Abstract
Introduction
Prolonged fasting in northern elephant seals (NES) is characterized by a reliance on lipid metabolism, conservation of protein, and reduced plasma insulin. During early fasting, glucose infusion previously reduced plasma free fatty acids (FFA); however, during late-fasting, it induced an atypical elevation in FFA despite comparable increases in insulin during both periods suggestive of a dynamic shift in tissue responsiveness to glucose-stimulated insulin secretion.
Objective
To better assess the contribution of insulin to this fasting-associated shift in substrate metabolism.
Methods
We compared the responses of plasma metabolites (amino acids (AA), FFA, endocannabinoids (EC), and primary carbon metabolites (PCM)) to an insulin infusion (65 mU/kg) in early- and late-fasted NES pups (n = 5/group). Plasma samples were collected prior to infusion (T0) and at 10, 30, 60, and 120 min post-infusion, and underwent untargeted and targeted metabolomics analyses utilizing a variety of GC-MS and LC-MS technologies.
Results
In early fasting, the majority (72%) of metabolite trajectories return to baseline levels within 2 h, but not in late fasting indicative of an increase in tissue sensitivity to insulin. In late-fasting, increases in FFA and ketone pools, coupled with decreases in AA and PCM, indicate a shift toward lipolysis, beta-oxidation, ketone metabolism, and decreased protein catabolism. Conversely, insulin increased PCM AUC in late fasting suggesting that gluconeogenic pathways are activated. Insulin also decreased FFA AUC between early and late fasting suggesting that insulin suppresses triglyceride hydrolysis.
Conclusion
Naturally adapted tolerance to prolonged fasting in these mammals is likely accomplished by suppressing insulin levels and activity, providing novel insight on the evolution of insulin during a condition of temporary, reversible insulin resistance.
Keywords: Endocannabinoids, Fatty acids, Lipidomics, Metabolomics, Substrate metabolism
Introduction
The classical actions of insulin are to stimulate cellular glucose uptake and promote anabolic processes (Sonksen and Sonksen 2000). Thus, during periods of starvation or extended food deprivation, insulin and insulin signaling are suppressed to help preserve circulating glucose to support the energetic burdens imposed by the caloric restriction/reduction (DeFronzo et al. 1978). Therefore, brief and acute bouts of reversible insulin resistance are adaptive in most vertebrates to ameliorate the stress of short-term or abbreviated periods of food deprivation. However, prolonged (2–3 months) fasting is a natural component of the life history of the northern elephant seal (NES) that is characterized by reliance on lipid oxidation, conservation of protein, and reversible adipose-specific insulin resistance (Adams and Costa 1993; Houser and Costa 2001; Viscarra et al. 2013; Crocker et al. 1998; C. L. Ortiz 1978).
Glucose-stimulated insulin secretion, similar to what is achieved during an oral glucose tolerance test (oGTT), typically reduces plasma non-esterified free fatty acids (NEFA) (Karpe et al. 2011), and this response is consistent during early fasting in NES pups (Viscarra et al. 2011a). However, during late fasting, an oGTT induced an atypical elevation in NEFA suggesting that prolonged fasting in seals initiates a dynamic shift in substrate utilization, especially as it relates to lipid metabolism. This phenotype in late-fasted seal pups is consistent with the development of adipose-specific insulin resistance and demonstrates the remarkable plasticity of their metabolic network (being able to switch between extreme stages of body mass gain and prolonged fasting independent of consequences on energy balance). Furthermore, a detailed acylcarnitine profile in early- and late-fasted NES pups revealed that total plasma acylcarnitine and acyl:free carnitine ratio increased, indicative of increased accumulation of plasma fatty acylcarnitines and of a higher degree of incomplete β-oxidation with fasting-induced insulin resistance (Viscarra et al. 2012). The fasting-associated increase in plasma acyl:free carnitine ratio suggests that a temporal change in the dynamics of mitochondrial β-oxidation occurs with fasting duration in NES pups (Adams et al. 1992). This acylcarnitine profile also revealed higher levels of medium-chain fatty acid acylcarnitine derivatives that are consistent with those observed in insulin-resistant mice (Koves et al. 2008) and type 2 diabetic humans (Adams et al. 2009). These patterns of acylcarnitine derivatives are also indicative of increased lipid metabolism and decreased protein catabolism (Viscarra et al. 2012). Thus, high-throughput analyses such as acylcarnitine profiles are extremely useful in helping to elucidate potential cellular mechanisms and more thoroughly describe dynamic shifts in substrate utilization and metabolism.
Extensive metabolomic studies in diabetic subjects (Rhee et al. 2011; Wang et al. 2011; Mihalik et al. 2012; Floegel et al. 2013; Liu et al. 2015; Palmer et al. 2015) and animals (Koves et al. 2008; Li et al. 2010), and the metabolomic response to an oGTT in humans have been performed (Wopereis et al. 2009; Zhao et al. 2009; Lanza et al. 2010; Spégel 2010). These studies demonstrate distinct alterations in amino acid and branched-chain fatty acid metabolism between normoglycemic and insulin resistant/diabetic subjects (Liu et al. 2015; Rhee et al. 2011; Wang et al. 2011; Mihalik et al. 2012; Du et al. 2013; Floegel et al. 2013; Palmer et al. 2015). Furthermore, the difference in metabolomic response to an oGTT between normal weight/glycemic subjects and their obese/diabetic counterparts is strikingly distinct and can offer significant insight as to which cellular mechanisms are perturbed (i.e., TCA cycle, mitochondrial β-oxidation, gluconeogenesis, lipolysis etc.). However, during an oGTT, circulating insulin is elevated via glucose-stimulated insulin secretion, making it difficult to differentiate the direct glucose from the insulin-mediated effects. Unfortunately, to the best of our knowledge, the metabolomic and lipidomic responses to insulin infusion have not been reported. Therefore, to better assess this insulin-associated shift in substrate metabolism, we compared the response of plasma metabolites to insulin infusion in early- and late-fasted NES pups, which exhibit a unique insulin resistance-like metabolic phenotype with fasting duration.
Methods
Animals
All procedures were reviewed and approved by the Institutional Animal Care and Use Committees of the University of California, Merced and California State University, Sonoma. Northern elephant seal pups constituting two different cohorts at Año Nuevo State Reserve were studied at two postweaning periods (n = 5/period): early (1–2 weeks postweaning; 127 ± 1 kg) and late (6–7 weeks postweaning; 93 ± 4 kg). Pups were weighed, sedated, and infused in the field as previously described (Viscarra et al. 2011b; Viscarra et al. 2011a; Viscarra et al. 2013). Briefly, pups were sedated with 1mg/kg Telazol (tiletamine/zolazepam HCl, Fort Dodge Labs, Ft Dodge, IA) administered intramuscularly. Once immobilized, an 18 gauge, 3.5 inch spinal needle was inserted into the extradural vein. Blood samples were obtained, and infusions performed from this site. Continuous immobilization was maintained with ~100mg bolus intravenous injections of ketamine as needed (Champagne et al. 2012).
Insulin infusion protocol
To determine the metabolomic and lipidomic responses to insulin as a function of fasting-induced, adipose-specific insulin resistance, fasting seal pups were infused (i.v.) with a mass-specific dose (0.065 U insulin/kg) (Humulin; Eli Lilly, Indianapolis, IN) as previously described (Viscarra et al. 2013). Time-course responses to insulin by comparing the differences of the area under the curve (AUC) values yield information about the metabolic plasticity of the animals in the context of fasting duration. Prior to each infusion, a pre-infusion blood sample (i.v.) was collected (baseline or T0). Following the bolus infusion of insulin, blood samples collected at 10, 30, 60, and 120 min were used for this suite of metabolomic/lipidomic analyses. Procedures were terminated at 120 min out of concern for the safety of the animals. Immediately following the collection of the 120 min samples, glucose was infused slowly to assist in the restoration of pre-infusion levels. Blood samples were centrifuged on site for 15 min at 3000g, and the plasma was transferred to cryo-vials, frozen by immersion in liquid nitrogen, and stored at −80°C.
Analysis of primary metabolites
The MiniX database (Scholz and Fiehn 2007) was used as a Laboratory Information Management System (LIMS) and for sample randomization prior to all analytical procedures.
Sample preparation
Aliquots of plasma (30μL) stored at −80°C were thawed, extracted, derivatized, and the metabolite abundances quantified by gas chromatography time-of-flight (GCTOF) mass spectrometry (MS) as previously described (Fiehn 2008). Briefly, the aliquots were extracted with 1mL of degassed acetonitrile/isopropanol/water (3:3:2) solution at −20°C, centrifuged, the supernatant removed, and solvents evaporated to dryness under reduced pressure. To remove membrane lipids and triglycerides, dried samples were reconstituted with acetonitrile/water (1:1), decanted and taken to dryness under reduced pressure. Internal standards, C8–C30 fatty acid methyl esters, were added to samples and derivatized with methoxyamine hydrochloride in pyridine and subsequently by N-methyl-N-(trimethylsilyl) trifluoroacetamide (Sigma-Aldrich) for trimethylsilylation of acidic protons.
