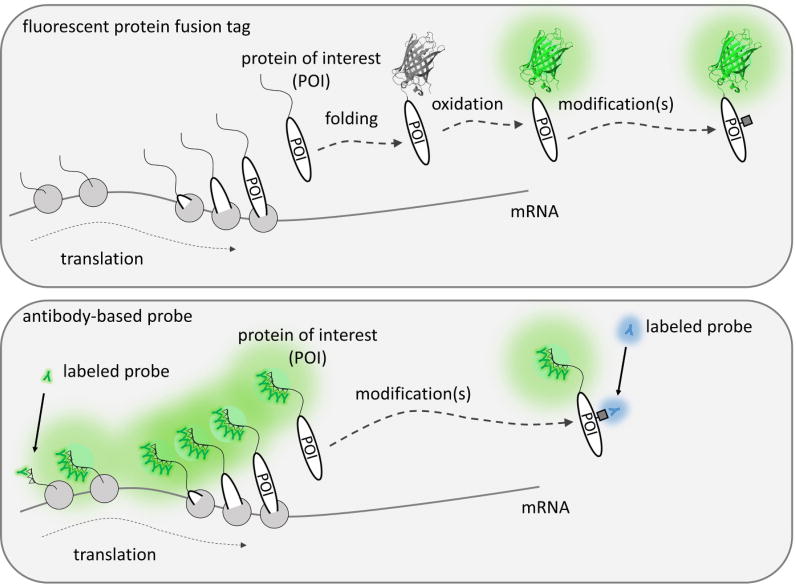
In this review, we will describe an alternative live-cell imaging modality that is beginning to shed new light on even the darkest recesses of the central dogma. The new imaging modality replaces the permanence of a fluorescent tag genetically fused to a protein with a more transient antibody-based probe that is engineered to bind its target with high specificity and affinity, yet minimal interference. The beauty of these probes is they bring pre-existing fluorescence to a protein rather than relying on the protein itself to fluoresce. This simple principle makes it possible to image proteins without restriction, from their births to their deaths, and in all their modified forms in between (Key Figure, Figure 1). In what follows, we will describe the basic design principles behind these live-cell probes, discuss how they are being used to image translational and post-translational gene regulatory dynamics in living cells, summarize ongoing challenges, and envision how these probes will be improved and applied in the future.
Figure 1. (Key Figure). Visualizing translational and post-translational dynamics with antibody-based probes.
Unlike fluorescent fusion tags like GFP (shown as a glowing beta-barrel structure; PDBID: 4KW4), which take time to fluoresce, antibody-based probes (the green “Y” shapes) can bring pre-formed fluorescence to epitopes (triangles) fused to a protein of interest (POI) still being translated (gray circles represent ribosomes). Furthermore, antibody-based probes can distinguish post-translational modifications (gray squares), whereas GFP cannot.
Fab and scFv: useful antibody-based probes for imaging protein dynamics
To image both translational and post-translational gene regulatory dynamics in living cells, a probe must be able to distinguish both unmodified and modified peptides, irrespective of whether or not they are fully folded or mature. Antibody fragments (Fab) and single chain variable fragments (scFv) fit this criteria and have been successfully used for these purposes [17–21]. Both Fab and scFv can bind short unmodified or modified peptide epitopes with high specificity, like the full antibodies from which they are derived. In addition, Fab and scFv have two key advantages over full antibodies for live-cell imaging purposes: (1) their small size and (2) their monovalency [22,23]. First, their small size allows them to quickly and efficiently access target epitopes in the complex and crowded cellular environment. For example, Fabs in living cells can pass through the nuclear pore and immediately bind target proteins within the nucleus [22]. Full antibodies, in contrast, cannot pass through the nuclear pore and therefore require cell division and nuclear envelope breakdown to access the nucleus [22]. Second, their monovalency prevents aggregation and interference. This is because Fab and scFv have a single binding domain that transiently binds only one target epitope at a time. Full antibody, in contrast, are multivalent and can therefore bind multiple target epitopes at a time with high avidity. This means that target epitopes can not only be blocked for an extended period, but may also form aggregate chains of -target-antibody-target-antibody- [24].
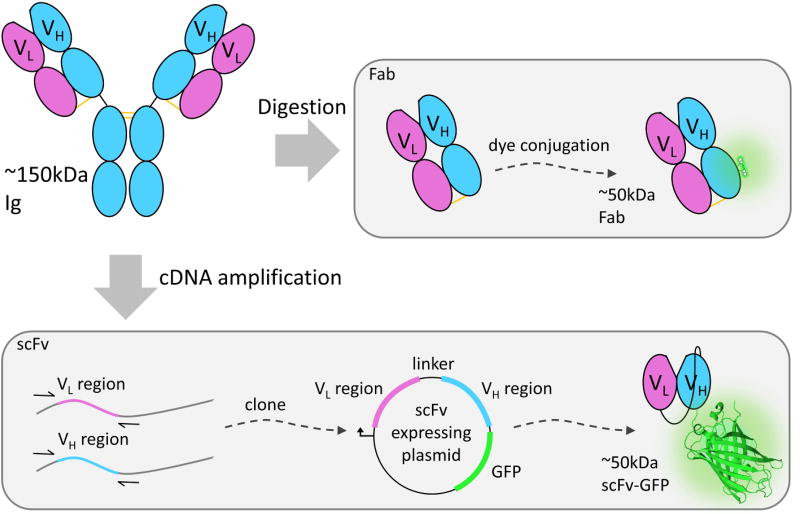
While Fab and scFv are both derived from full antibodies, there are key differences in their production and form (Figure 2). Briefly, Fab are generated directly from an antibody [25], whereas scFv are genetically encoded Fab “mimics” [26]. To generate Fab, the constant Fc base of a Y-shaped immunoglobulin (Ig) is digested away. This leaves two separate and identical “sticky” epitope-binding arms – the Fab -each weighing around 50 kDa (or roughly one-third the weight of the full Ig). There are commercial kits for generating Fab from Ig [27], so it is relatively straightforward to make Fab, albeit expensive. After the digestion is complete, each Fab is a heterodimer consisting of one constant and one variable portion of the heavy and light chains from the parental Ig. These chains are held together by pre-formed disulfide bonds and residual electrostatic interactions [28]. For live-cell imaging, Fab are typically conjugated with small synthetic dyes [29].
Figure 2. Fab and scFv both originate from immunoglobins.
Immunoglobins (Igs) are made up of heavy (blue ovals) and light (pink ovals) chains. IgG binding to a target is facilitated by the heavy (VH) and light chain variable regions (VL). Fragmented antigen-binding (Fabs) fragments can be generated by digesting any commercially available IgG. Fabs retain their ability to bind the IgG's targets and can be conjugated with a small synthetic dye. Fab with a conjugated dye is roughly 50kDa in size. By starting with the cDNA of the heavy and light variable regions of an IgG they can be cloned into a plasmid vector to create a single chain variable fragment. This allows variable regions to be connected by a polypeptide linker and even fused to a fluorescent protein such as GFP (glowing beta-barrel structure; PDBID: 4KW4). Once scFv is fused to a GFP their collective size is roughly 50kDa too.
