Abstract
The intestinal microbiome in early life influences development of the mucosal immune system and predisposition to certain diseases. Because less is known about the microbiome in the stomach and its relationship to disease, we characterized the microbiota in the stomachs of 86 children and adults and the impact of H. pylori infection on the bacterial communities. The overall composition of the gastric microbiota in children and adults without H. pylori infection was similar, with minor differences in only low abundance taxa. However, the gastric microbiota in H. pylori-infected children, but not infected adults, differed significantly in the proportions of multiple high abundance taxa compared with their non-infected peers. The stomachs of H. pylori-infected children also harbored more diverse microbiota, smaller abundance of Firmicutes, and larger abundances of non-Helicobacter Proteobacteria and several lower taxonomic groups than stomachs of H. pylori-infected adults. Children with restructured gastric microbiota had higher levels of FOXP3, IL10 and TGFβ expression, consistent with increased T regulatory cell responses, compared with non-infected children and H. pylori-infected adults. The gastric commensal bacteria in children is altered during H. pylori infection in parallel with more tolerogenic gastric mucosae, potentially contributing to the reduced gastric disease characteristic of H. pylori-infected children.
INTRODUCTION
The consortia of bacteria that colonize the intestinal tract shortly after birth shape the development of the mucosal immune system and impact susceptibility to and protection against an array of mucosal as well as non-mucosal diseases.1 Studies in both mice and humans, for example, have implicated alterations in the intestinal microbiome during early life in the development of obesity,2 type 1 diabetes,3–5 atherosclerosis6 and Crohn’s disease.7,8 Emerging evidence indicates that the intestinal microbiota plays a fundamental role in modulating inflammatory responses through the ability of certain bacteria to elaborate products such as short chain fatty acids9–12 and capsular polysaccharide13–15 that induce the differentiation of local regulatory T cells (Tregs) capable of suppressing inflammatory responses. However, the commensal microbiota in the upper gastrointestinal tract and the impact of early life events on microbial composition and immune cell responses in the upper tract mucosa have received little investigative attention, particularly in humans.16
To begin to understand the dynamics of the microbial landscape in the upper gastrointestinal tract, we focused on the stomach and the impact of an early life process, Helicobacter pylori infection, on the composition of the residential bacteria. H. pylori is usually acquired in early childhood, but the immunobiology of H. pylori infection in children, in contrast to that of adults, especially in the context of the commensal bacteria, is poorly understood.17 Worldwide, H. pylori infection is the leading cause of inflammation, ulceration and neoplasia of the stomach, but children have significantly less H. pylori-associated inflammatory disease than adults.18 Having shown that H. pylori burden, virulence factor prevalence and genotype distribution are similar in infected children and adults,19,20 we reasoned that H. pylori infection in children might impact the microbial composition of the stomach,21 potentially enhancing gastric Treg responses capable of down-regulating T helper 1 (TH1)- and TH17-mediated H. pylori inflammatory disease in infected children. To address this issue, we characterized the gastric microbiota in H. pylori-infected and non-infected children and adults in a region of Latin America where H. pylori is endemic and H. pylori-associated gastric cancer is the second leading cause of cancer-related mortality.
RESULTS
Commensal gastric bacteria in children and adults is similar in the absence of H. pylori infection
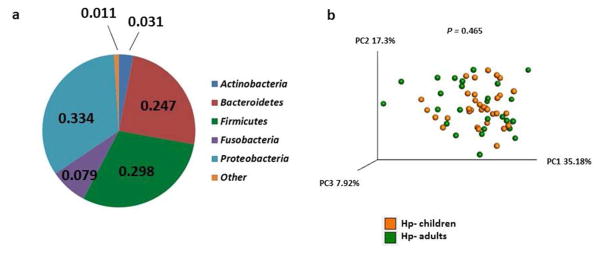
The study cohort included 86 children and adults with and without chronic H. pylori infection in Santiago, Chile (Table 1). Based on 16S rRNA bacterial gene sequencing, the microbiome profiles in the gastric fluids of the four groups of subjects were dominated by the phyla Actinobacteria, Bacteroidetes, Firmicutes, Fusobacteria and Proteobacteria (Figure 1a and Supplementary Tables S1 and S2). These five phyla also have been shown to dominate the gastric microbiota in U.S. adults.22 Principal coordinate analysis (PCoA) of weighted UniFrac distance matrices indicated that the overall composition of the gastric bacterial communities in children and adults without H. pylori infection was not significantly different (Figure 1b ), even though a small number of specific taxa were present at significantly different frequencies (Supplementary Figure S1). Thus, in the absence of H. pylori infection, young age alone did not predispose subjects residing in an H. pylori endemic region to a distinct consortium of gastric bacteria.
Table 1.