GCTOF data acquisition and processing
Derivatized samples were analyzed on an Agilent 7890A gas chromatograph (Santa Clara, CA) with a 30m long, 0.25mm i.d. Rtx5Sil-MS column with 0.25μm 5% diphenyl film with an additional 10m integrated guard column (Restek, Bellefonte PA) (Fiehn 2008; Kind et al. 2007). An aliquot (0.5μL) was injected at 50°C (ramped to 250°C) in splitless mode with a 25 sec splitless time. The chromatographic gradient consisted of a constant flow of 1mL/min, ramping the oven temperature from 50°C to 330°C over 22 min. Mass spectrometry was performed using a Leco Pegasus IV time-of-flight mass spectrometer, 230°C transfer line temperature, electron ionization at −70V, and an ion source temperature of 250°C. Mass spectra were acquired at 1800V detector voltage at m/z 85–500 with 17 spectra/sec. Acquired spectra were further processed using the BinBase database (Fiehn 2005; Scholz and Fiehn 2007). Briefly, output results were filtered based on multiple parameters to exclude noisy or inconsistent peaks (Kind et al. 2007). Detailed criteria for peak reporting included mass spectral matching, spectral purity, signal-to-noise ratio, and retention time (Fiehn 2005). All entries in BinBase were matched against the Fiehn mass spectral library of 1,200 authentic metabolite spectra using retention index and mass spectrum information or the NIST11 commercial library. All samples were analyzed in one batch, and data quality and instrument performance were constantly monitored using quality control and reference plasma samples (National Institute of Standards and Technology 2011). Quality controls were comprised of a mixture of standards and analyzed every 10 samples, were monitored for changes in the ratio of the analyte peak heights, and used to ensure equivalent conditions within the instrument (p>0.05, t-Test comparing observed to expected ratios of analyte response factors) over the duration of the sample acquisition (Fiehn 2008). Pooled plasma samples (n=9) were included and served as additional quality controls to assess normalization efficiency. Metabolites were reported if present in at least 50% of the samples. Data reported as quantitative ion peak heights were normalized by the sum intensity of all annotated metabolites and used for further statistical analysis.
Targeted metabolomics analyses of non-esterified fatty acids and endocannabinoids: Non-esterified fatty acid sample preparation, data acquisition, and processing
Plasma NEFA were isolated as previously described (Smedes 1999; Gladine et al. 2014). Specifically, plasma aliquots (100mL) were enriched with 5mL 0.2mg/mL butylated hydroxytoluene/EDTA in 1:1 methanol/water, and a suite of extraction surrogates, which included deuterated-tri-palmitoyl glycerol (d31-16:0-TG; CDN Isotopes, Pointe-Claire, Quebec, Canada), deuterated distearoylphosphotidylcholine (d35-18:0-PC; Avanti Polar Lipids, Alabaster, Alabama), dodeca-(9E)-enoyl cholesterylesters (22:1n9-CE; NuChek Prep, Elysian MN) and dodecatrienoic acid (22:3n3; NuChek Prep). Lipids were then extracted with cyclohexane/2-propanol/ammonium acetate (10:8:11). Enriched samples were mixed with cyclopropane/2-propanol (10:8:11) and the phases split with ammonium acetate. The organic phase was isolated and the aqueous phase was re-extracted with cyclohexane. The combined organic total lipid extract was reduced to dryness and reconstituted in 200μL of methanol/toluene (1:1), and the total lipid extract was used to quantify plasma fatty acids as methyl esters by GC-MS. Extracted samples were spiked with 15:1n5 free acid to track methylation efficiency, brought to a final volume of 200mL with 90:10 methanol/toluene (v/v), and left at room temperature for 30 min before being dried. The remaining fatty acid methyl esters were re-constituted in a hexane (300mL)/44mM tricosanoate methyl ester (23:0; NuChek Prep) (10μL) solution (30,000:1) and vortexed. A 100μL aliquot was transferred to a GC-MS vial for analysis (Agilent 6890/5973N MSD, Agilent Technologies, San Jose, CA) with electron impact ionization and in simultaneous-selected, ion monitoring/full scan mode. Analytes were separated on a 30m/0.25mm/0.25μm DB-225ms column. Analytes were quantified with ChemStation vE.02.14 (Agilent Technologies) using internal standard methodologies against 5 to 8 point calibration curves.
Endocannabinoid sample preparation, data acquisition, and processing
Endocannabinoids were isolated by solid phase extraction on 10mg Waters Oasis-HLB cartridges (Milford, MA) as previously described (Luria et al. 2007). Prior to extraction, cartridges were washed with 1 column volume ethyl acetate followed by 2 column volumes methanol, and conditioned with 2mL of 95:5 (v/v) water/methanol (MeOH) with 0.1% acetic acid. The column reservoir was spiked with 5μL anti-oxidant solution, (0.2mg/mL BHT/EDTA in 1:1 MeOH/water), and 10μL 1000nM analytical surrogates. Sample aliquots (250μL media) were then introduced to the column reservoir and diluted with 1 column volume wash solution (5% MeOH, 0.1% acetic acid). The sample was gravity extracted and the sorbent bed was washed with 1 column volume of 20% methanol and 0.1% acetic acid. The solid-phase extraction cartridges were dried by vacuum (@ -7.5 in. Hg for 20 min). Analytes were then eluted by gravity with 0.2mL MeOH, followed by 0.5mL acetonitrile, followed by 0.5mL ethyl acetate into 2mL autosampler vials containing 10μL of a 20% glycerol/MeOH solution. Eluent was dried by vacuum evaporation for 35 min, and residues were re-constituted with 100μL of 100nM internal standard solution containing 1-cyclohexylureido, 3-dodecanoic acid (CUDA), in 50:50 MeOH/acetonitrile. Vials were vortexed for 1 min to dissolve residues, chilled 15 min on wet ice, and extracts transferred to a centrifugal filter (0.1μm Durapore, Millipore, Billerica, MA). After centrifugation (3 min at <4500g and 6°C), the extracts were transferred to 150μL glass inserts in 2mL amber vials, capped, and stored at −20°C until analysis by UPLC-MS/MS. The internal standard was used to quantify the recovery of the deuterated extraction surrogates by ratio response.
Endocannabinoid analysis
Analytes in a 10μL injection of extract were separated with an Aquity C18 Ethylene Bridged Hybrid (BEH) 1.7μm, 150mm × 2.1mm column utilizing a Waters Acquity UPLC (Waters, Milford, MA). The solvent gradient is described in Table 1 with a slight modification from Shearer et al (Shearer et al. 2010). The autosampler was maintained at 10°C. Resolved analytes were detected by positive mode electrospray ionization and multiple reaction monitoring on an API 4000 QTrap (AB Sciex, Framingham, MA, USA) using the following operating parameters: curtain gas = 20.0 psi, temperature = 500°C, ion-spray voltage = 5500.00V, collision gas = high, ion source gas 1 & 2 = 40.0 psi, collision cell exit potential = 10.0V, and entrance potential = 10.0V. Analyte retention times, mass transitions, optimized collision and declustering potential voltages, dwell times, and analytical surrogate associations for each analyte are shown in Supplemental Table 1. Analytes were quantified using isotope dilution and internal standard methodology with 5 to 7 point calibration curves (R2 ≥ 0.997). Calibrants and internal standards were either synthesized [CUDA] or purchased from commercial sources (Cayman Chemical, Ann Arbor, MI or Avanti Polar Lipids Inc., Alabaster, AL), unless otherwise indicated. Data was processed utilizing AB Sciex Analyst version 1.6.2. Surrogate recoveries are in Supplemental Table 1.
Table 1.
Mean area under the curves (AUC; ± SD) for significantly different (p < 0.05) metabolites from the plasma metabolite classes (endocannabinoids, fatty acids, amino acids, carbohydrates, ketone bodies, and organic acids) measured for early- and late-fasted northern elephant seal pups. The metabolite’s pathway and metabolite class are identified.
| Metabolite | Pathway | Metabolite class | Early | Late |
|---|---|---|---|---|
| Anandamide (AEA) | Endocannabinoid metabolism | Endocannabinoid | 27 ± 71 | −73 ± 27 |
| Oleoylethanolamide (OEA) | Endocannabinoid metabolism | Endocannabinoid | 269 ± 690 | −627 ± 490 |
| Myristic acid (C14:0) | Lipid metabolism | Fatty acid | 36600 ± 7.2e+05 | −2.4e+06 ± 1.7e+06 |
| Palmitic Acid (C16:0) | Lipid metabolism | Fatty acid | −3320 ± 6000 | −13700 ± 6400* |
| Isoheptadecanoic acid (C17:0) | Lipid metabolism | Fatty acid | −7910 ± 110000 | −137000 ± 47000 |
| Vaccenic Acid (C18:1n7) | Lipid metabolism | Fatty acid | −304 ± 1700 | −3380 ± 2400 |
| Elaidic acid (C18:1n9) | Lipid metabolism | Fatty acid | 4.2e+06 ± 1.6e+06 | −5.5e+06 ± 2.3e+06* |
| Oleic Acid (C18:1n9) | Lipid metabolism | Fatty acid | −3350 ± 11000 | −29300 ± 12000* |
| Linoleic Acid (C18:2n6) | Lipid metabolism | Fatty acid | −93 ± 540 | −1290 ± 830 |
| Alanine | Amino acid metabolism | Amino acid | −9.7e+06 ± 4.6e+06 | 5e+06 ± 6.5e+06 |
| Beta-alanine | Pantothenate and pyrimidine metabolism | Amino acid | −5740 ± 16000 | 21400 ± 8100 |
| Isoleucine | Amino acid metabolism | Amino acid | −1670000 ± 1e+06 | −3.6e+06 ± 1.2e+06 |
| N-acetylglutamate | Amino acid metabolism | Amino acid | −9120 ± 9000 | 9730 ± 13000 |
| Erythritol | Carbohydrate metabolism | Carbohydrate | 607 ± 75000 | 125000 ± 75000 |
| Hexuronic acid | Carbohydrate metabolism, pentose phosphate pathway, ascorbic acid metabolism | Carbohydrate | 18600 ± 31000 | 81800 ± 43000 |
| Isothreonic acid | Carbohydrate metabolism, ascorbic acid metabolism | Carbohydrate | 28400 ± 35000 | 103000 ± 38000 |
| D-arabitol | Carbohydrate metabolism, pentose phosphate pathway | Carbohydrate | 9500 ± 28000 | 52000 ± 15000 |
| Maltose | Carbohydrate metabolism | Carbohydrate | −78600 ± 51000 | 34300 ± 87000 |
| D-Xylose | Carbohydrate metabolism | Carbohydrate | 21100 ± 90000 | 179000 ± 78000* |
| 3-Hydroxybutyric acid | Ketogenesis, Ketolysis | Ketone body | 3.3e+06 ± 5.7e+06 | −1.4e+07 ± 7.2e+06 |
| Hydroxybutyric acid | Ketogenesis, Ketolysis | Ketone body | 82600 ± 35000 | 194000 ± 85000* |
| Indole-3-lactate | Amino acid metabolism | Organic acid | −2110 ± 31000 | 58200 ± 24000* |
| Pyruvic acid | Glycolysis, gluconeogenesis, pyruvate metabolism | Organic acid | 6.5e+05 ± 4.9e+05 | 2.1e+06 ± 7.6e+05* |
Symbol
FDR adjusted p-value <0.05
Statistical analyses
All statistical analyses were performed using R version 3.0.1. The area under the curve (AUC) for t= 0, 10, 30, 60, and 120 min for each metabolite as a function of time post-insulin infusion at each fasting period was calculated based on trapezoidal rule integration. Calculated AUC values were used to summarize the relative change in metabolite concentrations as a function of time for each sample. Variance in AUC values accurately reflects the variance in sample-wise metabolite trends over time. Therefore, the AUC values represent efficient mathematical representations of the original data and enable comparisons between samples’ metabolite concentrations over time. However, there is no assumed biochemical context encoded by the AUC method. Independent sample t-tests were conducted with adjustment for unequal variance. The probability of the test statistics (p-values) were adjusted for multiple hypotheses tested (adjusted p-value) (Benjamini 1995) and the false discovery rate was independently estimated (q-value).