In an scFv, the variable regions of the heavy and light chains of a Fab are genetically linked N- to C-terminus in a single chain [30]. The flexible linker acts in place of both the constant regions and the disulfide bond in Fab. For live-cell imaging, scFv are typically fused to fluorescent proteins [31], resulting in a probe that is comparable in size to a fluorescently conjugated Fab. One point to keep in mind is that scFv are often unstable and insoluble in the cytoplasm of cells, in part because antibodies are naturally folded in a different environment, the endoplasmic reticulum. In combination with the unnatural linker and fluorescent fusion tag, this makes it challenging to engineer a functional scFv for intracellular expression [32–34]. For this reason, there are only a limited number of good scFv for live-cell imaging purposes [31,32,35]. Nevertheless, once a good scFv is generated, it is a more convenient, inexpensive, and reliable (no lot-to-lot variability [36]) probe than Fab because the parental antibodies are no longer required.
Before imaging, Fab and scFv have to first be introduced into living cells. Since scFv are genetically encoded, they can be transiently or stably expressed inside living cells or even whole organisms. In contrast, since Fab are not genetically encoded, they need to be physically loaded. The easiest and most efficient way to do this without wasting Fab is bead-loading [22,37–39]. With this technique, thousands of cells can be loaded in a few seconds using just a fraction of a microgram of Fab. Other ways to get Fab into cells include microinjection [19], electroporation [40], and, most recently, treatment with the pore-forming bacterial enzyme streptolysin O (SLO) [41]. All of these techniques are variations on the same theme: permeabilizing the membrane of cells so that purified Fab can get in. Care should therefore be taken to ensure cells remain healthy after loading.
Probing translational gene regulatory dynamics in living cells with Fab and scFv
Although translation of mRNA into protein is one of the most basic and essential processes of life, it had never been directly imaged in a living system until last year [42]. Before then, only limited aspects of the translation process could be imaged. For example, the FlAsH and ReAsH dyes have been used to rapidly bind and light up tagged nascent proteins and to detect the sites of translation in living cells [43]. However, photobleaching of dyes and non-specific binding limited signal-to-noise and prevented real-time imaging. Some of these issues may be addressable using a 3× repeat tag for signal amplification [44]. More recently, the TRICK assay was used to image and distinguish translated mRNA from untranslated mRNA [10]. In this assay, RNA fluorescence is lost upon the first round of translation, so it is not possible to track multiple rounds of translation for extended periods of time.
Without the ability to directly image translation in vivo, dissecting complex translational regulatory dynamics has been challenging [9,10]. Not only has it been difficult to distinguish different co-translational regulatory mechanisms from each other but – more importantly – it has been difficult to distinguish all forms of co-translational regulation from post-translational regulation. To illustrate, if the level of a particular protein in a cell was measured to be low, it was not clear if this was due to less mRNA translation, more protein degradation, or some combination of the two.
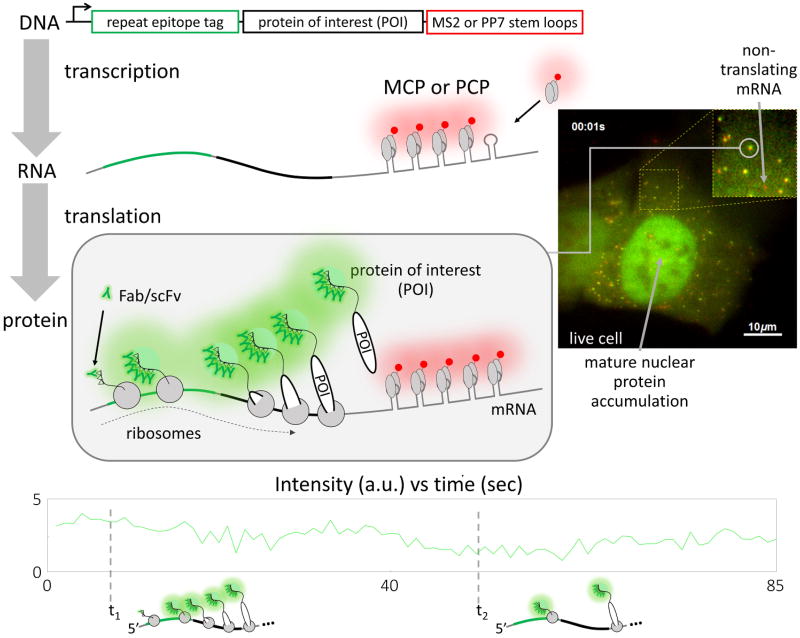
Recently, five research groups have independently figured out how to address this problem by directly imaging and quantifying the translation process in living cells [17–21]. Remarkably, all solved the problem in essentially the same way: by using antibody-based probes that bind to repeat epitope tags. This burst of papers attests to the robustness of the approach, which allowed each group to bring pre-formed fluorescence to the growing nascent peptide chain without the need for the chain to fold, mature, or fluoresce on its own (Figure 3).
Figure 3. Imaging gene regulatory dynamics in live cells using antibody-based probes.
To successfully image live cell translation dynamics, 3′UTR stem loops and a repeat epitope tag need to be encoded into the gene of interest. During transcription, the stem loops will form (gray loops) and quickly be bound by a coat protein (MS2 or PP7; gray ovals) fused to a fluorescent protein tag (red circle). This enables the visualization of mature mRNA molecules in living cells. When these marked mRNA are translated, the repeat epitopes (triangles) will emerge from the ribosome and be quickly bound by a fluorescent probe (Fab or scFv; glowing “Y”). This will quickly bring light to the growing nascent chain of the protein of interest (POI) while it is being translated. The co-migration of the red mRNA signal and the green nascent chain signal will mark sites of translation. A sample live-cell image is shown, where the green signal corresponds to Fab labeling spaghetti monster tagged lysine demethylase (KDM5B) and the red signal corresponds to MS2-labeled mRNA. These signals colocalize to form yellow spots that mark the sites of translation. The green fluorescence time series from the circled yellow spot in the inset is shown below, along with cartoons depicting how ribosomes might be distributed along the mRNA at the two specified time points. A full movie of this cell can be seen in Supplementary Movie 1. Scale bars, 10 μm.
In general, each group amplified signal from translation sites by fusing tags encoding tandem repeats of short, linear epitopes to the N-termini of proteins-of-interest (POIs), as shown in Figure 3. This makes it possible to recruit multiple fluorescent antibody-based probes to a single elongating nascent chain. Since more than one ribosome can translate an mRNA at a time (in polysomes), the amplified signal from a single chain is multiplied by the number of elongating ribosomes, yielding amplification of amplification. To visualize this, four of the groups used the SunTag system [17,18,20,21]. Here, the probe is an scFv derived from a GCN4 antibody and evolved via ribosome display to tightly bind a 33 aa epitope [33]. This was later adapted for intracellular stability within yeast [34]. To prevent aggregation in mammalian cells, a protein G (GB1) domain was further fused to the C-terminus of the scFv and the 33 aa epitope was shortened to just 19 aa (in version 4) [35]. These epitopes are repeated in the SunTag and separated from one another by short linkers (either 24 repeats [17,18,20] or 56 repeats [21], totaling 571 aa or 1419 aa, respectively). These linkers allow every epitope in the SunTag to be bound by an scFv, resulting in very bright fluorescence signals. In a similar but distinct manner, our group used the spaghetti monster system to visualize translation [19]. Here, the antibody-based probes are Fab that were generated from classic antibodies, either FLAG or HA [45,46]. The spaghetti monster tag (325 aa) is composed of ten FLAG (8 aa) or HA (9 aa) epitopes distributed throughout a 3D protein scaffold to prevent steric hindrance between bound Fab or full antibody [47]. In addition to the repeated peptide epitope tag, a repeated mRNA stem loop tag (MS2 [48] or PP7 [49]) was also used by all groups to label and track the POI-encoding mRNA. This was necessary to distinguish nascent peptide chains still being translated from mature protein, since only the former would colocalize with mRNA.