Demographic and clinical features of the Chilean subjects
| Groupa | A (n=12) | B (n=14) | C (n=33) | D (n=27) | P valuec |
|---|---|---|---|---|---|
| Mean age ± SD | 10.8 ±2.6 | 51.8 ±14 | 7.9 ±4.9 | 52 ± 18.2 | 0.1; 0.9 |
| Gender (%) | |||||
| Female | 6 (50) | 9 (64) | 15 (45) | 16 (59) | 0.5 |
| Male | 6 (50) | 5 (36) | 18 (54) | 11 (41) | |
|
| |||||
|
Indication for endoscopy (%)
| |||||
| Recurrent abdominal pain | 10 (83) | 8 (57) | 10 (30) | 13 (48) | 0.09; 0.6; 0.02; 0.4 |
| Dyspepsia | 0 (0) | 0 (0) | 1 (3) | 1 (4) | |
| Peptic ulcer | 1 (8) | 0 (0) | 1 (3) | 0 (0) | |
| Acid reflux symptoms | 0 (0) | 3 (21) | 4(11) | 2 (7) | |
| Epigastric pain | 1 (8) | 0 (0) | 1 (3) | 0 (0) | |
| Other | 0 (0) | 3 (21) | 16 (46) | 11 (38) | |
|
| |||||
|
Endoscopic finding (%)
| |||||
| Normal | 1 (8) | 1 (7) | 23 (70) | 7 (26) | 0.9; 0.0007; 0.0003; 0.15 |
| Erosive esophagitis | 0 (0) | 1 (7) | 3 (9) | 7 (26) | 0.3; 0.09; 0.3; 0.18 |
| Erosive gastropathy | 1 (8) | 4 (29) | 0 (0) | 6 (22) | 0.2; 0.005; 0.08; 0.6 |
| Nodular gastropathy | 11 (92) | 4 (29) | 1 (3) | 1 (4) | 0.001; 0.9; 0.00001; 0.02 |
| Gastric ulcer | 0 (0) | 1 (7) | 0 (0) | 1 (4) | 0.3; 0.1; nd; 0.1 |
| Duodenal ulcer | 1 (8) | 0 (0) | 0 (0) | 0 (0) | 0.3; nd; 0.08; nd |
| Non-specific duodenal changes | 0 (0) | 3 (21) | 6 (18) | 1 (4) | 0.08; 0.08; 0.1; 0.06 |
| Otherb | 0 (0) | 4 (29) | 0 (0) | 7 (26) | 0.04; 0.005; 0.5; 0.9 |
A, H. pylori-infected children; B, H. pylori-infected adults; C, Non-infected children; D, Non-infected adults.
Other: hiatal hernia, gastric polyp(s), esophageal varices, hypertensive portal gastropathy and duodenal lymphangiectasia.
P values: Mean age, first P value, A vs C; second P value, B vs D; P values for other variables represent the following comparisons: A vs B; C vs D; A vs C; B vs D.
Figure 1.
Gastric microbiota in urban children and adults in Chile. (a) Average proportion of the five most abundant phyla of gastric bacteria in children and adults with and without H. pylori infection (n=86 total subjects). (b) PCoA plot of the gastric microbiota in children (n=33) and adults (n=27) without H. pylori infection. Weighted Unifrac was used as the distance measure with each dot in the PCoA plot representing one subject. P value was determined using the permanova test in QIIME.
Gastric microbiota in children is reshaped during H. pylori infection
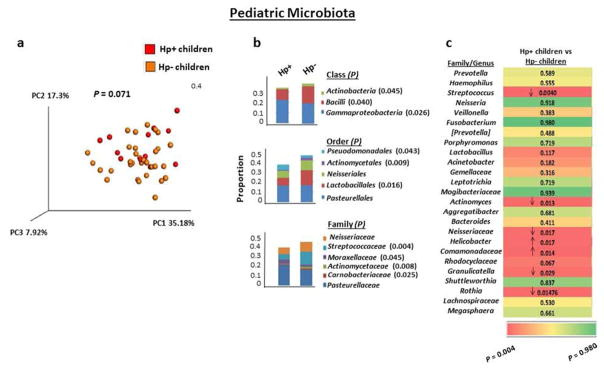
To assess the influence of H. pylori infection on the community of commensal bacteria in the stomachs of children, we next analyzed the gastric fluid microbiota in H. pylori-infected and non-infected children residing in the same endemic region, with similar demographics and without recent antibiotic or antacid therapy. The overall taxonomic composition of the microbiota in infected children was not significantly different from that of non-infected children based on beta diversity, although a strong trend toward significance was observed (Figure 2a). Among the major phyla of gastric bacteria harbored by the children, H. pylori-infected children contained a significantly smaller mean proportion of Actinobacteria than non-infected children (Supplementary Figure S2); the proportions of the other phyla were not significantly different in the infected and non-infected children. The reduced abundance of Actinobacteria in the gastric fluids of H. pylori-infected urban children, but not infected adults, in Chile extends the finding that H. pylori infection was associated with a reduction in gastric Actinobacteria in rural adult Amerindians in Venezuela.23 We next analyzed the gastric microbiota for lower taxonomic groups, including class, order and family, when the mean abundance of a member of a taxa was greater than 1% in at least one of the four groups of subjects. The fluids of H. pylori-infected children harbored significantly reduced proportions of three bacterial classes (Actinobacteria, Bacilli and Gammaproteobacteria; P=0.045, 0.040 and 0.026, respectively), three orders (Pseudomonadales, Actinomycetales, and Lactobacillales; P=0.043, 0.009 and 0.016, respectively) and four families (Streptococcaceae, Moraxellaceae, Actinomycetaceae and Carneobacteriaceae; P=0.004,0.045, 0.008 and 0.025, respectively) compared with fluids from non-infected children (Figure 2b).
Figure 2.