Cluster analysis was conducted using k-means on a self-organizing map (SOM). SOM was calculated on a 10 × 10 hexagonal grid from mean time course patterns for early or late fasting groups. The combined data set of early and late metabolite time course patterns were used to identify similarities between early and late groups. Cluster analysis was conducted using k-means on the 100 generated self-organizing map codebooks. The k-means cluster number was selected based on an evaluation of within-cluster distance for 2 to 50 clusters, with 7 clusters identified as the optimal cluster number. Metabolite to k-means cluster assignments were recovered based on their assignment to SOM codebooks.
Multivariate analyses, principal component analysis (PCA) and orthogonal partial least squares discriminant analysis (O-PLS-DA) were conducted on combined metabolite baseline and AUC values, which were mean centered and scaled to unit variance. PCA was calculated based on the singular value decomposition (Stacklies et al. 2007). PCA sample leverage (distance to center of mass in the PCA plane) and DmodX (projected distance to the PCA plane) were used to evaluate potential extreme and moderate outliers, respectively. O-PLS-DA was used to build a classification model to discriminate between early and late fasting animal baseline and AUC patterns and to identify the top 10% of all variable contributions to the observed class discrimination between the two classes (feature selection). Leave-one-out cross-validation was used to fit a preliminary 2 latent variable (LV) O-PLS-DA model.
The top 10% of all AUC and baseline discriminants (features) were selected based on fulfilling two criteria: (1) correlation with model scores (Wiklund et al. 2008) (Spearman’s rho, P ≤ 0.1) and (2) the absolute value of the model loading on the first latent variable 1 (LV1) ≥ 90th quantile (Palermo et al. 2009), where LV1 is the model component capturing the maximum difference between early and late fasting groups. The classification performance of the selected and excluded feature models was validated and compared using Monte Carlo cross-validation (MCCV) and permutation testing. MCCV was carried out by randomly selecting 2/3 of the animals as a training set (to build models) and using 1/3 of the animals to test the models, while maintaining the proportion of late and early samples in the full dataset. This procedure was repeated 100 times and used to estimate distributions for the model performance statistics. Permutation testing (prediction of randomly assigned phenotype labels) was combined with the described MCCV model cross-validation and used to estimate the probability of achieving the model’s predictive performance by chance, through comparison of the actual model Q2, AUC, sensitivity, and specificity to those of the NULL hypothesis as defined by the permuted models. Permutation p-values (Phipson and Smyth 2010) were calculated to describe the proportion of cross-validation results showing favorable (less than or greater than, depending on the specific metric) performance for the actual model compared to the permuted (random early or late class labels) and selected vs. excluded feature models.
Metabolic trajectory analysis (Keun et al. 2004) was carried out to display geometric differences in early and late fasting animals’ response to insulin infusion. PCA was calculated on baseline (T0) adjusted, centered, and scaled to unit variance metabolomic measurements for t = 0, 10, 30, 60 and 120 min. Separate models were calculated based on all measurements or only the top 10% of all O-PLS-DA selected metabolic discriminants between early and late fasting animals. PCA scores were annotated with median scores and standard errors for each group/time-point pair.
Network analysis was used to assess statistically significant results within a biochemical context. A biochemical and chemical similarity network (Barupal et al. 2012) was calculated for all measured metabolites with KEGG (Kanehisa et al. 2016) and PubChem CID (Kim et al. 2016) identifiers using MetaMapR (Grapov et al. 2015). Enzymatic interactions were determined based on product-precursor relationships defined in the KEGG RPAIR database. Molecules not directly involved in biochemical transformations, but sharing structural properties, based on PubChem Substructure Fingerprints (Cao et al. 2008) were connected at a Tanimoto similarity threshold ≥ 0.7. Pathway enrichment analysis was conducted using Metaboanalyst 3.0 (Xia and Wishart 2011). Significantly different metabolites between early and late-fasting mammals were matched against the Homo sapiens pathway library and analyzed using hypergeometric tests with out-degree centrality.
Results
Prolonged fasting increases β-oxidation and spares lean tissue
We identified 41 out of 171 known metabolites that changed significantly in baseline (T0) values between early and late fasting (p<0.05) (Figure 1 and Supplemental Table 1). These metabolites clustered into seven distinct biochemical categories for analysis: (1) amino acids, (2) endocannabinoids, (3) fatty acids, (4) glucose, (5) ketone bodies, (6) organic acids, and (7) primary carbon metabolites (Figure 1). Prolonged fasting was associated with the most profound changes in FFA. The increases in baseline fatty acid concentrations ranged from 28% (palmitic acid) to 144% (elaidic acid). Similarly, the ketones acetoacetate and 3-hydroxybutyric acid increased 133% and 347% in late fasting, respectively.
Figure 1.
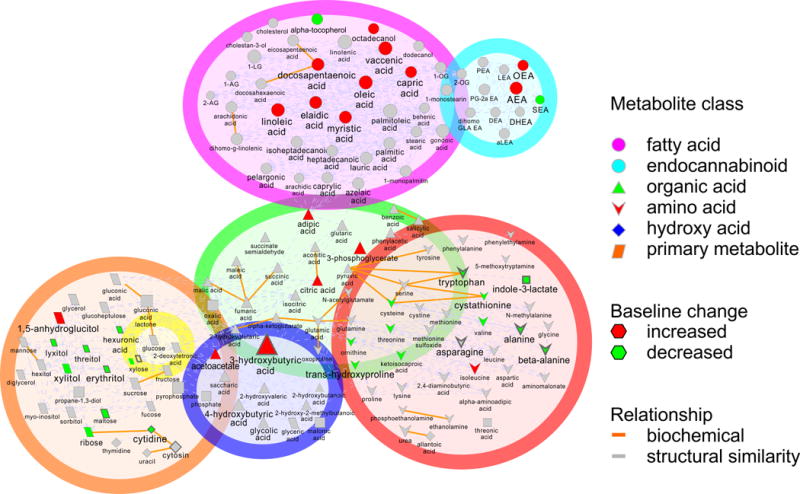
Map of the seven plasma metabolite classes (amino acids, endocannabinoids, fatty acids, glucose, hydroxy acids, organic acids, and primary carbon metabolites) measured illustrating specific metabolites that were significantly different with adjusted p-values below 0.05 at baseline (T0) between early and late fasting periods in northern elephant seal pups.
The metabolite’s pathway and metabolite class are identified
aMean AUC ± standard deviation
bEndocannabinoid concentrations are in nM
cFatty acids concentrations are in μM
dFDR adjusted p-value < 0.05
Conversely, there were mixed changes with other primary metabolites and endocannabinoids over this time (Fig. 1 and Supplemental Table 1). The primary metabolite 1,5-anhydroglucitol and endocannabinoid anandamide increased by 37% and 74%, respectively. Several of these metabolites are downstream products of lipid oxidation, providing additional evidence of upregulated lipid oxidation in late fasting. In contrast, endocannabinoid-like DHEA and SEA and glucogenic/ketogenic amino acids alanine, asparagine, cysteine, tryptophan, tyrosine and valine decreased.
When baseline levels were normalized to the animal’s plasma volume (estimated by body mass) to correct for changes associated with fasting duration (Somo et al. 2015), this correction had only a subtle effect on our results. The effect of this correction was observed in a few metabolites related to lipid oxidation. However, since many lipid oxidation products were significant without plasma volume adjustment, this did not change our interpretation of the results. Pathway enrichment analysis of significantly perturbed metabolites (p < 0.05) indicated 9 biochemical pathways that were significantly deregulated between early and late fasting in seals (p<0.05) (Supplemental Table 2). The most perturbed pathways included ketone and branched-chain amino acid metabolism.
Collectively, the changes in baseline concentrations and path analyses demonstrate an increase in fatty acid and ketone pool size, coupled with a decrease in amino acids and primary metabolites in late-fasted seals indicating that fasting duration shifts substrate metabolism toward an increase in lipolysis, β-oxidation, and ketone metabolism as the primary sources of energy, associated with a robust conservation of protein (lean tissue) (Figure 1).