To quantify the kinetics of translation, each group employed a wide variety of experimental approaches. Despite these differences, there was consistency between the studies, as shown in Table 1. An important parameter was the translation elongation rate. This rate can be measured in a number of different ways, exactly analogous to how transcription elongation rates are measured in living cells using the MS2 RNA labeling system [3,50,51]. Whether measured by run-off inhibitor treatments [18,20,21], photobleaching [17,19,21], fluorescence correlation analysis [17,19], or the tracking of mRNA being translated by single ribosomes [18], all studies concluded that the translation elongation rate was on the order of 10 aa/sec. Importantly, this rate is fairly close to what has been measured in vitro and also via ribosome profiling [52,53].
Table 1. A comparison of live-cell measurements of single RNA translation kinetics.
| Feature | Morisaki et al. | Wu et al. | Yan et al. | Wang et al. | Pichon et al. |
|---|---|---|---|---|---|
| Cell Line | U2OS | U2OS and primary hippocampal neurons | U2OS and HEK293 | HeLa and primary hippocampal HeLa neurons | HeLa |
| Tags | Spaghetti monster 10× FLAG or 10× HA | 24× SunTag | 5×, 10×, and 24× SunTag | 24× SunTag | 32× and 56× SunTag |
| Probes | Bead-loaded FLAG-Cy3 or HA-A488 Fab | Stably expressed GCN4 scFv-GFP | Stably expressed GCN4 scFv-GFP | Stably expressed Stably expressed GCN4 scFv-GFP | Stably expressed GCN4 scFv-GFP |
| Elongation Rate | ∼10 aa/sec | ∼5 aa/sec | ∼3-6 aa/sec | ∼4 aa/sec | ∼13-18 aa/sec |
| Initiation Rate (sec/ribosome) | ∼30 sec | ∼50 sec (U2OS) ∼30 sec (PHN) | ∼30 sec | ∼20 sec | ∼15 sec |
| Nucleotides per ribosome | ∼200-900 | ∼700 | ∼300 | ∼250 | ∼600-800 |
Another key measurable was the number of nascent chains (i.e. ribosomes) per mRNA. Each group measured this by calibrating the intensity of individual translation sites. Although this number varied more from group to group than the elongation rate, the variability diminishes when considering the density of ribosomes along mRNA. In this case, all groups measured about one ribosome every 200-900 nucleotides. By combining this density estimate with the measured elongation rate, it is possible to calculate the translation initiation rate. Again, there was excellent agreement between studies, being somewhere around 30 seconds. A caveat to all these estimates is that not all steps of translation can be distinguished from the fluorescence signal alone (Box 1).
Box 1. Dissecting the steps of translation with antibody-based probes.
Most of what we know about translation has been gathered from experiments either in vitro with purified protein components [54,55], in populations of lysed (dead) cells [56– 58], or indirectly in living cells by examining the translational end product: mature proteins [59]. These studies have revealed that translation is an energy intensive process [60] that can be divided into four key steps [61,62]: ribosome search and recruitment, initiation, nascent peptide elongation, and termination. Each of these steps is subject to tight regulation [63,64]. In eukaryotes, the small (40S) subunit of the ribosome binds to and scans along mRNA during the search and recruitment phase [62]. Once an appropriate start codon is located, it assembles with other factors into a large (80S) initiation complex capable of decoding mRNA [62]. Elongation occurs as the ribosome decodes the mRNA codon by codon [61], creating a growing nascent peptide chain that protrudes from the ribonucleoprotein complex [61]. As synthesis of the chain is completed, the ribosome terminates and the nascent protein is finally released [65].
Considering these four steps, which ones can be discriminated by live-cell imaging with fluorescent antibody-based probes? It is important to keep in mind that the only measurable is the fluctuating fluorescence signal, which is a proxy for the fluctuating number of nascent chains being translated. From this signal, elongation rates are estimated by dividing the length of the tagged protein by the dwell time of the fluorescence. Using photobleaching [17,19] or run-off [18,20,21] assays, the dwell time is approximately the time it takes fluorescence to recover after photobleaching or disappear after initiation inhibition, respectively. For correlation analysis [17,19], the dwell time is calculated from the autocorrelation of the fluorescence, analogous to measurements of transcription [51]. All of these estimates assumes three things: (1) the probe is quickly recruited to the newly synthesized epitopes in the nascent chain; (2) the timescale of probe unbinding is distinguishable from the timescale of translation; and (3) the termination time is negligible. In general, if termination is too slow, then the dwell time will be some combination of elongation and termination. Initiation rates are calculated by dividing the dwell time by the number of ribosomes per transcript. This number can be found by dividing the intensity of a polysome by that of a single mature protein. Since fluorescence cannot be detected until elongation begins, the initiation rate will fundamentally be a combination of the ribosome scanning, assembling into the larger 80S subunit at the start codon, and elongating through a portion of the repeatepitope tag (to be accessible to probes, the nascent chain must emerge from the ribosomal tunnel, meaning around 30 amino acids or more of the tag need to be translated [66]).
Beyond the basic translation initiation and elongation kinetics that were quantified, different groups also observed a variety of other unique translational dynamics. One surprising observation was that mRNA being translated can by highly mobile and move about the cell just as rapidly as mRNA not being translated [17,19,21]. Since polysomes are so massive, this would suggest the mobility is active rather than passive. Indeed, in neurons, many motoring mRNA were observed to be actively translated [17], in opposition to the earlier belief that transcripts are repressed when they are being motored to their destination [67]. Another interesting observation that emerged from different groups was that translation can be organized in higher-order structures consisting of two or more polysomes clustered into small groups [19] or large translation factories [21]. Other forms of spatial organization were also observed, including ER-bound transcripts, which displayed greatly restricted mobility [17], and neuronal transcripts, which localized to the far edge of distal dendrites [17].
All groups also perturbed translation using a variety of different treatments, including (1) puromycin [68], which caused premature release of the fluorescently labeled nascent peptide chains; (2) cycloheximide [17,19], which stalled translation (although the number of ribosomes per transcript did not notably increase, as anticipated by earlier work in fixed cells [69]); (3) harringtonine [20,21], which prevented new ribosome initiation (permitting the run-off assays); and (4) DTT and arsenic stress, which dynamically altered start codon selection in a burst [20]. The origin of these diverse observations is not yet clear. Some possibilities include differences in 3′ and 5′ untranslated regions (UTRs), codon usage, or the local cellular environment.
Although the SunTag scFv and Fab-based translation imaging systems are similar, they have distinct advantages and disadvantages. The SunTag scFv can be stably expressed in cells and even put into an endogenous locus using CRISPR [21]. Thus, imaging could in principle be done throughout whole transgenic animals or within sensitive cell types (such as neurons [17,20]) that are difficult to load with Fab. On the other hand, Fab currently offer more flexibility for multi-color imaging applications because Fab can be generated from a wide range of commercially available monoclonal antibodies [19]. It is therefore easy to make orthogonal probes (such as FLAG and HA), whereas an orthogonal scFv probe that can be used in combination with the SunTag system has yet to be developed.