The gastric microbiota of H. pylori-infected children differs from that of non-infected children. (a) PCoA plot of the gastric microbiota in H. pylori-infected (n=12) and non-infected (n=33) children with weighted Unifrac as the distance measure. P value was determined using the permanova test in QIIME. (b) Frequencies of gastric bacteria by class, order and family in H. pylori-infected and non-infected children. The indicated class, order or family of bacteria was included when its 16S rDNA sequences were at least 1% of the total bacterial DNA content in at least one of the four groups of subjects (children and adults with and without H. pylori infection) and when the frequency was significantly different between any two groups. Stacked bar graphs show the average frequency for each indicated subject group. P values were determined using the Kruskal Wallis test with multiple comparison correction by applying false discovery rate (FDR) analysis. (c) Gastric microbiota by genera in H. pylori-infected and non-infected children. The top 25 most abundant genera in the cohort of 86 H. pylori-infected and non-infected children and adults are listed from most to least abundant. The ranking of genera was based on the average abundance of a given genus for the four groups of subjects. Numbers correspond to P values, and the direction of the arrow indicates an increase or decrease in the frequency of a given genus in the infected children relative to the non-infected children. When a genus could not be assigned, the family is listed. Color scheme shows red on the left representing the lowest P value and green on the right representing the highest P value. P values were determined using the Kruskal Wallis test with multiple comparison correction by applying false discovery rate (FDR) analysis.
At the genus level, we expanded the analysis to include the 25 most abundant genera. As shown in Figure 2c, the abundance of five genera (Streptococcus, Actinomyces, an undefined genus in family Neisseriaceae, Granulicatella and Rothia) was decreased significantly in H. pylori-infected children compared with non-infected children (P=0.004–0.029), and the abundance of an undefined genus in family Comamonadaceae was increased significantly in H. pylori-infected versus non-infected children (P=0.014). Predictably, H. pylori-infected children contained a higher abundance of Helicobacter sequences than non-infected children (P=0.017). Together, these findings indicate that H. pylori infection promotes a restructured gastric microbiota, at least during the infection, at multiple taxonomic levels in children.
Gastric microbiota in adults is not altered by H. pylori infection
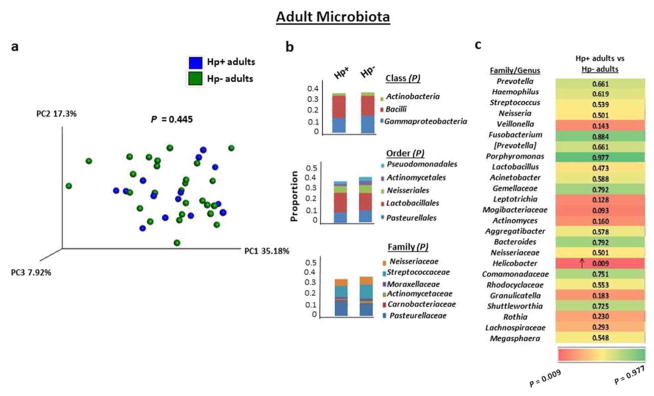
The overall composition of the gastric microbiota of H. pylori-infected adults was similar to that of non-infected adults, as assessed by PCoA (Figure 3a) and reported for U.S. adults.22 In contrast to the differences in the gastric microbiota of H. pylori-infected and non-infected children, H. pylori-infected and non-infected adults also harbored similar proportions of the major phyla, including Actinobacteria. At the class, order and family level, the mean proportion of bacterial taxa in infected and non-infected adults was not significantly different (Figure 3b). In addition, H. pylori-infected and non-infected adults harbored similar proportions of the 25 most abundant bacteria at the genus level, except for a predictable increase in Helicobacter sequences in H. pylori-infected adults compared with non-infected adults (P=0.009) (Figure 3c). Thus, in contrast to the impact of H. pylori infection on multiple gastric bacterial communities in children, the infection in adults was not associated with any changes in the abundance of the detected sequences, indicating that H. pylori infection had a much stronger impact on the composition and abundance of commensal bacteria in stomachs of children than adults.
Figure 3.
The gastric microbiota of H. pylori-infected adults does not differ from that of non-infected adults. (a) PCoA plot of the gastric microbiota in H. pylori-infected (n=14) and non-infected (n=27) adults with weighted Unifrac as the distance measure. P value was determined using the permanova test in QIIME. (b) Frequencies of bacteria by class, order and family in H. pylori-infected and non-infected adults. Analysis was performed as described in Figure 2b. (c) Gastric microbiota by genera in H. pylori-infected and non-infected adults. Analysis was performed as described in Figure 2c.