Despite protracted fasting, insulin facilitates a shift toward fatty acid metabolism
The time course trajectories following insulin infusion identified 23 of 171 metabolites with significantly (p<0.05) perturbed AUC values between early and late fasting (Table 1) indicating that fasting duration alters the tissue’s responsiveness to insulin. Fatty acid metabolism in response to insulin is characterized by decreased AUC values between early and late fasting, with all AUC values of FFA in late fasting being negative (Table 1). The responses of ketones and EC to the insulin infusion were similar suggesting that lipolytic and endocannabinoid pathways share common insulin-mediated processes (Figure 2). In early fasting, these metabolite levels decreased, reaching a nadir at 30 min post-infusion. For FFA, levels returned to baseline by 60 min and remained so for the rest of the measurement period, whereas ketones and EC peaked at 60 min before returning to baseline at 120 min (Figure 2). Initially, insulin increased primary metabolites and amino acids before levels reached a nadir at 60 min and returned to baseline by 120 min (Figure 2).
Figure 2.
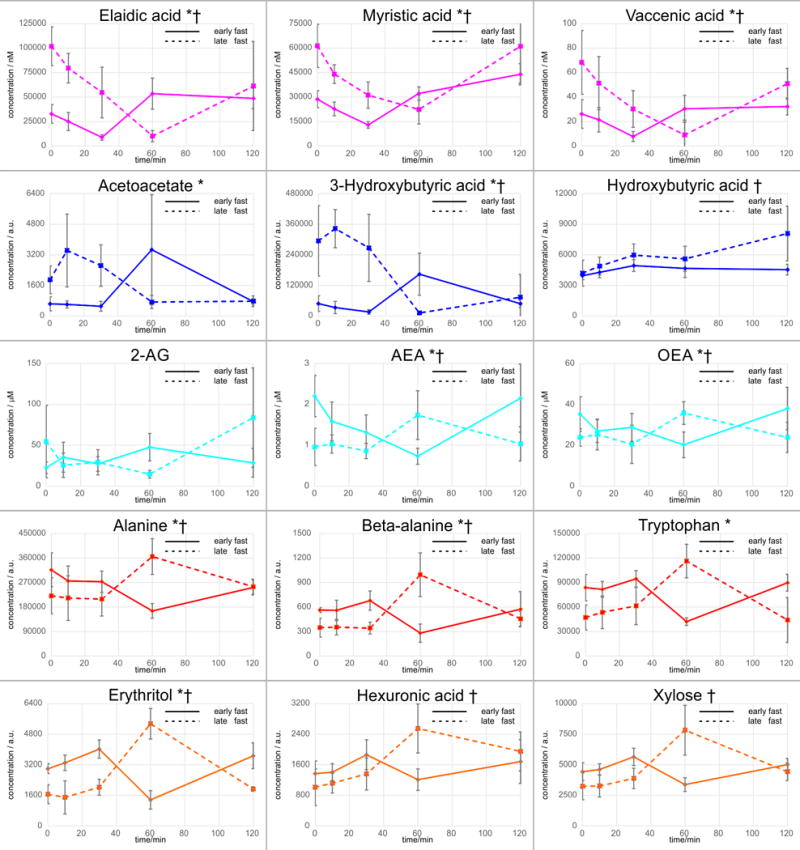
Mean abundances ± standard deviation of metabolic trajectories (t = 0 to 120 min) for those metabolites which indicated significantly (raw p-value < 0.05) different AUCs (Table 1) during insulin infusion between early to late fasting mammals is shown. †Endocannabinoid and Endocannabinoid-like compounds are presented as nM; *free fatty acids are presented as μM
In late fasting, FFA and EC decreased similarly, reaching a nadir at 60 min before returning to baseline by 120 min (Figure 2). Ketones displayed a transient increase before a nadir at 60 min and returning to baseline by 120 min (Figure 2). Primary metabolites and amino acids AUC values increased (with all values being positive with the exception of isoleucine). In contrast to the early fasting response, insulin increased amino acid levels reaching a peak at 60 min before decreasing to baseline at 120 min (Figure 2).
Principal component analysis (PCA) revealed that the maximum geometric distance occurred at 60 min for both early and late fasting, regardless of trajectory (Figure 3A, C). In the early fast the majority (72%) of metabolite trajectories return to baseline (+/− 30% of baseline) at 120 min. In contrast, the majority of metabolite concentrations in the late fast (53%) did not return to baseline (+/− 30% of baseline), and present a residual difference at 120 min post-infusion. Collectively, the integrated insulin responses (AUC) and PCA data reveal that in late-fasted animals (characterized by insulin resistance) the available pool of free fatty acids is depleted quickly, and likely shuttled into the TCA cycle. Conversely, the insulin-induced increases in primary metabolites (AUCs) are indicative of activation of gluconeogenic precursors (Figures 3C–D, 4C–D).
Figure 3.
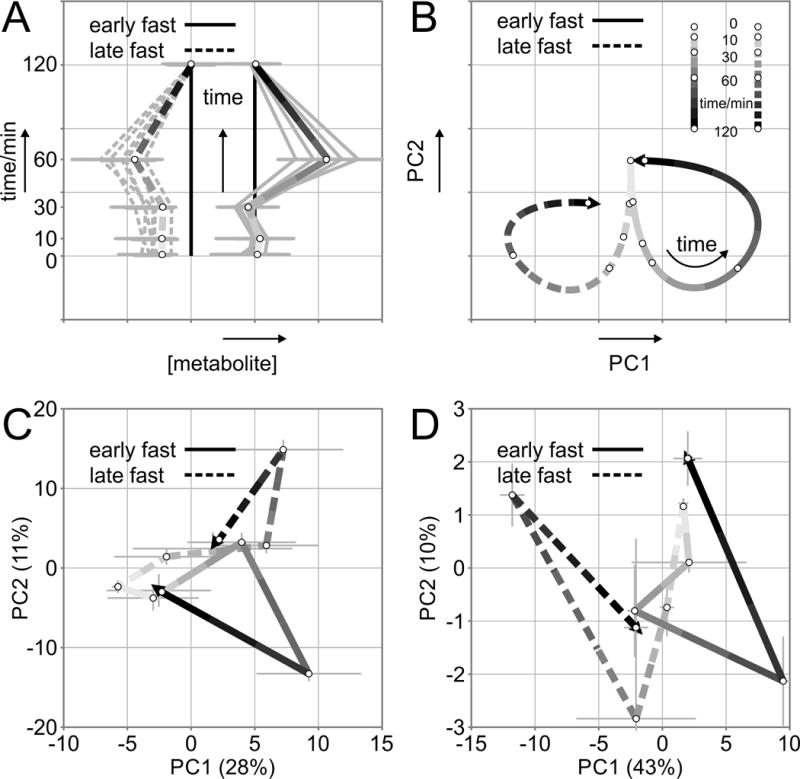
Schematic representation of a metabolite trajectories, and b principal component analysis plot explaining difference in outcome and direction of early- (solid line) and late-fasted (dashed line) response over time. Principal component analysis scores displaying the median points (±standard error) for early- and late-fasted northern elephant seal pups in response to exogenous insulin infusion for c all measured metabolites and d orthogonal partial least squares discriminant analysis selected top 10% of all discriminating metabolites. The line color becomes darker as the time course runs from the first point (t = 0) to the end point (t = 120 min, arrowhead). Solid lines = early fast, dashed lines = late fast
Figure 4.
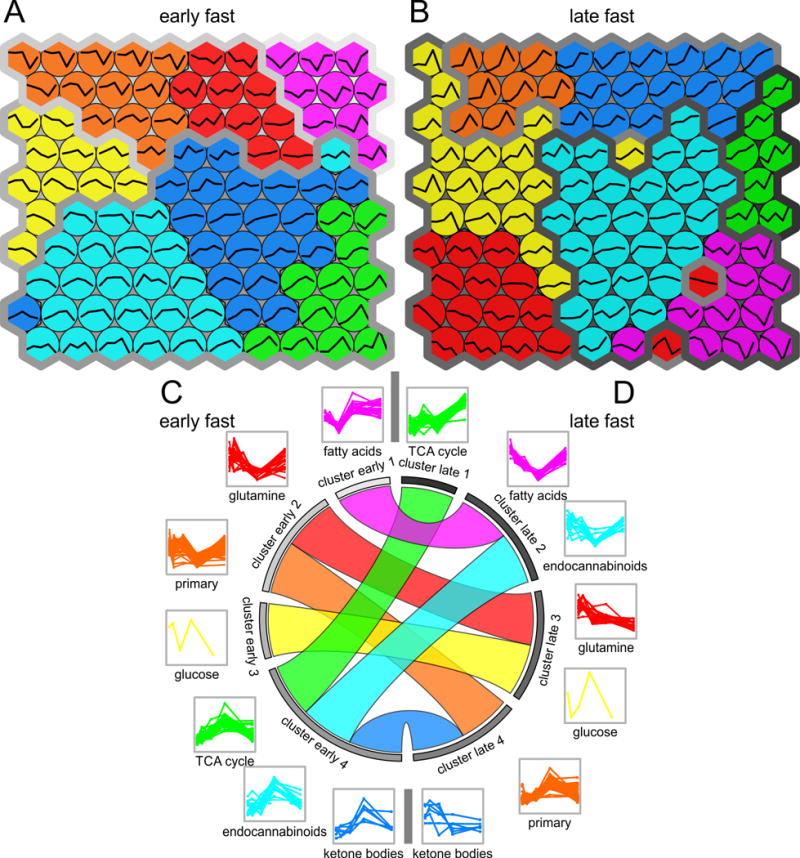
Self-organizing maps of the seven plasma metabolite classes (amino acids, endocannabinoids, fatty acids, glucose, hydroxy acids, organic acids, and primary carbon metabolites) measured for a early- and b late-fasted northern elephant seal pups following an exogenous insulin infusion. Metabolite trajectories are shown within the map nodes. The seven metabolite classes were grouped by area under the curve (AUC) using hierarchical clustering into four clusters in both c early and d late fast. The circular diagram shows how each metabolite class transitions into a new cluster from early fast (left hemisphere) to late fast (right hemisphere). The metabolite classes are arranged according to their response to insulin with increasing values for AUC from top to bottom
Fasting duration alters the metabolic response to insulin
Hierarchical clustering identified groups of metabolites with similar trajectories in both early and late fasting as well as in a joined cohort of both early and late fast. The seven main classes of metabolites grouped into four distinct clusters in early and 4 clusters in late fasting (Figure 4C–D). Clustering based on hierarchical (Figure 4C–D) and non-hierarchical methods on self-organizing maps (Figure 4A–B) aligned well with each other. In early fasting, ketone, EC, and TCA cycle metabolite profiles demonstrated a common peak and clustered together. Trajectories for amino acids and primary metabolites were closely related. The FFA profiles were characterized by a rapid, initial decrease recovering by 60 min, which was distinctive from the profiles for amino acids and primary metabolites.