Probing post-translational gene regulatory dynamics in living cells with Fab and scFv
While it is now well documented that post-translational modifications (PTMs; Box 2) significantly enhance the repertoire of proteins and the complexity of the genome [70], the study of PTM dynamics and their role in gene regulation is still a maturing field. Much like the study of translation, PTMs have mainly been studied in fixed cells or bulk assays. This has greatly limited the temporal resolution of their measured behavior [11,13]. A big part of the problem is that PTMs are difficult to image using standard fluorescent fusion tags [11,71]. Since these tags are permanently fused to a protein at the translation step, they cannot distinguish any events that occur after translation, such as PTMs.
Box 2. : Post-translational protein modifications.
Proteins have unique identities that depend on their histories and that change depending on their future encounters. One way these identities are established is via post-translational modifications (PTMs) to evolutionarily conserved residues within a protein [16,75–77]. There are a wide variety of modifications [78], including chemical ones such as phosphorylation, methylation, acetylation, … etc. Once applied, PTMs can drastically change the structure and behavior of a protein [79,80]. For example, proteins can be activated, as is the case for the tumor suppressor p53 when it's acetylated [81]. PTMs can also flag different stages of a protein's life. A good example of this is the unstructured C-terminal tail of rpb1 [82,83], the catalytic subunit of RNA polymerase II. This tail is sequentially phosphorylated at specific residues during distinct phases of the transcription cycle, thereby demarcating transcriptional activation, initiation, and elongation [82–84]. Arguably the most well studied PTMs are the ones that constitute the histone code [16,85–87]. These modifications accumulate in a combinatorial fashion along the N-terminal tails of core histones. In specific combinations, they are thought to alter the accessibility of chromatin to transcription factors, thereby promoting or blocking the transcription of underlying genes [12,88–90].
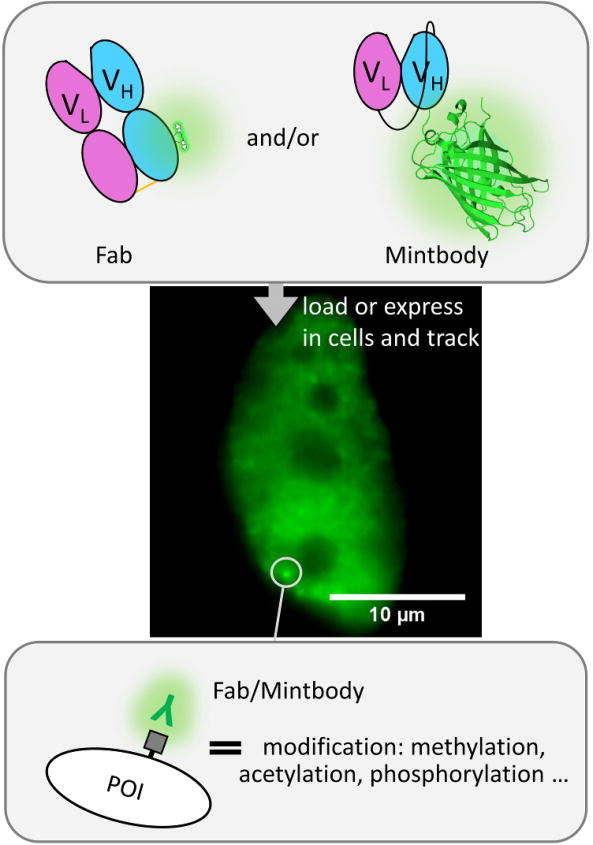
To get around this problem, fluorescent Fab and scFv have recently been used to directly target and label specific PTMs in living cells. A leading developer and proponent of this technology is the Kimura lab, which has now created a host of excellent Fab and scFv for imaging histone and RNA polymerase II modification dynamics in vivo [23]. A key to their success has been the development of a panel of high-quality antibodies against histones [72,73] and RNA polymerase II [29,74] that were stringently screened for their ability to mark and distinguish specific modifications in vitro as well as in vivo. Fab generated from these were used to establish the FabLEM technique (Fab-based imaging of Live, Endogenous Modifications) [22]. With FabLEM, regions enriched with target modifications will recruit Fab, whose fluorescence can then be tracked to quantify the dynamics of modifications in or around that area (Figure 4). For FabLEM to work well, the bound fraction of Fab should be high; that is, the time to find and bind a target should be significantly shorter than the time the Fab remains bound to the target [22]. Provided this is satisfied, signal-to-noise will be good. Ideally, Fab should also not remain bound for too long to their endogenous targets, so as not to interfere with natural target dynamics. As a rule of thumb, binding times should be between 10 and 30 seconds or so, as estimated by photobleaching experiments [22,74].
Figure 4. Marking endogenous modifications with antibody-based probes.
By loading fluorescently conjugated Fab (green and pink ovals with small glowing molecule) and/or an scFv-GFP (pink and blue ovals with glowing beta-barrel structure; PDBID: 4KW4), modified proteins can be dynamically labeled in living cells. In this example a Fab/Mintbody (glowing “Y”) is targeting residue-specific modifications (gray square) of an endogenous protein of interest (POI). The modifications accumulate in the nucleus, so Fab/Mintbody accumulate there as well. The fluctuations in the accumulations of the Fab/Mintbody in specific sub-nuclear regions can be used to quantify modification dynamics. The sample cell shown shows the accumulation of Fab (green) labeling endogenous histone H3 Lysine 27 acetylation. Scale bar, 10 μm.
So far, FabLEM has been applied in a number of different live-cell settings. In adherent immortal cell lines, such as HeLa and U2OS, the response of endogenous histone and RNA polymerase II modifications levels were monitored upon drug treatments [22,38,74] and from one generation of cells to the next [29]. In female cell lines, the movement and dynamic of inactive X was quantified [22]. In cell lines with tandem gene arrays, the precise timing and causality of histone acetylation and methylation in gene activation by RNA polymerase II was demonstrated [74]. Finally, Fab have also been loaded into living animals for imaging, including mouse preimplantation embryos [23] and Drosophila embryos [91] to quantify histone modification dynamics during development. In these studies, Fab-loaded cells continued to behave and divide normally and animals remained healthy, demonstrating that the rapid binding dynamic of Fab to endogenous modifications does not necessarily interfere with natural processes.
More recently, the Kimura lab has begun to produce fluorescently tagged scFv to mimic their Fab, probes they call mintbodies (modification-specific intracellular antibodies) [23,31]. The first mintbody was made to specifically target histone H3K9 acetylation [31]. Its genetic encodability was exploited to create stable cell lines that express the mintbody as well as genetically modified zebrafish and Drosophila. Since then, another mintbody has been introduced to target histone H4 monomethylation and track dosage compensated X-chromosomes in mouse and nematode cells [32]. In this study, the structure of the H4 monomethylation specific mintbody was determined and the sequence compared to other similar mintbodies with varying degrees of specificity for target inside living cells. This analysis identified several critical residues that contribute to the proper folding and stability of mintbody that will aid future design of functional intracellular scFv.