H. pylori-infected children and infected adults harbor different bacterial communities in their stomachs
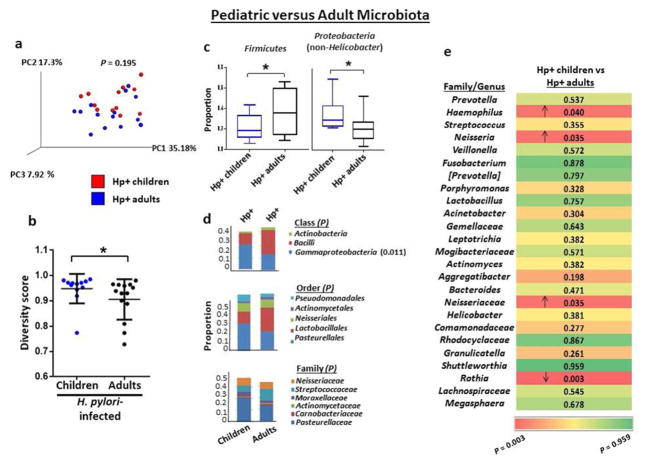
Early life exposure to commensal bacteria in the lower gastrointestinal tract is associated with the prevention of colonic inflammation in mice through microbe induction of protective immune responses.24 Commensal bacteria accelerate H. pylori-induced gastric inflammation and the development of neoplasia in mice,25 underscoring the importance of characterizing the shift in microbial composition during H. pylori infection in humans. Therefore, to further explore potential alterations in the microbial composition in the gastric compartment in early life, we next compared the taxonomic composition of the stomach associated with H. pylori infection in children with that of infected adults. The PCoA plot of microbial composition in the stomachs of H. pylori-infected children and infected adults showed no significant differences in overall composition (Figure 4a). However, since the diversity of microbes in a specific ecological niche can impact disease outcome,26 we assessed the diversity of the gastric bacteria in children and adults with H. pylori infection, based on the Simpson index, which reflects both the total number and distribution of phylotypes. In contrast to the association between increased bacterial diversity in the lower gastrointestinal tract and increased age,27 H. pylori-associated bacterial communities in the stomach were significantly more diverse in children than adults (P=0.027; Figure 4b). Importantly, the diversity of gastric bacteria in non-infected children and non-infected adults was not significantly different (Supplementary Figure S3). Thus, H. pylori infection confers upon children in an endemic area a greater diversity in gastric microbiota than infection in adults.
Figure 4.
The gastric microbiota in H. pylori-infected children differs from that of infected adults. (a) PCoA plot of the gastric microbiota of H. pylori-infected children (n=12) and infected adults (n=14) with weighted Unifrac as the distance measure. P value was determined using the permanova test in QIIME. (b) Richness and abundance distribution of the gastric microbiota in children and adults with H. pylori infection. Alpha diversity was determined using the Simpson index, and horizontal lines indicate mean ±standard deviation (SD). (c) The proportions of Firmicutes and non-Helicobacter Proteobacteria were significantly different in the stomachs of H. pylori-infected children and infected adults. Box-and-whisker plots display the median, maximum and minimum values for the indicated group. (d) Frequencies of gastric bacteria by class, order and family in H. pylori-infected children and infected adults. Analysis was performed as described in Figure 2b. (e) Gastric microbiota by genera in H. pylori-infected children and infected adults. Analysis was performed as described in Figure 2c. *P<0.05, Student’s t-test (b) or the Kruskal Wallis test (c).
Consistent with a more diverse gastric microbiota, H. pylori-infected children harbored a smaller mean abundance of Firmicutes and larger mean abundance of non-Helicobacter Proteobacteria than infected adults (P=0.034 and 0.041, respectively; Figure 4c). Infected children also harbored significantly increased proportions of the class Gammaproteobacteria (P=0.011; Figure 4d). At the genus level, the stomachs of H. pylori-infected children harbored a significantly reduced abundance of the genus Rothia among the top 25 most prevalent genera compared with infected adults (P=0.003; Figure 4e). Additionally, H. pylori-infected children harbored a significantly increased abundance of Haemophilus, Neisseria, and an unidentified genus in family Neisseriaceae (P=0.040, 0.035, and 0.035, respectively). Notably, non-infected children also harbored a significantly increased abundance of Neisseria compared with non-infected adults (P=0.029; Supplementary Figure S1), suggesting that young age is associated with higher levels of Neisseria independent of H. pylori infection status. No other differences in the microbiome between H. pylori-infected children and infected adults were recapitulated in the comparison between non-infected children and non-infected adults. Thus, H. pylori infection was associated with significant differences in the gastric microbial communities in children compared with infected adults, although fewer differences than in H. pylori-infected children compared with non-infected children.
Altered gastric microbiota in H. pylori-infected children is associated with increased Treg responses
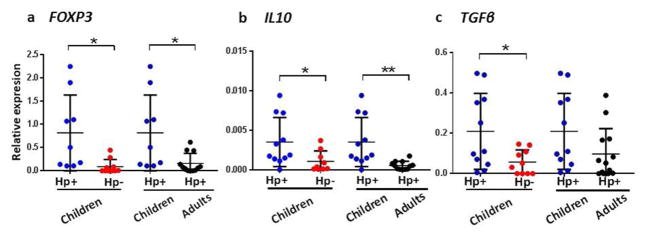
Studies in mice have implicated commensal bacteria in the expansion and differentiation of Tregs capable of suppressing inducible inflammation in colonic mucosa.28–31 In the stomach, Tregs control TH2-type autoimmune gastritis in mice32 and are associated with reduced TH1- and TH17-mediated gastritis in humans.19,20 However, to our knowledge, the role of the microbiota in the induction of Tregs in the upper gastrointestinal tract has not been investigated in mice or humans. Therefore, in parallel with characterization of the gastric microbiota and when sufficient amounts of RNA were available, we examined the gastric mucosa for Treg responses based on mRNA expression. Children with H. pylori infection, who also had an altered gastric microbial composition compared with non-infected children and infected adults, expressed significantly more FOXP3 transcripts in their gastric mucosa than non-infected children and H. pylori-infected adults (P<0.05; Figure 5a). These findings suggest that the H. pylori-associated microbiota, rather than the H. pylori, contributes to the increased gastric Treg response in the children. The enhanced FOXP3 response in H. pylori-infected children with altered gastric microbial composition was accompanied by significantly increased levels of Il10 expression compared with both non-infected children (P<0.05) and H. pylori-infected adults (P<0.01) (Figure 5b). Consistent with Treg-mediated suppression of mouse small intestinal and colonic inflammation through the release of IL-10,33–35 the increased gastric Treg and IL10 responses in H. pylori-infected children suggests a mechanism for the down-regulation of H. pylori-induced inflammation in children.19,20 Further, the increased TGFβ expression in the gastric mucosa of H. pylori-infected children compared with non-infected children and infected adults (Figure 5c) is consistent with the known contribution of TGF-β to Treg induction and maintenance.36–38 Together, these findings suggest an association between altered gastric microbial composition and the induction of mucosal Treg responses in children.