In late fasting, FFA and EC were correlated and clustered together with a decrease at 60 min (Figure 4C–D). A comparison of the metabolomic profiles between early- and late-fasted animals in response to the insulin infusion demonstrated profound shifts in cellular metabolism and biochemical processes as a function of fasting duration. Cluster analysis allowed for assessment of the transition from negative to positive AUC values with fasting duration.
Discussion
The reliance of the elephant seal on lipid oxidation (RQ = 0.73) (C. L. Ortiz 1978; Houser et al. 2012) to meet its energetic demands during their prolonged bouts of fasting, which are a natural and evolved component of the animal’s life history, is unorthodox, if not unique. Animals, including humans, shift from reliance on one substrate (usually glucose initially) to another (lipids followed by protein) over the course of food deprivation or scarcity (Cahill 1970), but rarely is the RQ fixed over a prolonged period of food deprivation. What makes this evolved behavior even more fascinating is that the animals develop reversible, tissue-specific insulin resistance (Viscarra et al. 2011a; Viscarra et al. 2011b; Viscarra et al. 2012; Viscarra et al. 2013). Furthermore, fasting duration is associated with a decrease in plasma insulin (R. M. Ortiz et al. 2003; Viscarra et al. 2011a; Viscarra et al. 2011b)) suggesting that target tissues may be desensitized to insulin during this time to help maintain circulating glucose levels over the protracted fast. Despite exhibiting an insulin resistance-like phenotype and tolerating circumstances that would otherwise be considered detrimental, the fact that elephant seal pups developmentally thrive is truly remarkable. Here we show that: (1) despite an evolved mechanism to suppress circulating insulin with fasting duration, tissues remain responsive to insulin, (2) despite the reliance on lipid oxidation to fulfill their energetic needs, insulin has profound effects on endocannabinoids, ketones, and primary carbon metabolites, and (3) fasting duration (and thus the development of insulin resistance) has remarkable effects on modulating insulin-mediated metabolism of endocannabinoids, fatty acids and TCA cycle components. These findings are significant because they shed light on the evolution of insulin actions on substrate metabolism in an animal model of reversible insulin resistance at a time when the identification of alternative models for the study of metabolic disorders is at a premium.
An important and significant component of the current study was the use of a combination of untargeted and targeted metabolomics with the aims of identifying metabolic alterations associated with fasting status and insulin resistance in a large mammal. Furthermore, a thorough examination of the metabolic responses of early- and late-fasted seals to exogenous insulin provides previously unrealized insight into the metabolic programming or biochemical shifts induced by insulin.
Fasting Duration Shifts the Metabolic Trajectories to Insulin Infusion
An important and very compelling finding stemming from the insulin infusion studies is the extensive metabolomic characterization of the effects of fasting duration (and thus IR-like condition) on the cellular responses to insulin. Early- and late-fasted seals exhibit different metabolic profiles in responses to insulin indicating that fasting duration shifts the cellular sensitivity to insulin. The most striking distinctions were the responses in circulating FFAs, endocannabinoid and endocannabinoid-like metabolites, and ketones. In late-fasted seals, FFAs and the endocannabinoids, AEA and OEA, decreased during the first 60 min of insulin infusion followed by a return to baseline values. In hibernating bears, the growth hormone-induced increase in insulin was associated with nearly a fourfold increase in FFA 7 days post-infusion suggestive of increased lipolysis (Blumenthal et al. 2011). The corresponding reductions in AEA and OEA are consistent with previous data demonstrating that endocannabinoid and N-acylethanolamides reflect changes in FFA concentrations (Joosten et al. 2010). On the contrary, 3-hydroxybutyric acid decreased during the first 60 min of insulin infusion and remained suppressed suggesting that under normal conditions insulin promotes the complete metabolism of ketones so that they do not accumulate. In contrast, the growth hormone-induced increase in insulin stimulated over a 6-fold increase in plasma β-hydroxy butyrate in hibernating bears (Blumenthal et al. 2011). The differential responses compared to hibernating bears most likely reflect differences in energetic demands between the two animals as hibernating bears are hypothermic and metabolically quiescent while fasting seal pups are metabolically active and normothermic.
A consequence of diabetes is the accumulation of ketones, which may be the result of increased hepatic production and/or incomplete metabolism by extrahepatic tissues (Jefferies et al. 2015). Furthermore, the corresponding trajectories in FFAs were inversely mirrored by carbohydrates suggesting that exogenous insulin in late-fasted seals facilitates a preferential switch from lipid oxidation to glucose utilization and the promotion of glycolysis consistent with the Randle cycle (Hue and Taegtmeyer 2009). This is further supported by an increase in pyruvic acid with insulin infusion in late-fasted seals, which was not observed in early-fasted seals, suggesting that this shift toward a Randle cycle may be a significant metabolic shift to facilitate the adipose-specific insulin resistance (McGarry 1998)(McGarry 2002). Many of the TCA intermediates exhibited a similar response to insulin suggesting that these intermediates are derived from carbohydrate metabolism rather than fatty acid oxidation or amino acid catabolism. If insulin has the potential to preferentially shift substrate utilization as suggested by the data, then this may help explain why insulin levels remain relatively low and decrease with fasting duration, ultimately, helping to support a fixed RQ of 0.73. Conversely, when the animals begin to feed in cold-water environments, a shift toward a reduced metabolic dependence on lipid oxidation would allow the seals to conserve fat stores (insulation), and increase carbohydrate metabolism. Nonetheless, these data demonstrate the very dynamic effects of insulin that would otherwise be masked by attempts to reconcile based solely on plasma levels.
The switch to glucose and related-carbohydrate metabolism in late-fasted seals during insulin infusion may also explain the observed trajectories in most amino acids, particularly alanine. During baseline, late-fasted seals exhibited marked reductions in several amino acids compared to early-fasted seals, likely due to a combination of increased anaplerosis of ketogenic and gluconeogenic precursors, and a decrease in proteolysis (protein catabolism/ lean-tissue sparing). This is consistent with what has been reported previously (Houser and Crocker 2004; Crocker et al. 1998; Houser and Costa 2001). However, during insulin infusion amino acids, particularly alanine, mimicked the metabolic trajectories of carbohydrates. Alanine is particularly relevant given its importance in protein synthesis (mainly in the muscles) as well as nitrogen disposal from peripheral tissue to the liver (where it is transaminated back to pyruvate). Pyruvate can then either be oxidized or converted to glucose via gluconeogenesis (Rosen et al. 1959). Collectively, the aforementioned changes in late-fasted seals during insulin infusion suggest that: (1) the reliance on fatty acid oxidation is decreased, (2) the utilization of carbohydrates is increased, (3) circulating amino acids for protein synthesis is altered, and (4) nitrogen disposal is increased, all of which may be a consequence of the buildup of nitrogenous waste associated with amino acid catabolism that occurs with food deprivation or starvation (Cahill 1970; Goodman et al. 1980).
Also of note is the apparent lag in response to insulin infusion in late- compared to early- fasted seals. During early fasting, insulin infusion most often resulted in rapid, acute changes in metabolite concentrations that returned to baseline within the measurement period. Conversely, in late-fasted seals the responses were mostly either inverse, in cases where preferential usage of metabolic pathways changed, or their behaviors were similar with the exception that there was a lag affect in which concentrations did not return to baseline over the measurement period. This difference in the timing of responses to insulin between early- and late-fasted seals suggests that fasting duration alters the initiation of compensatory biochemical mechanisms that contribute to the maintenance of substrate homeostasis, particularly glucose. Given insulin’s profound effects on perturbing metabolic pathways in elephant seal pups, this may partially explain the natural decrease in plasma insulin as an evolved adaptation to prolonged fasting as a means to: (1) alleviate the insulin-induced suppression of lipolysis, (2) abate increased cellular metabolism and the subsequent energetic burden (ie, insulin-induced anabolism) in the absence of caloric intake, and (3) facilitate cellular homeostasis. If so, this would help explain why impaired insulin signaling has such dire consequences on cellular metabolism and appropriate substrate metabolism in non-adapted mammals.