Besides antibody-based probes, FRET-based (förster resonance energy transfer) biosensors have also been developed to quantify the degree of protein modification activity within cells [92–94]. As these are sensors, they do not bind to endogenous modifications; instead, they contain a modifiable peptide domain fused to one fluorophore and a modification binding domain fused to another complementary fluorophore. Upon modification, these two domains bind, bringing the fluorophores together so that FRET can occur. In this way, they remotely monitor global modification levels without interference. Similar to these FRET sensors, a histone methylation specific biosensor was also recently created that is based on a split-luciferase complementation system [95]. This biosensor was tested in both living cells and mice. One complication with FRET- or complementation-based sensors is that they may be modified differently than the endogenous substrates, so signals could be misinterpreted.
The difficulties of live-cell imaging with Fab and scFv
While Fab and scFv offer unique advantages for imaging translational and post-translational gene regulatory dynamics in living cells compared to fluorescent fusion tags, they also suffer from several unique limitations that can make quantification difficult. Most of these limitations arise from the background signal from freely diffusing probes that are not bound to targets. This is a necessary evil of antibody-based imaging since unbound probe is always needed to quickly label newly made targets, should they appear.
To mitigate artifacts that might arise from background signal, it is important to keep a good ratio of probe to target epitope. What a good ratio is will depend on the specific application. In the case of PTM imaging, it is generally desirable to keep the number of probes relatively low compared to the number of targets. If not, targets can become saturated, in which case the large excess of free probes quickly increases the background and lowers the imaging signal-to-noise [22,23,29]. In contrast, for translational imaging, it is generally desirable to keep the concentration of probes higher than the concentration of targets. An appropriate concentration can be estimated from the number of target epitopes per cell using quantitative western blots [74]. Here, high background signal is less a problem because it is easily overcome by adding more repeat epitopes in the tag to amplify signal above noise. Therefore, it is more important to ensure there is an excess of free probe to quickly label the nascent peptides being translated. To illustrate, when we imaged single mRNA translation with the spaghetti monster repeat epitope tag, imaging was best in the first ten hours after transient transfection of the construct. After 24 hours of expression, Fab was soaked up by all the mature tagged protein, so new nascent peptide chains being translated were more difficult to detect [19]. One way to address this issue is to fuse a degron to the tagged protein so that it is quickly degraded [17]. In this way, the ratio of probe to target can be maintained at high levels indefinitely.
A related problem with live-cell imaging using antibody-based probes is interference that might occur from either the probe, the epitope tag, or both. When imaging PTMs, interference is especially problematic because the probes may compete with other endogenous proteins that also recognize the PTMs. This can disrupt natural cellular signaling pathways. It is therefore important to ensure that cells loaded with probes continue to divide and behave normally. The degree of competitive interference can be estimated by perturbing cells with activators or inhibitors to ensure probes react as expected [22,74]. To minimize competitive interference, the concentration of probes should again be kept low compared to epitopes. It is also desirable to ensure the probes do not bind the endogenous epitopes for too long. One way to estimate how long fluorescent probes bind target epitopes – assuming the epitopes are relatively immobile – is to photobleach the probes and see how long it takes the fluorescence to recover [74]. When imaging translation, in contrast, competitive interference is not really an issue (since the epitopes are exogenous and therefore should not be bound by other endogenous proteins). However, the large numbers of probes that are recruited to the repeated epitope tag can create a huge macromolecular complex, on the order of megadaltons. It is easy to imagine that this large, unnatural complex would somehow interfere with normal translation. So far this does not seem to be the case, as there is relatively good agreement between translation rates measured with repeat epitope tags of different sizes, including a 5×, 10×, 24×, and 56× SunTag [17-21] and a 10× FLAG or HA spaghetti monster tag [19]. In fact, the largest (56× SunTag) and one of the smallest (10× spaghetti monster) tags both gave relatively fast elongation rates (15-18 aa/sec and 8-12 aa/sec, respectively), whereas medium sized tags had lower rates (3-6 aa/sec), suggesting little correlation between tag size and measured translation kinetics. Nevertheless, tags and probes may still interfere with normal transcript and/or protein behavior. As an example, when a 56× SunTag was inserted into the 5′ end of one of the endogenous alleles coding for RNA polymerase II, even though transcripts appeared and were translated, the tagged polymerases were unable to enter the nucleus [21]. Thus, depending on the specific application, it is important to check multiple tag sizes and their positioning within genes to properly assess whether or not tags interfere with the dynamics being measured.
Concluding Remarks and Future Perspectives
Antibody-based probes are now being used more than ever to push the limits of live-cell imaging of gene regulatory dynamics. Their ability to bind to and amplify signal from small epitopes makes them ideal tools for seeing traditionally “hard-to-see” gene products, particularly nascent polypeptide chains being translated and chemically modified protein sub-populations. Given these unique advantages, we anticipate the development and use of antibody-based probes will only continue to grow. Now that the experimental groundwork has been laid for live-cell imaging of single RNA translation dynamics, gene expression networks can finally be dissected with ultimate single gene resolution in vivo. This will lead to a better understanding of how transcription and translation dynamics are precisely coupled and controlled in space and time within living cells and how these coupled dynamics help establish specific cellular identities in the face of molecular noise. Similarly, the ability to image post-translational protein modification dynamics in vivo makes it possible to relate the biochemical histories of proteins to their unique behaviors. This will lead to a better understanding of how specific sub-populations of proteins work in an orchestrated fashion to fine-tune genetic networks and ultimately help define the phenotypes of cells and organisms.
To accelerate future applications, what is now needed is a concerted effort to develop more and better probes. A high priority is to increase the number of genetically-encoded antibody-based probes targeting small peptide epitopes, whether they be endogenous or exogenous. These probes would find broad application, not just for multicolor imaging in vivo, but also potentially for combinatorial genome editing purposes with CRISPR-Cas9 [96]. In fact, the SunTag was originally developed with this in mind [97,98]. Here it will be important to not only improve the efficiency of scFv production [32], but also move away from the scFv scaffold altogether. Other antibodymimetics that have potential for live-cell imaging include single chain camelid nanobodies and/or chromobodies [99], monobodies [100], affibodies [101], and DARPINs [102]. Among these and other peptide-binding pairs [103,104], it will be interesting to see which leads to the smallest possible live-cell imaging tag/probe combination (See Outstanding Questions). Another high priority is reducing the background problem inherent to antibody-based imaging. One possible solution would be the design of a probe that lights up only when bound to its epitope. Split GFP satisfies this requirement. Specifically, the small GFP11 fragment can be used as an epitope in a tandem repeat tag [105]. Multiple GFP1-10 fragments bind the GFP11 epitopes and fuse with them to create multiple mature GFPs that brightly fluoresce [106]. It remains unclear, however, if the fluorescence matures fast enough to capture the translation process. Finally, antibodies can bind lots of different substrates besides simple peptides. Thus, the general imaging strategy described here could be expanded to include other potential targets not amenable to standard GFP-tagging. Targets could include DNA, RNA, macro-molecular complexes, or even endogenous nascent polypeptide chains. Given this long list of things to do, the future is literally bright for antibody-based imaging of gene expression.
Supplementary Material
Trends Box.
- Antibody fragments (Fab) and single-chain variable fragments (scFv) are useful live-cell imaging probes.
- Fab and scFv can bring pre-formed fluorescence to unfolded or modified peptides in living cells, unlike standard fluorescent protein fusion tags.
- Fab need to be loaded into cells to image protein dynamics; scFv can be genetically expressed.
