Abstract
Microbial infections in plant leaves remain a major challenge in agriculture. Hence an understanding of disease mechanisms at the molecular level is of paramount importance for identifying possible intervention points for their control. Whole-transcriptome changes during early disease stages in susceptible plant species are less well-documented than those of resistant ones. This study focuses on the differential transcriptional changes at 24 hours post inoculation (hpi) in tomato leaflets affected by three pathogens: (1) Phytophthora infestans, (2) Botrytis cinerea, and (3) Oidium neolycopersici. Grey mould (B. cinerea) was the disease that had progressed the most by 24 hpi, both in terms of visible symptoms as well as differential gene expression. By means of RNA-seq, we identified 50 differentially expressed tomato genes specifically induced by B. cinerea infection and 18 specifically induced by P. infestans infection at 24 hpi. Additionally, a set of 63 genes were differentially expressed during all three diseases when compared by a Bayesian approach to their respective mock infections. And Gene expression patterns were found to also depend on the inoculation technique. These findings suggest a specific and distinct transcriptional response in plant leaf tissue in reaction to B. cinerea and P. infestans invasion at 24 hpi, indicating that plants may recognize the attacking pathogen.
Introduction
Plant-microbe interaction starts with the arrival of the pathogen’s dispersal and infection units on the host. Visual disease symptoms of fungal/oomycetal attack in plant leaves only appear after a certain incubation period, by which time the application of plant protection products (PPP) may already be ineffective. Research work is mostly focused on the incompatible plant-pathogen interaction and aims to understand tolerance or resistance mechanisms and to identify the responsible genes. Results of compatible interaction in crops are not often analysed and reported. Analysis of the very first physiological reactions and the underlying changes at gene transcriptional level during disease attack could lead to a better understanding of disease mechanisms and help to develop new approaches for early disease detection, which in turn could contribute to improved PPP application techniques.
The increasing market for fresh and processed tomatoes (Solanum lycopersicum) makes this fruit crop one of the most important grown worldwide1. Like all plant species tomatoes are challenged by a wide variety of pathogenic organisms. Grey mould, late blight and oidium are among the tomato leaf diseases with the greatest economic impact. These three diseases are distinct in terms of the pathogen’s mechanism for host tissue invasion, time when the first symptoms develop and the response triggered in/by the host tissue. Grey mould, caused by the generalist Botrytis cinerea, is a destructive disease attacking more than 1400 plant species2. The necrotroph invades leaf tissue through the stomata or directly through the cuticle by forming appressoria and penetration pegs3. The symptoms may even become visible within the first 24–48 hours post inoculation (hpi)4. The hemibiotroph Phytophthora infestans, which causes late blight, is a relatively specific pathogen that mainly attacks tomato and potato leaves and harvested organs5. P. infestans invades leaf tissue through the stomata or the cuticle and haustoria that grow into the cells6. The first stage of late blight in tomato leaves is characterized by a biotrophic life style followed by a necrotrophic phase7. The growth of oomycetes, including P. infestans, is characterized during the biotrophic phase by nutrient uptake via haustoria–plant-derived nutrient and molecule exchange interphases located at the plant cell plasma membrane6. The biotrophic phase (48 hpi) was estimated by macro and microscopical assessments, and comparative gene expression studies with so-called transition (96 hpi) and necrotrophic phases (144 hpi)8. Depending on the experimental conditions, first symptoms appeared on the potato and tomato leaves at approximately 48–72 hpi7, 9. Oidium in tomato is caused by the obligate biotrophic fungus Oidium neolycopersici, which invades the epidermal cells by means of appressoria formation10. After invading the host tissue with appressoria formation at 6–8 hpi11, the first symptoms (conidia) become visible after seven days12. An up-regulation of the mildew locus O SlMLO1, 4 and 14 has been reported as early as 10 hpi for O. neolycopersici in the tomato cultivar Moneymaker13.
Plants have evolved different defence layers that mainly involve three signalling molecules: (1) salicylic acid (SA), (2) jasmonic acid (JA), and (3) ethylene (ET). The SA-dependent defence pathway is mostly activated by biotrophic pathogens such as during the early phase of P. infestans infection. In contrast JA and ET are mainly involved in defence reactions triggered by necrotrophic pathogens such as B. cinerea 14, 15. The genes NPR1 and PDF1.2, THI2.1, HEL, and CHIB 14 are major components of the SA and JA/ET defence pathways. The cross-talk between the SA-JA-ET pathways is highly complex and the role of ET is to a certain extent inconsistent in terms of presence/absence, influence on resistance and virulence, and cross-communication with the JA and SA pathways10, 16.
Changes in gene expression are one of the first reaction levels that follow plant-environment interactions, including pathogen attack. These changes are known to occur at the infection site and in surrounding tissue. For instance, in B. cinerea-Arabidopsis thaliana interactions, differentially expressed (DE) genes have been reported at 12 and 24 hpi and at 0–6 and 6–12 mm distant from the infection site17 in high resolution with two-hours sampling intervals during a 48 hours experiment18. Comparable studies have been published for P. infestans 8, 19, 20 and in-depth information is available for host-O. neolycopersici interactions e.g.13, 21. To our knowledge, studies comparing more than one pathogen using same plant material, grown in tomato under equivalent greenhouse and laboratory conditions are not available. Knowing early and disease-specific reactions in susceptible plant may allow the establishment of reduced pesticide regimes and potentially site specific application techniques. A first attempt to identify such potential marker genes could either be based on comparable disease development stages or common time points. Since comparable stages of B. cinerea, P. infestans and O. neolycopersici invasion are not obvious due to their different strategies, we decided to use a fixed time point of 24 hpi as a base for comparison. Using an RNA-seq approach we aimed to identify S. lycopersicum genes that were differentially expressed at 24 hpi, specifically the following three diseases: grey mould, late blight and powdery mildew. The identified genes were characterized in silico using the commonly available databases.