Late fasted seals model an insulin resistance-like phenotype
The increase in baseline free fatty acid values in late fasting, along with the concomitant decrease in primary carbon metabolites, is indicative an insulin resistance-like (IR-like) phenotype. Temporary and reversible insulin resistance (IR) is a common response in mammals to conserve circulating glucose, but the fasting durations between those mammals and seals are not comparable. Additionally, the unique metabolic network constructed for seals here provides a framework to differentiate reversible, long-term IR-like phenotype and true insulin resistance. Similar to IR, the IR-like phenotype observed in seals is characterized by elevated plasma FFA and glucose levels. While the data clearly demonstrate that tissues remain responsive to insulin, the changes in plasma metabolites in response to insulin are shifted by approximately 30 minutes suggesting that the sensitivity to insulin in peripheral target tissues is decreased. Thus, in addition to relatively low plasma insulin concentrations in fasting seal pups (Viscarra et al. 2011a; Viscarra et al. 2013; Viscarra et al. 2011b; R. M. Ortiz et al. 2003; Champagne et al. 2012) a decrease in tissue sensitivity is an adaptation to facilitate the development of an IR-like condition, which collectively, is important for the maintenance of circulating glucose to support the metabolism of glucose-dependent tissues.
Additional comparisons of response curves to insulin reveal further unique differences. Notably, insulin increased plasma amino acid levels indicative of protein catabolism, which is not typically seen in other models of insulin resistance. However, this unique response is likely an evolved adaptation to facilitate the development of an IR-like phenotype that is not associated with detrimental outcomes. It is also important to note that the amino acid response to insulin is the opposite of their unstimulated, fasting metabolic state where protein is rigorously conserved. Collectively, these data define an IR-like phenotype that may characterize an intermediary phenotype and differentiate it from the human condition, which is associated with severe metabolic derangements and consequences, and the current condition, which is evolved and a natural component of the animal’s life history and absent of irrecoverable metabolic detriments.
Baseline Changes
Consistent with the findings by others (Tsatsoulis et al. 2013; Houser et al. 2013; Viscarra et al. 2013; Viscarra et al. 2012; Friedrich 2012) we observed an increase in baseline plasma free medium- (caprate and myristate) and long-chain (oleate, vaccinate, linoleate, EPA, and palmitate) fatty acids in late-fasted seals indicating that lipolysis increased, likely as a consequence of the increased demand for energy derived from the oxidation of FFA. Our findings are corroborated by previous data demonstrating that adipose lipases (LPL and ATGL) involved in regulation of non-esterified fatty acids (NEFA) are elevated with fasting in elephant seal pups (Viscarra et al. 2012). Additionally, circulating ketones (acetoacetate and 3-hydroxybutyric acid) increased in late compared to early fasting animals (Champagne et al. 2013). During food deprivation and insulin resistance, ketones are frequently elevated, a consequence of the increased reliance on beta-oxidation of FFA and amino acid catabolism (Stojanovic and Ihle 2011).
In addition to the increase in ketones and FFAs, the endocannabinoid AEA was increased in late-fasted pups. Insulin resistance or diabetes is known to impact endocannabinoid metabolism (D’Eon et al. 2008; Di Marzo et al. 2009). The regulation of the endocannabinoids in adipocytes is compromised during insulin resistance, leading to enhanced endocannabinoid signaling (D’Eon et al. 2008). Consequently, this dysregulation of ECs impairs metabolism resulting in changes in fatty acid synthesis and utilization, insulin sensitivity, and glucose utilization (Naughton et al. 2013). The impact of alterations in EC and EC-like metabolism on insulin sensitivity and metabolic syndrome is of particular interest and remains a focal point in future studies.
It has been postulated that up-regulation of the TCA cycle accommodates increased rates of FFA oxidation to prevent ketoacidosis via the accumulation of ketone bodies and to maintain endogenous glucose production (Champagne et al. 2012). Specifically, when glucose is limited or being preserved, anaplerotic reactions can supply the TCA cycle with intermediates (Champagne et al. 2012). In our study, ketogenic (tryptophan and threonine) and gluconeogenic precursors (alanine, asparagine, tryptophan, and threonine) decreased in late-fasted seals suggesting that the use of anaplerotic pathways increased with fasting (Owen et al. 2002). Interestingly, the characteristic increase in circulating branched-chain amino acids with the exception of isoleucine, commonly observed in insulin resistant, obese diabetics (McCormack et al. 2013; Friedrich 2012; Adams 2011), was not observed in fasting seals. On the contrary, valine decreased in late-fasted seals. However, similar to previously published observations of decreased glucose and insulin levels in late-fasted seals (Viscarra et al. 2013), not all metabolic changes resembled an obesity–related, insulin resistance phenotype. Thus, the observed alterations in branched-chain amino acid metabolism may have evolved differentially in elephant seals to provide a unique metabolic pathway during prolonged fasting that compensates for the fasting-associated insulin resistance described in fasting seal pups (Viscarra et al. 2012).
Consistent with the observed and previously reported (Viscarra et al. 2011b) decreases in glucose with fasting duration, other circulating sugars and sugar-alcohols were also reduced in late- versus early-fasted seals. This may suggest that utilization of other carbohydrate sources is increased to conserve circulating glucose levels as much as possible for use by glucose-dependent tissues. Circulating 1,5-anhydroglucitol, a marker of glycemic control (Yamanouchi et al. 1996; Buse et al. 2003; Champagne et al. 2013), was increased in late-fasted seals consistent with previously reported changes in both lactating and post-weaned pups (Champagne et al. 2013). The lack of a change in 1,5-anhydroglucitol in response to insulin suggests that any perturbations in carbohydrate metabolism (ie, glycogenolysis) by insulin were not sufficient to induce detectable changes in 1,5-anhydroglucitol, which is consistent with the fasting-associated increases in baseline levels in the presence of decreasing insulin levels (Ortiz et al. 2003; Champagne et al. 2013).
Perspectives
The present study describes the shifts in numerous metabolic pathways in fasted, insulin-infused northern elephant seal pups that are characterized by temporary, reversible insulin resistance (Viscarra et al. 2012). In particular, increases in fasting plasma ECs, FFAs, and ketones as well as decreased substrates for glutathione production were representative of this seemingly pathological, however, well-adapted condition. Nonetheless, the increase in 1,5-anhydroglucitol and decrease in AAs suggest that seal muscle may be comparatively more insulin sensitive than their adipose tissue, a contention supported by assessment of the phosphorylation of insulin receptor and other signaling proteins in adipose and muscle (Viscarra et al. 2012; Viscarra et al. 2013). Previously published data on gene expression, enzyme activity, and endocrine regulation as well as glucose and triglyceride metabolism support the metabolomics results described herein (Viscarra et al. 2011b; Viscarra et al. 2013; Champagne et al. 2012; Houser et al. 2012). Thus, metabolomics proved to be an insightful tool to more thoroughly characterize this metabolic phenotype and to complement results gathered previously. Likewise, the results further support the establishment of the northern elephant seal as a useful, tractable large mammalian model for the investigation of temporary, reversible insulin resistance. The use of metabolomic approaches to compliment the suite of other studies in seals clearly illustrates the dynamic effects of insulin and its potential importance in regulating substrate metabolism during prolonged food deprivation in a large mammal naturally adapted to such an extreme behavior.
Supplementary Material
Significance Statement.
Insulin resistance is a complicated biological process, but it can be temporary and reversible in many mammals. Here we employed a metabolomics approach to illustrate the shifts in numerous metabolic pathways in fasted, insulin-infused northern elephant seal pups that are characterized by temporary, reversible insulin resistance. The evolution of insulin effects appears to be extremely dynamic and potentially very perplexing especially in a large mammal that naturally exhibits relatively low levels. The present study highlights how the use of plasma levels alone to interpret the hormone’s function could be problematic, and the dynamic effects of insulin and its potential importance in regulating substrate metabolism during prolonged food deprivation in a large mammal naturally adapted to such an extreme behavior.
Acknowledgments
We thank M. Thorwald for assisting with sample collection.
Funding: J.A.V. was supported by a National Institutes of Health National Heart, Lung, and Blood Institute Supplement to Support Diversity (R01HL09176-S). R.M.O. was partially supported by N.I.H. N.H.L.B.I. Career Development Award (K02HL103787). F.V.F. is grateful for the support of grant CA154887 from the National Institutes of Health, National Cancer Institute, University of California, Cancer Research Coordinating Committee CRN-17-427258, National Science Foundation, University of California Senate Graduate Research Council, and Health Science Research Institute program grants. Research funded by N.I.H. N.H.L.B.I. (R01HL09176), and N.I.H. West Coast Metabolomics Center (U24 DK097154). Additional support was provided to J.W.N by the USDA (Intramural Project 2032-51530-022-00D). The USDA is an equal opportunity employer and provider.
Footnotes
Disclosures
None of the authors have any conflicts of interest to disclose.
Ethical approval
All procedures were reviewed and approved by the Institutional Animal Care and Use Committees of both the University of California Merced and Sonoma State University. All work was realized under the National Marine Fisheries Service marine mammal permit #87-1743.