- Fab and scFv have recently been used to image and quantify single-mRNA translation kinetics in living cells, yielding consistent estimates of average initiation and elongation rates.
- Fab and scFv have recently been used to image and quantify post-translation modifications to histones and RNA polymerase II in living cells, revealing their spatiotemporal co-regulation.
- For imaging translation, probes should outnumber targets; for imaging endogenous post-translational modifications, targets should outnumber probes.
Outstanding questions.
Besides Fab and scFv, what other types of probes can be used to image translational and post-translational gene regulatory dynamics? Will affibodies, monobodies, chromobodies, or DARPins work? How minimalistic can a probe/tag combo get for live-cell imaging purposes?
What is the underlying cause of the diversity of translation dynamics so far observed in living cells? Can new probes be developed to simultaneously quantify all steps of translation, including scanning, initiation, elongation, and termination?
Can scFv for live-cell imaging be produce more efficiently? Can the solubility of an scFv in living cells be predicted from its sequence?
Can the background problem from freely diffusing probes be solved? Is it possible to design probes that rapidly bind and light up epitopes for imaging translational and post-translation protein dynamics?
Acknowledgments
We thank all members of the Stasevich lab for their helpful discussion and comments. T.J.S. and K.L. are supported by the W. M. Keck Foundation and the NIH (R35GM119728). T.J.S is also supported by funds from the Boettcher Foundation's Webb-Waring Biomedical Research Program.
Glossary Terms
- Fab
An antibody fragment (antigen-binding fragment) composed of the variable light and heavy chains as well as the constant light and heavy chains of an immunoglobin
- Fluorescent fusion tags
Protein fluorophores (like the green fluorescent protein) that can be genetically fused to a protein of interest
- Mintbody
A modification specific intracellular antibody (scFv) fused to a fluorescent protein for live-cell imaging
- Polysome
Multiple ribosomes in complex with an mRNA they are translating
- Post-translational modification (PTM)
A modification to a protein after translation that can impact protein function. Common chemical modifications include phosphorylation, acetylation, and methylation
- scFv
A single-chain variable fragment in which the variable light and heavy regions of an immunoglobin are genetically fused to create an antibody-mimetic
- Synthetic dyes
Small molecular fluorophores that can be conjugated to purified proteins
- UTRs (5′ and 3′)
Untranslated regions of an mRNA that are outside of the protein coding region
- FlAsH and ReAsH
Fluorescein arsenical hairpin binder (FlAsH) and resorufin arsenical hairpin binder (ReAsh). Biarsenical dyes that can covalently bind to a tetracysetine motif (CCXXCC) and then become fluorescent
- TRICK
Translating RNA imaging by coat protein knock off (TRICK). A method that can distinguish populations of translated RNA from untranslated RNA. This method relies on two different fluorescent coat proteins (MS2 and PP7) that each bind a distinct RNA hairpin. Tandem repeats of PP7 hairpins are placed in the coding region of the RNA and tandem repeats of MS2 hairpins are placed in the 3′UTR. As the RNA is being translated, the PP7 coat proteins bound to the PP7 hairpins are knocked off, resulting in a loss in PP7 fluorescence at the translation site
- In vivo
In the context of this review, in vivo is defined as any experiments carried out inside of a living system, the smallest unit being a living cell in a culture dish
- In vitro
In the context of this review, in vitro is defined as any experiment carried out inside of a test tube, such as bulk cell lysate assays
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Crick F. Central Dogma of Molecular Biology. Nature. 1970;227:561–563. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- 2.Li GW, Xie XS. Central dogma at the single-molecule level in living cells. Nature. 2011;475:308–315. doi: 10.1038/nature10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Darzacq X, et al. Imaging transcription in living cells. Annu Rev Biophys. 2009;38:173–196. doi: 10.1146/annurev.biophys.050708.133728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hager GL, et al. Transcription dynamics. Mol Cell. 2009;35:741–753. doi: 10.1016/j.molcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larson DR, et al. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science. 2011;332:475–478. doi: 10.1126/science.1202142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shimomura O. The discovery of aequorin and green fluorescent protein. J Microsc. 2005;217:3–15. doi: 10.1111/j.0022-2720.2005.01441.x. [DOI] [PubMed] [Google Scholar]
- 7.Tsien RY. THE GREEN FLUORESCENT PROTEIN. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 8.Chalfie M. GFP: Lighting up life. Proc Natl Acad Sci U S A. 2009;106:10073–10080. doi: 10.1073/pnas.0904061106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chao JA, et al. Imaging Translation in Single Cells Using Fluorescent Microscopy. Cold Spring Harb Perspect Biol. 2012;4:a012310–a012310. doi: 10.1101/cshperspect.a012310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halstead JM, et al. An RNA biosensor for imaging the first round of translation from single cells to living animals. Science (80- ) 2015;347:1367–1671. doi: 10.1126/science.aaa3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henikoff S, Shilatifard A. Histone modification: cause or cog? Trends Genet. 2011;27:389–396. doi: 10.1016/j.tig.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 13.Ecker JR, et al. Genomics: ENCODE explained. Nature. 2012;489:52–55. doi: 10.1038/489052a. [DOI] [PubMed] [Google Scholar]
- 14.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 15.Zhao Y, et al. Histone acetylation regulates both transcription initiation and elongation of hsp22 gene in Drosophila. Biochem Biophys Res Commun. 2005;326:811–816. doi: 10.1016/j.bbrc.2004.11.118. [DOI] [PubMed] [Google Scholar]
- 16.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu B, et al. Translation dynamics of single mRNAs in live cells and neurons. Science. 2016;352:1430–5. doi: 10.1126/science.aaf1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan X, et al. Dynamics of Translation of Single mRNA Molecules In Vivo. Cell. 2016;165:976–989. doi: 10.1016/j.cell.2016.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morisaki T, et al. Real-time quantification of single RNA translation dynamics in living cells. Science. 2016;352:1425–9. doi: 10.1126/science.aaf0899. [DOI] [PubMed] [Google Scholar]
- 20.Wang C, et al. Real-Time Imaging of Translation on Single mRNA Transcripts in Live Cells. Cell. 2016;165:990–1001. doi: 10.1016/j.cell.2016.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pichon X, et al. Visualization of single endogenous polysomes reveals the dynamics of translation in live human cells. J Cell Biol. 2016;214:769–81. doi: 10.1083/jcb.201605024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayashi-Takanaka Y, et al. Tracking epigenetic histone modifications in single cells using Fab-based live endogenous modification labeling. Nucleic Acids Res. 2011;39:6475–6488. doi: 10.1093/nar/gkr343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimura H, et al. Visualizing posttranslational and epigenetic modifications of endogenous proteins in vivo. Histochem Cell Biol. 2015;144:101–109. doi: 10.1007/s00418-015-1344-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka KAK, et al. Membrane molecules mobile even after chemical fixation. Nat Methods. 2010;7:865–866. doi: 10.1038/nmeth.f.314. [DOI] [PubMed] [Google Scholar]