Figure 5.
Expression of (a) FOXP3, (b) Il10, and (c) TGFβ mRNA in the gastric tissue of H. pylori-infected children (n=11), non-infected children (n=10) and H. pylori-infected adults (n=13). Expression is normalized to GAPDH mRNA. *P<0.05 **P<0.01, one-way analysis of variance (ANOVA) with Tukey’s post-hoc test.
DISCUSSION
We present the first comparative analysis of the gastric microbiota in H. pylori-infected children and adults residing in an H. pylori endemic region of the world. We characterized the bacteria in gastric fluid, which was less difficult to obtain than tissue from the children and likely represents a combination of shed mucosal bacteria and luminal populations, analogous to fecal bacteria39. Based on 16S rDNA sequencing, the gastric microbiota of H. pylori-infected children was distinct from that of non-infected children in abundance of multiple bacterial classes, orders, families and genera. H. pylori-infected children also harbored a more diverse gastric microbiota, a smaller abundance of Firmicutes, larger abundance of non-Helicobacter Proteobacteria, and an altered abundance of a class and four genera of gastric bacteria compared with infected adults. In contrast, the composition of the gastric microbiota in both non-infected children compared with non-infected adults, as well as H. pylori-infected adults compared with non-infected adults, displayed relatively few differences in taxa abundance.
The lower gastrointestinal tract is host to an extraordinary array of diverse bacteria, including diversity among bacteria from different micro-anatomic niches 16,39,40. Considerably less is known about the composition of commensal bacteria in the upper gastrointestinal tract and the impact of an early life event such as an infection on upper tract commensal populations.41 We have speculated that the stomachs of children colonized with H. pylori might harbor a microbiota different from non-infected children and possibly H. pylori-infected adults.21 The findings reported here confirm this speculation and suggest that colonization by H. pylori impacts the microbial landscape in the stomachs of children, potentially provoking, or contributing to, the dendritic cell (DC)-induced Treg response that suppresses pediatric TH1- and TH17-mediated gastric inflammation19,20. DCs in the gastric epithelium and sub-epithelial space, as we have reported in humans42,43, are strategically positioned to monitor the gastric microbiota. In mice, certain members of the gut microbiota and their metabolites have been shown to induce mucosal Treg responses in the lower intestinal tract mucosa.10,28,31 Whether the gastric microbiota in H. pylori-infected children similarly generate short chain fatty acids or small molecules such as ribosomally synthesized, post-translationally modified peptides44 capable of modulating mucosal Treg responses is not known but is an intriguing possible mechanism for the enhanced gastric Treg response in the children.
As Chile has moved to high income status based on per capita income (World Bank Country Classification 2016: http://data.worldbank.org/about/country-classifications/countary-and-lending-groups), the standard of living has increased in parallel with the overall hygiene of the country. These changes have led to a decrease in the prevalence of H. pylori among school children to 18.1% in middle income families in Santiago.45 In contrast, the prevalence of H. pylori among children in low income families residing in the poor neighborhoods of suburban Santiago has remained high (55.9%) (Harris PR, et al. manuscript submitted). The pediatric and adult gastric samples in the study reported here were obtained from persons in middle income families residing in developed areas of Santiago.
In children, an array tissues, including the jejunum, ileum and colon, contain a significantly greater proportion of Tregs than the same tissues in adults.46 Consequently, infants and young children may be predisposed to more tolerogenic responses than adults. Our findings suggest that the intrinsic tolerance in certain tissues in early life may be increased in the gastric mucosa in the setting of H. pylori infection, promoting low-level colonization by the bacteria and protection against H. pylori-associated inflammation and precancerous lesions, as well as allergic airway disease, as reported in mice.36,47,48 The importance of age-related tolerance in protecting against precancerous pathology is underscored by the finding that Treg depletion in H. pylori-infected infant mice without preneoplastic pathology leads to prompt clearance of the bacteria and the development of preneoplastic gastric pathology,36 suggesting that tolerance plays a key role in protecting against H. pylori-induced precancerous pathology in the stomach.
Our finding that the altered gastric microbiota in H. pylori-infected children was associated with an increased gastric Treg response does not establish causality. However, H. pylori burden, virulence and genotype are similar in infected children and adults19,20. Also, in adults the distribution of non-cultivable phylotypes in the stomach is not impacted by gastric pH levels.22 To determine whether H. pylori infection in children might affect the gastric pH, we measured the gastric pH in a separate, demographically similar cohort of 48 prospectively evaluated children from the same area of Santiago referred for diagnostic endoscopy for chronic upper gastrointestinal tract symptoms. Among 29 infected (urease test positive) children (mean age 11.9 ±3 years) and 19 non-infected children (mean age 10.8 ±4.3 years), the mean gastric pH in the infected children was 2.3 ±1.1 and the non-infected children 2.2 ±0.7 (P=0.6), suggesting that H. pylori infection did not impact the mean gastric pH in the children. Also, in a separate study of Chilean children with H. pylori infection and iron deficiency18, infected children had a mean fasting gastric pH of 3.0 ±1.9 and non-infected children a pH of 2.4 ±1.3 (P=0.06). Thus, while not having gastric pH levels for our subjects may be a limitation of the study, our separate pH results and the earlier study by Harris, et al.18 showed no significant difference in mean gastric pH in children with and without H. pylori infection.