References
- Adams SH. Emerging perspectives on essential amino acid metabolism in obesity and the insulin-resistant state. Adv Nutr. 2011;2(6):445–456. doi: 10.3945/an.111.000737000737. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams SH, Costa DP. Water conservation and protein metabolism in northern elephant seal pups during the postweaning fast. J Comp Physiol B. 1993;163(5):367–373. doi: 10.1007/BF00265640. [DOI] [PubMed] [Google Scholar]
- Adams SH, Costa DP, Winter SC. Plasma carnitine in fasting neonatal and adult northern elephant seals. Am J Physiol. 1992;263(3 Pt 1):E570–574. doi: 10.1152/ajpendo.1992.263.3.E570. [DOI] [PubMed] [Google Scholar]
- Adams SH, Hoppel CL, Lok KH, Zhao L, Wong SW, Minkler PE, et al. Plasma acylcarnitine profiles suggest incomplete long-chain fatty acid beta-oxidation and altered tricarboxylic acid cycle activity in type 2 diabetic African-American women. J Nutr. 2009;139(6):1073–1081. doi: 10.3945/jn.108.103754jn.108.103754. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barupal DK, Haldiya PK, Wohlgemuth G, Kind T, Kothari SL, Pinkerton KE, et al. MetaMapp: mapping and visualizing metabolomic data by integrating information from biochemical pathways and chemical and mass spectral similarity. BMC Bioinformatics. 2012;13:99. doi: 10.1186/1471-2105-13-99. doi:1471-2105-13-99[pii]10.1186/1471-2105-13-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57(1):289–300. [Google Scholar]
- Blumenthal S, Morgan-Boyd R, Nelson R, Garshelis DL, Turyk ME, Unterman T. Seasonal regulation of the growth hormone-insulin-like growth factor-I axis in the American black bear (Ursus americanus) Am J Physiol Endocrinol Metab. 2011;301(4):E628–636. doi: 10.1152/ajpendo.00082.2011. 10.1152/ajpendo.00082.2011ajpendo.00082.2011[pii] [DOI] [PubMed] [Google Scholar]
- Buse JB, Freeman JL, Edelman SV, Jovanovic L, McGill JB. Serum 1,5-anhydroglucitol (GlycoMark): a short-term glycemic marker. Diabetes Technol Ther. 2003;5(3):355–363. doi: 10.1089/152091503765691839. [DOI] [PubMed] [Google Scholar]
- Cahill GF., Jr Starvation in man. N Engl J Med. 1970;282(12):668–675. doi: 10.1056/NEJM197003192821209. [DOI] [PubMed] [Google Scholar]
- Cao Y, Jiang T, Girke T. A maximum common substructure-based algorithm for searching and predicting drug-like compounds. Bioinformatics. 2008;24(13):i366–374. doi: 10.1093/bioinformatics/btn186btn186. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne CD, Boaz SM, Fowler MA, Houser DS, Costa DP, Crocker DE. A profile of carbohydrate metabolites in the fasting northern elephant seal. Comp Biochem Physiol Part D Genomics Proteomics. 2013;8(2):141–151. doi: 10.1016/j.cbd.2013.02.002S1744117X(13)00017-8. [pii] [DOI] [PubMed] [Google Scholar]
- Champagne CD, Houser DS, Fowler MA, Costa DP, Crocker DE. Gluconeogenesis is associated with high rates of tricarboxylic acid and pyruvate cycling in fasting northern elephant seals. Am J Physiol Regul Integr Comp Physiol. 2012;303(3):R340–352. doi: 10.1152/ajpregu.00042.2012ajpregu.00042.2012. [pii] [DOI] [PubMed] [Google Scholar]
- Crocker DE, Webb PM, Costa DP, Le Boeuf BJ. Protein catabolism and renal function in lactating northern elephant seals. Physiol Zool. 1998;71(5):485–491. doi: 10.1086/515971. [DOI] [PubMed] [Google Scholar]
- D’Eon TM, Pierce KA, Roix JJ, Tyler A, Chen H, Teixeira SR. The role of adipocyte insulin resistance in the pathogenesis of obesity-related elevations in endocannabinoids. Diabetes. 2008;57(5):1262–1268. doi: 10.2337/db07-1186db07-1186. [pii] [DOI] [PubMed] [Google Scholar]
- DeFronzo RA, Soman V, Sherwin RS, Hendler R, Felig P. Insulin binding to monocytes and insulin action in human obesity, starvation, and refeeding. J Clin Invest. 1978;62(1):204–213. doi: 10.1172/JCI109108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Di Marzo V, Verrijken A, Hakkarainen A, Petrosino S, Mertens I, Lundbom N, et al. Role of insulin as a negative regulator of plasma endocannabinoid levels in obese and nonobese subjects. Eur J Endocrinol. 2009;161(5):715–722. doi: 10.1530/EJE-09-0643EJE-09-0643. [pii] [DOI] [PubMed] [Google Scholar]
- Du F, Virtue A, Wang H, Yang XF. Metabolomic analyses for atherosclerosis, diabetes, and obesity. Biomark Res. 2013;1(1):17. doi: 10.1186/2050-7771-1-172050-7771-1-17. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O. Setup and Annotation of Metabolomic Experiments by Integrating Biological and Mass Spectrometric Metadata. Proc Lect Notes Bioinformatics. 2005:224–239. [Google Scholar]
- Fiehn O. Extending the breadth of metabolite profiling by gas chromatography coupled to mass spectrometry. Trends Analyt Chem. 2008;27(3):261–269. doi: 10.1016/j.trac.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floegel A, Stefan N, Yu Z, Muhlenbruch K, Drogan D, Joost HG, et al. Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes. 2013;62(2):639–648. doi: 10.2337/db12-0495db12-0495. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich N. Metabolomics in diabetes research. J Endocrinol. 2012;215(1):29–42. doi: 10.1530/JOE-12-0120JOE-12-0120. [pii] [DOI] [PubMed] [Google Scholar]
- Gladine C, Newman JW, Durand T, Pedersen TL, Galano JM, Demougeot C, et al. Lipid profiling following intake of the omega 3 fatty acid DHA identifies the peroxidized metabolites F4-neuroprostanes as the best predictors of atherosclerosis prevention. PLoS One. 2014;9(2):e89393. doi: 10.1371/journal.pone.0089393PONE-D-13-37598. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman MN, Larsen PR, Kaplan MM, Aoki TT, Young VR, Ruderman NB. Starvation in the rat. II. Effect of age and obesity on protein sparing and fuel metabolism. Am J Physiol. 1980;239(4):E277–E286. doi: 10.1152/ajpendo.1980.239.4.E277. [DOI] [PubMed] [Google Scholar]
- Grapov D, Wanichthanarak K, Fiehn O. MetaMapR: pathway independent metabolomic network analysis incorporating unknowns. Bioinformatics. 2015;31(16):2757–2760. doi: 10.1093/bioinformatics/btv194btv194. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houser DS, Champagne CD, Crocker DE. A non-traditional model of the metabolic syndrome: the adaptive significance of insulin resistance in fasting-adapted seals. Front Endocrinol (Lausanne) 2013;4:164. doi: 10.3389/fendo.2013.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houser DS, Costa DP. Protein catabolism in suckling and fasting northern elephant seal pups (Mirounga anglstirostris) J Comp Physiol B. 2001;171(8):635–642. doi: 10.1007/s003600100214. [DOI] [PubMed] [Google Scholar]
- Houser DS, Crocker DE. Age, sex, and reproductive state influence free amino acid concentrations in the fasting elephant seal. Physiol Biochem Zool. 2004;77(5):838–846. doi: 10.1086/422055. doi:PBZ030085[pii]10.1086/422055. [DOI] [PubMed] [Google Scholar]
- Houser DS, Crocker DE, Tift MS, Champagne CD. Glucose oxidation and nonoxidative glucose disposal during prolonged fasts of the northern elephant seal pup (Mirounga angustirostris) Am J Physiol Regul Integr Comp Physiol. 2012;303(5):R562–570. doi: 10.1152/ajpregu.00101.2012ajpregu.00101.2012. [pii] [DOI] [PubMed] [Google Scholar]
- Hue L, Taegtmeyer H. The Randle cycle revisited: a new head for an old hat. Am J Physiol Endocrinol Metab. 2009;297(3):E578–591. doi: 10.1152/ajpendo.00093.200900093.2009. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferies CA, Nakhla M, Derraik JG, Gunn AJ, Daneman D, Cutfield WS. Preventing Diabetic Ketoacidosis. Pediatr Clin North Am. 2015;62(4):857–871. doi: 10.1016/j.pcl.2015.04.002S0031-3955(15)00042-5. [pii] [DOI] [PubMed] [Google Scholar]
- Joosten MM, Balvers MG, Verhoeckx KC, Hendriks HF, Witkamp RF. Plasma anandamide and other N-acylethanolamines are correlated with their corresponding free fatty acid levels under both fasting and non-fasting conditions in women. Nutr Metab (Lond) 2010;7(49) doi: 10.1186/1743-7075-7-491743-7075-7-49. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44(D1):D457–462. doi: 10.1093/nar/gkv1070gkv1070. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpe F, Dickmann JR, Frayn KN. Fatty acids, obesity, and insulin resistance: time for a reevaluation. Diabetes. 2011;60(10):2441–2449. doi: 10.2337/db11-042560/10/2441. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keun HC, Ebbels TM, Bollard ME, Beckonert O, Antti H, Holmes E, et al. Geometric trajectory analysis of metabolic responses to toxicity can define treatment specific profiles. Chem Res Toxicol. 2004;17(5):579–587. doi: 10.1021/tx034212w. [DOI] [PubMed] [Google Scholar]
- Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, et al. PubChem Substance and Compound databases. Nucleic Acids Res. 2016;44(D1):D1202–1213. doi: 10.1093/nar/gkv951gkv951. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind T, Tolstikov V, Fiehn O, Weiss RH. A comprehensive urinary metabolomic approach for identifying kidney cancerr. Anal Biochem. 2007;363(2):185–195. doi: 10.1016/j.ab.2007.01.028. doi:S0003-2697(07)00043-7[pii]10.1016/j.ab.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Koves TR, Ussher JR, Noland RC, Slentz D, Mosedale M, Ilkayeva O, et al. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab. 2008;7(1):45–56. doi: 10.1016/j.cmet.2007.10.013S1550-4131-(07)00306-3. [pii] [DOI] [PubMed] [Google Scholar]