- 25.Coulter A, Harris R. Simplified preparation of rabbit fab fragments. J Immunol Methods. 1983;59:199–203. doi: 10.1016/0022-1759(83)90031-5. [DOI] [PubMed] [Google Scholar]
- 26.Huston JS, et al. Protein engineering of antibody binding sites: recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc Natl Acad Sci. 1988;85:5879–5883. doi: 10.1073/pnas.85.16.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mariant M, et al. A new enzymatic method to obtain high-yield F(ab)2 suitable for clinical use from mouse IgGl. Mol Immunol. 1991;28:69–77. doi: 10.1016/0161-5890(91)90088-2. [DOI] [PubMed] [Google Scholar]
- 28.Liu H, May K. Disulfide bond structures of IgG molecules. MAbs. 2012;4:17–23. doi: 10.4161/mabs.4.1.18347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stasevich TJ, et al. Quantifying histone and RNA polymerase II post-translational modification dynamics in mother and daughter cells. Methods. 2014;70:77–88. doi: 10.1016/j.ymeth.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 30.Yusakul G, et al. Effect of linker length between variable domains of single chain variable fragment antibody against daidzin on its reactivity. Biosci Biotechnol Biochem. 2016;80:1306–1312. doi: 10.1080/09168451.2016.1156482. [DOI] [PubMed] [Google Scholar]
- 31.Sato Y, et al. Genetically encoded system to track histone modification in vivo. Sci Rep. 2013;3:2436. doi: 10.1038/srep02436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sato Y, et al. A genetically encoded probe for live-cell imaging of H4K20 monomethylation. J Mol Biol. 2016;428:3885–3902. doi: 10.1016/j.jmb.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 33.Hanes J, et al. Ribosome display efficiently selects and evolves high-affinity antibodies in vitro from immune libraries. Proc Natl Acad Sci U S A. 1998;95:14130–5. doi: 10.1073/pnas.95.24.14130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Worn A, et al. Correlation between in Vitro Stability and in Vivo Performance of Anti-GCN4 Intrabodies as Cytoplasmic Inhibitors. J Biol Chem. 2000;275:2795–2803. doi: 10.1074/jbc.275.4.2795. [DOI] [PubMed] [Google Scholar]
- 35.Tanenbaum ME, et al. A Protein-Tagging System for Signal Amplification in Gene Expression and Fluorescence Imaging. Cell. 2014;159:635–646. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gandhi MJ, et al. Lot-to-lot variability in HLA antibody screening using a multiplexed bead-based assay. Transfusion. 2013;53:1940–1947. doi: 10.1111/trf.12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McNeil PL, Warder E. Glass beads load macromolecules into living cells. J Cell Sci. 1987;88(Pt 5):669–678. doi: 10.1242/jcs.88.5.669. [DOI] [PubMed] [Google Scholar]
- 38.Hayashi-Takanaka Y, et al. Visualizing histone modifications in living cells: spatiotemporal dynamics of H3 phosphorylation during interphase. 2009;187:781–790. doi: 10.1083/jcb.200904137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manders EM, et al. Direct imaging of DNA in living cells reveals the dynamics of chromosome formation. J Cell Biol. 1999;144:813–821. doi: 10.1083/jcb.144.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boukany PE, et al. Nanochannel electroporation delivers precise amounts of biomolecules into living cells. Nat Nanotechnol. 2011;6:747–754. doi: 10.1038/nnano.2011.164. [DOI] [PubMed] [Google Scholar]
- 41.Teng KW, et al. Labeling proteins inside living cells using external fluorophores for microscopy. Elife. 2016;5:1–13. doi: 10.7554/eLife.20378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iwasaki S, Ingolia NT. Seeing translation. Science (80- ) 2016;352:1391–1392. doi: 10.1126/science.aag1039. [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez AJ, et al. Visualization of mRNA translation in living cells. J Cell Biol. 2006;175:67–76. doi: 10.1083/jcb.200512137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Engelenburg SB, et al. FACS-Based selection of tandem tetracysteine peptides with improved ReAsH brightness in live cells. ChemBioChem. 2010;11:489–493. doi: 10.1002/cbic.200900689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hopp TP, et al. A Short Polypeptide Marker Sequence Useful for Recombinant Protein Identification and Purification. Bio/Technology. 1988;6:1204–1210. [Google Scholar]
- 46.Green N, et al. Immunogenic structure of the influenza virus hemagglutinin. Cell. 1982;28:477–87. doi: 10.1016/0092-8674(82)90202-1. [DOI] [PubMed] [Google Scholar]
- 47.Viswanathan S, et al. High-performance probes for light and electron microscopy. Nat Methods. 2015;12:568–576. doi: 10.1038/nmeth.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bertrand E, et al. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 49.Chao JA, et al. Structural basis for the coevolution of a viral RNA–protein complex. Nat Struct Mol Biol. 2008;15:103–105. doi: 10.1038/nsmb1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coleman RA, et al. Imaging Transcription: Past, Present, and Future. Cold Spring Harb Symp Quant Biol. 2015;80:1–8. doi: 10.1101/sqb.2015.80.027201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coulon A, Larson DR. Fluctuation Analysis Dissecting Transcriptional Kinetics with Signal Theory. 1st. Vol. 572. Elsevier Inc; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ingolia NT, et al. Genome-Wide Analysis in Vivo of Translation with Nucleotide Resolution Using Ribosome Profiling. Science (80- ) 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ingolia NT, et al. Ribosome Profiling of Mouse Embryonic Stem Cells Reveals the Complexity and Dynamics of Mammalian Proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Capece MC, et al. A simple real-time assay for in vitro translation. Rna. 2014;21:296–305. doi: 10.1261/rna.047159.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alkalaeva EZ, et al. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125:1125–36. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- 56.Barrett RM, et al. Cell-specific Profiling of Nascent Proteomes Using Orthogonal Enzyme-mediated Puromycin Incorporation. ACS Chem Biol. 2016;11:1532–1536. doi: 10.1021/acschembio.5b01076. [DOI] [PubMed] [Google Scholar]
- 57.Signer RAJ, et al. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature. 2014;509:49–54. doi: 10.1038/nature13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu J, et al. Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc Natl Acad Sci. 2012;109:413–418. doi: 10.1073/pnas.1111561108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Floor SN, Doudna JA. Tunable protein synthesis by transcript isoforms in human cells. Elife. 2016;5:1–25. doi: 10.7554/eLife.10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rolfe DF, Brown GC. Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiol Rev. 1997;77:731–58. doi: 10.1152/physrev.1997.77.3.731. [DOI] [PubMed] [Google Scholar]
- 61.Moldave K. Eukaryotic protein synthesis. Annu Rev Biochem. 1985;54:1109–49. doi: 10.1146/annurev.bi.54.070185.005333. [DOI] [PubMed] [Google Scholar]
- 62.Hinnebusch AG. The Scanning Mechanism of Eukaryotic Translation Initiation. Annu Rev Biochem. 2014;83:779–812. doi: 10.1146/annurev-biochem-060713-035802. [DOI] [PubMed] [Google Scholar]