Thus, the changes in gastric microbial composition in H. pylori-infected children reported here suggests a previously unrecognized potential link between microbial composition in the stomach during early life and an increased gastric Treg response that may contribute to the reduced inflammatory sequelae of H. pylori infection in children.19,20 Whether the alternation in the gastric microbiota during childhood impacts gastric disease in adults is not known but is an important issue in view of the recent report that different gastric microbial compositions are associated with high and low gastric cancer risk in adults in Colombia.49 Conceivably, persons who become infected as children develop Treg-mediated tolerance to H. pylori, which allows the bacteria to persist, while suppressing the effector T-cell responses in most, but certainly not all, infected subjects.
In conclusion, the findings reported here indicate that H. pylori infection in children, but not adults, in an urban setting in South America is associated with changes in the commensal microbiota and an increased gastric Treg response. The gastric microbiota is an underappreciated potential factor that could be involved in promoting a more tolerant anti-inflammatory gastric environment in early life, possibly through the induction of Treg responses that down-regulate gastric inflammation in H. pylori-infected children.
MATERIALS AND METHODS
Subjects
The study included 45 children (<15 years of age) and 41 adults (>18 years of age) residing in Santiago, Chile (Table 1) undergoing evaluation for chronic symptoms suggestive of peptic disease, including dyspepsia and recurrent abdominal discomfort and pain in institutional review board (IRB)-approved protocols (X080508004 and 12–236). Epidemiologic analysis indicates that the genetic ancestry of the population residing in the Santiago region is >55% European, native American (predominantly Mapucha and Aymara) 41–45% and African 2–3%.50 Exclusion criteria included a history of acute onset of symptoms, acute or chronic vomiting, or the use of antibiotic, antacid, H2 blocker, proton-pump inhibitor, bismuth-containing compounds, or non-steroidal anti-inflammatory drugs within the preceding four weeks. All subjects were of the same low to medium socioeconomic level based on local and validated criteria provided by the Chilean Ministry of Economy, Development and Tourism, Department of Economy.51
Gastric fluid samples and H. pylori testing
After an overnight fast, subjects underwent esophagogastroduodenoscopy at Pontificia Universidad Católica de Chile. The sterilized endoscope was introduced directly into the stomach, avoiding the aspiration of fluid in the oral cavity and esophagus and the introduction of water into the stomach. In the stomach, up to 1 mL of gastric fluid was aspirated and, when used for DNA sequencing, was placed immediately in Xpedition lysis and stabilization buffer (Zymo Research, CA) and frozen at −70°C until DNA extraction. An antral biopsy from each subject was tested for the presence of H. pylori by rapid urease test (Pronto Dry, Ecifarma, Chile), which has an accuracy of >95%.52
16S rDNA sequencing
DNA was isolated from a minimum of 110 μL of gastric fluid using the Fecal DNA MiniPrep kit (Zymo Research). Barcoded PCR amplification of the V4 region of the rRNA gene was accomplished using previously designed primers,53 and the PCR was carried out under conditions described by Caporaso53 with modifications.54,55 PCR products were resolved on agarose gels, and the DNA was isolated, purified, and quantified.55 All samples were multiplexed using barcodes, and 101 base single end reads were sequenced using the Illumina platform. Sequence reads were demultiplexed and the raw sequencing reads were generated in FASTQ format.
Bioinformatic analysis
Quality assessment of the FASTQ files was performed using FASTQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and quality filtering was performed using the FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). Due to the low quality of bases toward the 3′ ends of the read, 11 bases were trimmed from the 3′ end for all reads, making the read length 90 bases. Any read with an average base quality (Q) score <20 was discarded. In addition, any read with more than 10% of the bases exhibiting a Q score <30 was discarded. The total number of reads was 20,237,011 with a mean of 235,314 reads per sample. The remainder of the steps (described next) were performed with the Quantitative Insight into Microbial Ecology (QIIME), version 1.9 and in-house developed Perl scripts, as we have described.55 Sequences were grouped into operational taxonomic units (OTUs) using the clustering program UCLUST at a similarity threshold of 100%.56 OTUs, for which total read count across all samples were ≤10 reads, were considered rare and filtered out. The taxonomic assignments (to the genus level) for all OTUs were performed using Ribosomal Database Project (RDP) classifier (http://www.ncbi.nlm.nih.gov.pubmed/17586664?dopt=AbstractPlus), available in QIIME against Greengenes database (v13_8).57,58 The OTU table is rarefied at read count 30,000 (four samples were excluded due to low number of reads). OTUs, for which total read abundance across all samples were 0.0005% reads, were considered rare and filtered out. OTUs then were grouped together to summarize taxon abundance at different hierarchical levels of classification (phylum, class, order, family, genus). Multiple sequence alignment of OTUs was performed with PyNAST.59 Alpha diversity was analyzed using the Simpson metric, as implemented in QIIME, and beta diversity (between sample diversity) among different samples was measured using Unifrac analysis.