- Lanza IR, Zhang S, Ward LE, Karakelides H, Raftery D, Nair KS. Quantitative metabolomics by H-NMR and LC-MS/MS confirms altered metabolic pathways in diabetes. PLoS One. 2010;5(5):e10538. doi: 10.1371/journal.pone.0010538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LO, Hu YF, Wang L, Mitchell M, Berger A, Coleman RA. Early hepatic insulin resistance in mice: a metabolomics analysis. Mol Endocrinol. 2010;24(3):657–666. doi: 10.1210/me.2009-0152me.2009-0152. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Feng R, Guo F, Li Y, Jiao J, Sun C. Targeted metabolomic analysis reveals the association between the postprandial change in palmitic acid, branched-chain amino acids and insulin resistance in young obese subjects. Diabetes Res Clin Pract. 2015;108(1):84–93. doi: 10.1016/j.diabres.2015.01.014S0168-8227(15)00022-4. [pii] [DOI] [PubMed] [Google Scholar]
- Luria A, Weldon SM, Kabcenell AK, Ingraham RH, Matera D, Jiang H, et al. Compensatory mechanism for homeostatic blood pressure regulation in Ephx2 gene-disrupted mice. J Biol Chem. 2007;282(5):2891–2898. doi: 10.1074/jbc.M608057200. doi:M608057200[pii]10.1074/jbc.M608057200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack SE, Shaham O, McCarthy MA, Deik AA, Wang TJ, Gerszten RE, et al. Circulating branched-chain amino acid concentrations are associated with obesity and future insulin resistance in children and adolescents. Pediatr Obes. 2013;8(1):52–61. doi: 10.1111/j.2047-6310.2012.00087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry JD. Glucose-fatty acid interactions in health and disease. Am J Clin Nutr. 1998;67(3 Suppl):500S–504S. doi: 10.1093/ajcn/67.3.500S. [DOI] [PubMed] [Google Scholar]
- McGarry JD. Banting lecture 2001: dysregulation of fatty acid metabolism in the etiology of type 2 diabetes. Diabetes. 2002;51(1):7–18. doi: 10.2337/diabetes.51.1.7. [DOI] [PubMed] [Google Scholar]
- Mihalik SJ, Michaliszyn SF, de las Heras J, Bacha F, Lee S, Chace DH, et al. Metabolomic profiling of fatty acid and amino acid metabolism in youth with obesity and type 2 diabetes: evidence for enhanced mitochondrial oxidation. Diabetes Care. 2012;35(3):605–611. doi: 10.2337/DC11-1577DC11-1577. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naughton SS, Mathai ML, Hryciw DH, McAinch AJ. Fatty Acid modulation of the endocannabinoid system and the effect on food intake and metabolism. Int J Endocrinol. 2013;2013:361895. doi: 10.1155/2013/361895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz CL. Water and energy flux in elephant seal pubs fasting under natural conditions. Physiol Zool 1978 [Google Scholar]
- Ortiz RM, Noren DP, Ortiz CL, Talamantes F. GH and ghrelin increase with fasting in a naturally adapted species, the northern elephant seal (Mirounga angustirostris) J Endocrinol. 2003;178(3):533–539. doi: 10.1677/joe.0.1780533. [DOI] [PubMed] [Google Scholar]
- Owen OE, Kalhan SC, Hanson RW. The key role of anaplerosis and cataplerosis for citric acid cycle function. J Biol Chem. 2002;277(34):30409–30412. doi: 10.1074/jbc.R200006200R200006200. [pii] [DOI] [PubMed] [Google Scholar]
- Palermo G, Piraino P, Zucht HD. Performance of PLS regression coefficients in selecting variables for each response of a multivariate PLS for omics-type data. Adv Appl Bioinform Chem. 2009;2:57–70. doi: 10.2147/aabc.s3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer ND, Stevens RD, Antinozzi PA, Anderson A, Bergman RN, Wagenknecht LE, et al. Metabolomic profile associated with insulin resistance and conversion to diabetes in the Insulin Resistance Atherosclerosis Study. J Clin Endocrinol Metab. 2015;100(3):E463–468. doi: 10.1210/jc.2014-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipson B, Smyth GK. Permutation P-values should never be zero: calculating exact P-values when permutations are randomly drawn. Stat Appl Genet Mol Biol. 2010;9 doi: 10.2202/1544-6115.1585. Article39. [DOI] [PubMed] [Google Scholar]
- Rhee EP, Cheng S, Larson MG, Walford GA, Lewis GD, McCabe E, et al. Lipid profiling identifies a triacylglycerol signature of insulin resistance and improves diabetes prediction in humans. J Clin Invest. 2011;121(4):1402–1411. doi: 10.1172/JCI4444244442. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen F, Roberts NR, Nichol CA. Glucocorticosteroids and transaminase activity. I. Increased activity of glutamicpyruvic transaminase in four conditions associated with gluconeogenesis. J Biol Chem. 1959;234(3):476–480. [PubMed] [Google Scholar]
- Scholz M, Fiehn O. SetupX–a public study design database for metabolomic projects. Pac Symp Biocomput. 2007:169–180. [PubMed] [Google Scholar]
- Shearer GC, Harris WS, Pedersen TL, Newman JW. Detection of omega-3 oxylipins in human plasma and response to treatment with omega-3 acid ethyl esters. J Lipid Res. 2010;51(8):2074–2081. doi: 10.1194/M900193-JLR200M900193-JLR200. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedes F. Determination of total lipid using non-chlorinated solvents. Analyst 1999 [Google Scholar]
- Somo DA, Ensminger DC, Sharick JT, Kanatous SB, Crocker DE. Development of Dive Capacity in Northern Elephant Seals (Mirounga angustirostris): Reduced Body Reserves at Weaning Are Associated with Elevated Body Oxygen Stores during the Postweaning Fast. Physiol Biochem Zool. 2015;88(5):471–482. doi: 10.1086/682386. [DOI] [PubMed] [Google Scholar]
- Sonksen P, Sonksen J. Insulin: understanding its action in health and disease. Br J Anaesth. 2000;85(1):69–79. doi: 10.1093/bja/85.1.69. [DOI] [PubMed] [Google Scholar]
- Spégel P. Metabolomic analysis of a human oral glucose tolerance test reveals fatty acids as reliable indicators of regulated metabolism. Metabolomics. 2010;6:56–66. [Google Scholar]
- Stacklies W, Redestig H, Scholz M, Walther D, Selbig J. pcaMethods–a bioconductor package providing PCA methods for incomplete data. Bioinformatics. 2007;23(9):1164–1167. doi: 10.1093/bioinformatics/btm069. doi:btm069[pii]10.1093/bioinformatics/btm069. [DOI] [PubMed] [Google Scholar]
- Stojanovic V, Ihle S. Role of beta-hydroxybutyric acid in diabetic ketoacidosis: a review. Can Vet J. 2011;52(4):426–430. [PMC free article] [PubMed] [Google Scholar]
- Tsatsoulis A, Mantzaris MD, Bellou S, Andrikoula M. Insulin resistance: an adaptive mechanism becomes maladaptive in the current environment - an evolutionary perspective. Metabolism. 2013;62(5):622–633. doi: 10.1016/j.metabol.2012.11.004S0026-0495(12)00438-6. [pii] [DOI] [PubMed] [Google Scholar]
- Viscarra JA, Champagne CD, Crocker DE, Ortiz RM. 5′AMP-activated protein kinase activity is increased in adipose tissue of northern elephant seal pups during prolonged fasting-induced insulin resistance. J Endocrinol. 2011a;209(3):317–325. doi: 10.1530/JOE-11-0017JOE-11-0017. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viscarra JA, Rodriguez R, Vazquez-Medina JP, Lee A, Tift MS, Tavoni SK, et al. Insulin and GLP-1 infusions demonstrate the onset of adipose-specific insulin resistance in a large fasting mammal: potential glucogenic role for GLP-1. Physiol Rep. 2013;1(2):e00023. doi: 10.1002/phy2.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viscarra JA, Vazquez-Medina JP, Crocker DE, Ortiz RM. Glut4 is upregulated despite decreased insulin signaling during prolonged fasting in northern elephant seal pups. Am J Physiol Regul Integr Comp Physiol. 2011b;300(1):R150–154. doi: 10.1152/ajpregu.00478.2010ajpregu.00478.2010. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viscarra JA, Vazquez-Medina JP, Rodriguez R, Champagne CD, Adams SH, Crocker DE, et al. Decreased expression of adipose CD36 and FATP1 are associated with increased plasma non-esterified fatty acids during prolonged fasting in northern elephant seal pups (Mirounga angustirostris) J Exp Biol. 2012;215(Pt 14):2455–2464. doi: 10.1242/jeb.069070215/14/2455. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17(4):448–453. doi: 10.1038/nm.2307nm.2307. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiklund S, Johansson E, Sjostrom L, Mellerowicz EJ, Edlund U, Shockcor JP, et al. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal Chem. 2008;80(1):115–122. doi: 10.1021/ac0713510. [DOI] [PubMed] [Google Scholar]
- Wopereis S, Rubingh CM, van Erk MJ, Verheij ER, van Vliet T, Cnubben NH, et al. Metabolic profiling of the response to an oral glucose tolerance test detects subtle metabolic changes. PLoS One. 2009;4(2):e4525. doi: 10.1371/journal.pone.0004525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Wishart DS. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat Protoc. 2011;6(6):743–760. doi: 10.1038/nprot.2011.319nprot.2011.319. [pii] [DOI] [PubMed] [Google Scholar]
- Yamanouchi T, Ogata N, Tagaya T, Kawasaki T, Sekino N, Funato H, et al. Clinical usefulness of serum 1,5-anhydroglucitol in monitoring glycaemic control. Lancet. 1996;347(9014):1514–1518. doi: 10.1016/s0140-6736(96)90672-8. [DOI] [PubMed] [Google Scholar]
- Zhao X, Peter A, Fritsche J, Elcnerova M, Fritsche A, Haring HU, et al. Changes of the plasma metabolome during an oral glucose tolerance test: is there more than glucose to look at? Am J Physiol Endocrinol Metab. 2009;296(2):E384–393. doi: 10.1152/ajpendo.90748.200890748.2008. [pii] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