- 63.Brar GA. Beyond the Triplet Code: Context Cues Transform Translation. Cell. 2016;167:1681–1692. doi: 10.1016/j.cell.2016.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stumpf C, et al. The Translational Landscape of the Mammalian Cell Cycle. Mol Cell. 2013;52:574–582. doi: 10.1016/j.molcel.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhouravleva G, et al. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995;14:4065–72. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ito K, Chiba S. Arrest Peptides: Cis -Acting Modulators of Translation. Annu Rev Biochem. 2013;82:171–202. doi: 10.1146/annurev-biochem-080211-105026. [DOI] [PubMed] [Google Scholar]
- 67.Doyle M, Kiebler MA. Mechanisms of dendritic mRNA transport and its role in synaptic tagging. EMBO J. 2011;30:3540–3552. doi: 10.1038/emboj.2011.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nathans D. PUROMYCIN INHIBITION OF PROTEIN SYNTHESIS: INCORPORATION OF PUROMYCIN INTO PEPTIDE CHAINS. Proc Natl Acad Sci U S A. 1964;51:585–92. doi: 10.1073/pnas.51.4.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schneider-Poetsch T, et al. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat Chem Biol. 2010;6:209–217. doi: 10.1038/nchembio.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Beck-Sickinger AG, Mörl K. Posttranslational Modification of Proteins. Expanding Nature's Inventory. In: Christopher T Walsh., editor. Angew Chemie Int Ed. Vol. 45. 2006. pp. 1020–1020. [Google Scholar]
- 71.Zhou VW, et al. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 72.Kimura H, et al. The organization of histone H3 modifications as revealed by a panel of specific monoclonal antibodies. Cell Struct Funct. 2008;33:61–73. doi: 10.1247/csf.07035. [DOI] [PubMed] [Google Scholar]
- 73.Hayashi-Takanaka Y, et al. Distribution of histone H4 modifications as revealed by a panel of specific monoclonal antibodies. Chromosom Res. 2015;23:753–766. doi: 10.1007/s10577-015-9486-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stasevich TJ, et al. Regulation of RNA polymerase II activation by histone acetylation in single living cells. Nature. 2014;516:272–275. doi: 10.1038/nature13714. [DOI] [PubMed] [Google Scholar]
- 75.Bah A, Forman-Kay JD. Modulation of Intrinsically Disordered Protein Function by Post-translational Modifications. J Biol Chem. 2016;291:6696–6705. doi: 10.1074/jbc.R115.695056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bailey AO, et al. Identification of the Post-translational Modifications Present in Centromeric Chromatin. Mol Cell Proteomics. 2016;15:918–931. doi: 10.1074/mcp.M115.053710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Strzyz P. Post-translational modifications: Extension of the tubulin code. Nat Rev Mol Cell Biol. 2016;17:609–609. doi: 10.1038/nrm.2016.117. [DOI] [PubMed] [Google Scholar]
- 78.Mann M, Jensen ON. Proteomic analysis of post-translational modifications. Nat Biotechnol. 2003;21:255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- 79.Shogren-Knaak M. Histone H4-K16 Acetylation Controls Chromatin Structure and Protein Interactions. Science (80- ) 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 80.Roque A, et al. Post-translational modifications of the intrinsically disordered terminal domains of histone H1: effects on secondary structure and chromatin dynamics. Chromosoma. 2016 doi: 10.1007/s00412-016-0591-8. [DOI] [PubMed] [Google Scholar]
- 81.Gu W, Roeder RG. Activation of p53 Sequence-Specific DNA Binding by Acetylation of the p53 C-Terminal Domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 82.Buratowski S. Progression through the RNA Polymerase II CTD Cycle. Mol Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 84.Egloff S et al. Updating the RNA polymerase CTD code: Adding gene-specific layers. Trends in Genetics. 2012 Jul 28;:333–341. doi: 10.1016/j.tig.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 85.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 86.Chi P, et al. Covalent histone modifications — miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- 88.Zhang R, et al. Histone Acetylation Regulates Chromatin Accessibility: Role of H4K16 in Inter-nucleosome Interaction. Biophys J. 2016;015 doi: 10.1016/j.bpj.2016.11.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Segala G, et al. Monoubiquitination of Histone H2B Blocks Eviction of Histone Variant H2A.Z from Inducible Enhancers. Mol Cell. 2016;64:334–346. doi: 10.1016/j.molcel.2016.08.034. [DOI] [PubMed] [Google Scholar]
- 90.Kimura H. Histone modifications for human epigenome analysis. J Hum Genet. 2013;58:439–445. doi: 10.1038/jhg.2013.66. [DOI] [PubMed] [Google Scholar]
- 91.Yuan K, O'Farrell PH. TALE-light imaging reveals maternally guided, H3K9me2/3-independent emergence of functional heterochromatin in Drosophila embryos. Genes Dev. 2016;30:579–593. doi: 10.1101/gad.272237.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sasaki K, Yoshida M. Genetically encoded FRET indicators for live-cell imaging of histone acetylation. Methods Mol Biol. 2014;1071:151–161. doi: 10.1007/978-1-62703-622-1_12. [DOI] [PubMed] [Google Scholar]
- 93.Sasaki K, et al. Development of live-cell imaging probes for monitoring histone modifications. Bioorg Med Chem. 2012;20:1887–1892. doi: 10.1016/j.bmc.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 94.Sasaki K, et al. Real-time imaging of histone H4 hyperacetylation in living cells. Proc Natl Acad Sci. 2009;106:16257–16262. doi: 10.1073/pnas.0902150106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sekar TV, et al. Genetically Encoded Molecular Biosensors To Image Histone Methylation in Living Animals. Anal Chem. 2015;87:892–899. doi: 10.1021/ac502629r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bruce VJ, et al. Minimalist Antibodies and Mimetics: An Update and Recent Applications. ChemBioChem. 2016;17:1892–1899. doi: 10.1002/cbic.201600303. [DOI] [PubMed] [Google Scholar]
- 97.Tanenbaum MEE, et al. A Protein-Tagging System for Signal Amplification in Gene Expression and Fluorescence Imaging. Cell. 2014;159:635–646. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gilbert LA, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Panza P, et al. Live imaging of endogenous protein dynamics in zebrafish using chromobodies. Development. 2015;142:1879–1884. doi: 10.1242/dev.118943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Batori V, et al. Exploring the potential of the monobody scaffold: effects of loop elongation on the stability of a fibronectin type III domain. Protein Eng. 2002;15:1015–20. doi: 10.1093/protein/15.12.1015. [DOI] [PubMed] [Google Scholar]
- 101.Gao J, et al. Affibody-based nanoprobes for HER2-expressing cell and tumor imaging. Biomaterials. 2011;32:2141–2148. doi: 10.1016/j.biomaterials.2010.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mironova KE, et al. Highly specific hybrid protein DARPin-mCherry for fluorescent visualization of cells overexpressing tumor marker HER2/neu. Biochem. 2014;79:1391–1396. doi: 10.1134/S0006297914120141. [DOI] [PubMed] [Google Scholar]
- 103.Zhao N, et al. Hyperthermostable binding molecules on phage: Assay components for point-of-care diagnostics for active tuberculosis infection. Anal Biochem. 2017 doi: 10.1016/j.ab.2016.12.021. [DOI] [PubMed] [Google Scholar]
- 104.Zhao N, et al. Phage display selection of tight specific binding variants from a hyperthermostable Sso7d scaffold protein library. FEBS J. 2016;283:1351–1367. doi: 10.1111/febs.13674. [DOI] [PubMed] [Google Scholar]
- 105.Kent KP, et al. Deconstructing Green Fluorescent Protein. J Am Chem Soc. 2008;130:9664–9665. doi: 10.1021/ja803782x. [DOI] [PubMed] [Google Scholar]
- 106.Kamiyama D, et al. Versatile protein tagging in cells with split fluorescent protein. Nat Commun. 2016;7:11046. doi: 10.1038/ncomms11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.