Real-time RT-PCR analysis for Treg gene and cytokine expression
Gastric antral biopsies, available from some but not all subjects, were snap frozen in liquid nitrogen and kept at −80°C until processed. Total RNA was extracted and, after reverse transcription, real-time PCR was performed on duplicate cDNA samples for FOXP3, IL10 and TGFβ, and mRNA levels were analyzed by comparing the differences in fold change in mRNA normalized to GAPDH mRNA.
Statistical analysis
Beta diversity of the gastric microbiota of H. pylori-infected and non-infected children and adults was determined using the permanova test implemented in QIIME, with weighted Unifrac as a distance measure. Differences in alpha diversity were determined by Student’s t-test. Relative expression differences of FOXP3, IL10 and TGFβ were determined using a 1-way analysis of variance (ANOVA) with Tukey’s post-hoc test. Significant differences in the frequencies of phyla, classes, orders, families and genera were determined using the Krukal Wallis test with multiple comparison correction using false discovery rate (FDR) analysis. Statistical significance was defined as P<0.05. Unless noted otherwise, statistics were calculated using GraphPad Prism 7 software.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants AI083127, RR-20136; the DeGregorio Family Foundation; the Research Service of the Veterans Administration; and Chilean Fondecyt grants 11140232 and 1130387. KMB was supported by NIH training grant T32-AI007493. The UAB Microbiome Resource is supported by the School of Medicine, Comprehensive Cancer Center (P30AR050948), Center for AIDS Research (5P30AI027767), Center for Clinical Translational Science (UL1TR001417) and the Heflin Center. The authors declare no conflict of interest.
Footnotes
The authors have no conflict of interests.
References
- 1.Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cho I, Yamanishi S, Cox L, Methe BA, Zavadil J, Li K, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621–626. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown CT, Davis-Richardson AG, Giongo A, Gano KA, Crabb DB, Mukherjee N, et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS One. 2011;6:e25792. doi: 10.1371/journal.pone.0025792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158:705–721. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Goffau MC, Luopajarvi K, Knip M, Ilonen J, Ruohtula T, Harkonen T, et al. Fecal microbiota composition differs between children with beta-cell autoimmunity and those without. Diabetes. 2013;62:1238–1244. doi: 10.2337/db12-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z, Roberts AB, Buffa JA, Levison BS, Zhu W, Org E, et al. Non-lethal Inhibition of Gut Microbial Trimethylamine Production for the Treatment of Atherosclerosis. Cell. 2015;163:1585–1595. doi: 10.1016/j.cell.2015.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kostic AD, Gevers D, Siljander H, Vatanen T, Hyotylainen T, Hamalainen AM, et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe. 2015;17:260–273. doi: 10.1016/j.chom.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garrett WS, Gordon JI, Glimcher LH. Homeostasis and inflammation in the intestine. Cell. 2010;140:859–870. doi: 10.1016/j.cell.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 12.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 14.Shen Y, Giardino Torchia ML, Lawson GW, Karp CL, Ashwell JD, Mazmanian SK. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe. 2012;12:509–520. doi: 10.1016/j.chom.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu H, Khosravi A, Kusumawardhani IP, Kwon AH, Vasconcelos AC, Cunha LD, et al. Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science. 2016;352:1116–1120. doi: 10.1126/science.aad9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donaldson GP, Lee MS, Mazmanian K. Gut biogeopgraphy of the bacterial microbiota. Nat Rev Microbiol. 2015 doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salama NR, Hartung ML, Muller A. Life in the human stomach: persistence strategies of the bacterial pathogen Helicobacter pylori. Nat Rev Microbiol. 2013;11:385–399. doi: 10.1038/nrmicro3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris PR, Serrano CA, Villagran A, Walker MM, Thomson M, Duarte I, et al. Helicobacter pylori-associated hypochlorhydria in children, and development of iron deficiency. J Clin Pathol. 2013;66:343–347. doi: 10.1136/jclinpath-2012-201243. [DOI] [PubMed] [Google Scholar]
- 19.Harris PR, Wright SW, Serrano C, Riera F, Duarte I, Torres J, et al. Helicobacter pylori gastritis in children is associated with a regulatory T-cell response. Gastroenterology. 2008;134:491–499. doi: 10.1053/j.gastro.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 20.Serrano C, Wright SW, Bimczok D, Shaffer CL, Cover TL, Venegas A, et al. Downregulated Th17 responses are associated with reduced gastritis in Helicobacter pylori-infected children. Mucosal Immunology. 2013;6:950–959. doi: 10.1038/mi.2012.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brawner KM, Morrow CD, Smith PD. Gastric microbiome and gastric cancer. Cancer J. 2014;20:211–216. doi: 10.1097/PPO.0000000000000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, et al. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci U S A. 2006;103:732–737. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maldonado-Contreras A, Goldfarb KC, Godoy-Vitorino F, Karaoz U, Contreras M, Blaser MJ, et al. Structure of the human gastric bacterial community in relation to Helicobacter pylori status. ISME J. 2011;5:574–579. doi: 10.1038/ismej.2010.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lofgren JL, Whary MT, Ge Z, Muthupalani S, Taylor NS, Mobley M, et al. Lack of commensal flora in Helicobacter pylori-infected INS-GAS mice reduces gastritis and delays intraepithelial neoplasia. Gastroenterology. 2011;140:210–220. doi: 10.1053/j.gastro.2010.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489:231–241. doi: 10.1038/nature11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geuking MB, Cahenzli J, Lawson MA, Ng DC, Slack E, Hapfelmeier S, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 31.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 32.Harakal J, Rival C, Qiao H, Tung KS. Regulatory T Cells Control Th2-Dominant Murine Autoimmune Gastritis. J Immunol. 2016;197:27–41. doi: 10.4049/jimmunol.1502344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rubtsov YP, Rasmussen JP, Chi EY, Fontenot J, Castelli L, Ye X, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 34.Chaudhry A, Samstein RM, Treuting P, Liang Y, Pils MC, Heinrich JM, et al. Interleukin-10 signaling in regulatory T cells is required for suppression of Th17 cell-mediated inflammation. Immunity. 2011;34:566–578. doi: 10.1016/j.immuni.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huber S, Gagliani N, Esplugues E, O’Connor W, Jr, Huber FJ, Chaudhry A, et al. Th17 cells express interleukin-10 receptor and are controlled by Foxp3(−) and Foxp3+ regulatory CD4+ T cells in an interleukin-10-dependent manner. Immunity. 2011;34:554–565. doi: 10.1016/j.immuni.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N, et al. Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fantini MC, Becker C, Tubbe I, Nikolaev A, Lehr HA, Galle P, et al. Transforming growth factor beta induced FoxP3+ regulatory T cells suppress Th1 mediated experimental colitis. Gut. 2006;55:671–680. doi: 10.1136/gut.2005.072801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maynard CL, Harrington LE, Janowski KM, Oliver JR, Zindl CL, Rudensky AY, et al. Regulatory T cells expressing interleukin 10 develop from Foxp3+ and Foxp3− precursor cells in the absence of interleukin 10. Nat Immunol. 2007;8:931–941. doi: 10.1038/ni1504. [DOI] [PubMed] [Google Scholar]
- 39.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lim ES, Zhou Y, Zhao G, Bauer IK, Droit L, Ndao IM, et al. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat Med. 2015;21:1228–1234. doi: 10.1038/nm.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bimczok D, Grams JM, Stahl RD, Waites KB, Smythies LE, Smith PD. Stromal regulation of human gastric dendritic cells restricts the Th1 response to Helicobacter pylori. Gastroenterology. 2011;141:929–938. doi: 10.1053/j.gastro.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bimczok D, Mannon P, Peter S, Wilcox CM, Harris PR, Smith PD, et al. Human gastric epithelial ceels contribute to gastric immune regulation by providing retinoic acid to dendritic cells. Mucosal Immunology. 2015;8:533–544. doi: 10.1038/mi.2014.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Donia MS, Fischbach MA. Human Microbiota. Small molecules from the human microbiota. Science. 2015;349:1254766. doi: 10.1126/science.1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jaime F, Villagran A, Serrano C, Cerda J, Harris PR. Frequency of Helicobacter pylori infection in 144 school age Chilean children. Rev Med Chil. 2013;141:1249–1254. doi: 10.4067/S0034-98872013001000003. [DOI] [PubMed] [Google Scholar]
- 46.Thome JJ, Bickham KL, Ohmura Y, Kubota M, Matsuoka N, Gordon C, et al. Early-life compartmentalization of human T cell differentiation and regulatory function in mucosal and lymphoid tissues. Nat Med. 2016;22:72–77. doi: 10.1038/nm.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arnold IC, Dehzad N, Reuter S, Martin H, Becher B, Taube C, et al. Helicobacter pylori infection prevents allergic asthma in mouse models through the induction of regulatory T cells. J Clin Invest. 2011;121:3088–3093. doi: 10.1172/JCI45041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oertli M, Sundquist M, Hitzler I, Engler DB, Arnold IC, Reuter S, et al. DC-derived IL-18 drives Treg differentiation, murine Helicobacter pylori-specific immune tolerance, and asthma protection. J Clin Invest. 2012;122:1082–1096. doi: 10.1172/JCI61029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang I, Woltemate S, Piazuelo MB, Bravo LE, Yepez MC, Romero-Gallo J, et al. Different gastric microbiota compositions in two human populations with high and low gastric cancer risk in Colombia. Sci Rep. 2016;6:18594. doi: 10.1038/srep18594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fuentes M, Pulgar I, Gallo C, Bortolini MC, Canizales-Quinteros S, Bedoya G, et al. Gene geography of Chile: regional distribution of American, European and African genetic contributions. Rev Med Chil. 2014;142:281–289. doi: 10.4067/S0034-98872014000300001. [DOI] [PubMed] [Google Scholar]
- 51.Adimark. Manual de Aplicación. Department of Economy; 2006. El Nivel Socioeconómico Esomar. [Google Scholar]
- 52.Malfertheiner P, Megraud F, O’Morain C, Bazzoli F, El-Omar E, Graham D, et al. Current concepts in the management of Helicobacter pylori infection: the Maastricht III Consensus Report. Gut. 2007;56:772–781. doi: 10.1136/gut.2006.101634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumar R, Eipers P, Little RB, Crowley M, Crossman DK, Lefkowitz EJ, et al. Getting started with microbiome analysis: sample acquisition to bioinformatics. Current Protocols in Human Genetics. 2014;82:18.18.11–18.18.29. doi: 10.1002/0471142905.hg1808s82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 57.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.