Results
Infection with B. cinerea is evident at 24 hpi
Nine pathogen-infected samples (PI: P. infestans, BC: B. cinerea and ON: O. neolycopersici) and their respective mock-inoculated variants (PIm, BCm and ONm) were subjected to Illumina HiSeq sequencing of the transcribed RNA at 24 hpi, which resulted in more than 1.13 billion total read pairs with a Q30 of 96% (project available at http://www.ebi.ac.uk/ena/data/view/PRJEB21223). Approximately 63 million read pairs were generated per sample. The reads were mapped against the available genomes of S. lycopersicum Heinz 1706, B. cinerea T4 and P. infestans. For O. neolycopersici, there was no genome sequence available. On average 80% of the reads per sample mapped to the host S. lycopersicum. 28 million read pairs (13.7%) of the B. cinerea and 1.4 (0.7%) of the P. infestans inoculated samples mapped to their respective genomes. One mock-inoculated sample of P. infestans infections yielded only 35% reads for S. lycopersicum and a Penicillium spp contamination was identified. Nevertheless the S. lycopersicon-derived reads clustered with the other mock-infections and therefore the sample was considered healthy. Pairwise comparison using edgeR22 on all the samples suggests similar patterns for all the mock-inoculated samples as well as for those inoculated with P. infestans and O. neolycopersici. In contrast, samples inoculated with B. cinerea clustered separately (Fig. 1), indicating a more advanced progression of the disease within the first 24 hpi.
Figure 1.
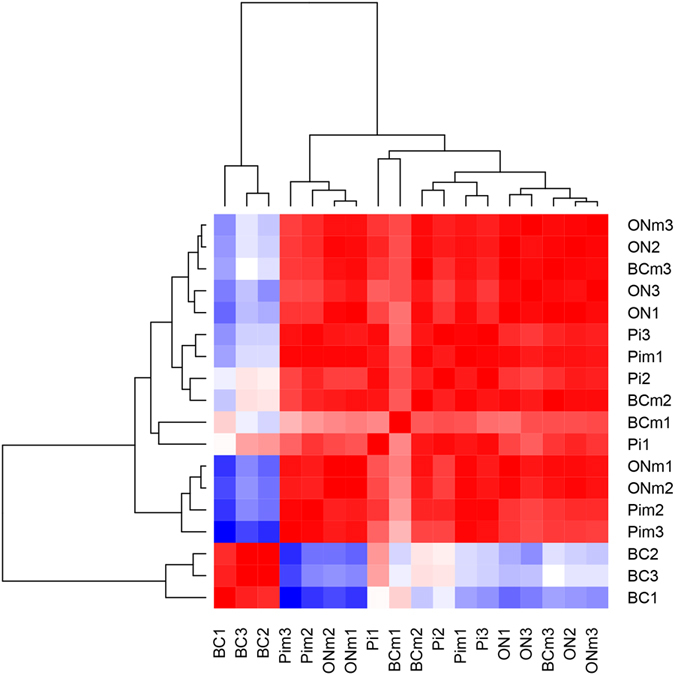
Pairwise comparison of read counts from the sequenced tomato leaf samples inoculated with either B. cinerea (BC), P. infestans (Pi) or O. neolycopersici (ON) and their corresponding mock (m) inoculations in three repetitions (1–3). Red, high similarity; blue, low similarity.
Eleven genes that were differentially expressed in pairwise comparison of the pathogen and its corresponding mock inoculation were used to validate the RNA-seq expression. Trends in up- and down-regulated RNA-seq-logFC (log2 fold change) correspond to the mock sample-normalized expression measured by means of qPCR (quantitative real-time PCR; Supplementary Table S1). Three genes with no indicated differential expression in RNA-seq and a FDR > 0.69 were up-regulated in a qPCR evaluation. The R2 of the linear correlation in the RNA-seq and qPCR data was 0.92 (Supplementary Figure S1). Time-course experiments with sampling in three-hours intervals from 0–24 hpi and then at 48 hpi lead to a correlation at 24 hpi with the RNA-seq expressions of R2 = 0.8 (Supplementary Figure S2 and S3).
Inoculation techniques alter gene expression
Nine patterns of cluster type [PI&PIm] vs. [BC&BCm&ON&ONm] comprising 668 genes were identified by summarizing genes that were differentially regulated between abaxial (PI and PIm) and adaxial (BC, BCm, ON and ONm) drop inoculation (all patterns can be found in Supplementary Data S1 with an explanatory file in Supplementary Data S2). Since B. cinerea was inoculated in half-strength grape juice, we tested if genes were clustering together in the pattern types [BC&BCm] vs. [PI&PIm&ON&ONm] and were able identify four patterns comprising 609 genes. Each sequenced gene can only be present in one pattern, hence there is no overlap between the genes expressed by different inoculation positions, inoculation media, and the ones identified as disease specific candidate genes that are described below. This suggests that both the location of the droplet deposition and the medium used for inoculation may impact gene expression.
Genes are regulated by disease in general
Using the R package EBSeq23 we identified one gene expression pattern (Fig. 2) that separates 63 genes by their differential regulation when diseased or healthy (Supplementary Table S2).
Figure 2.
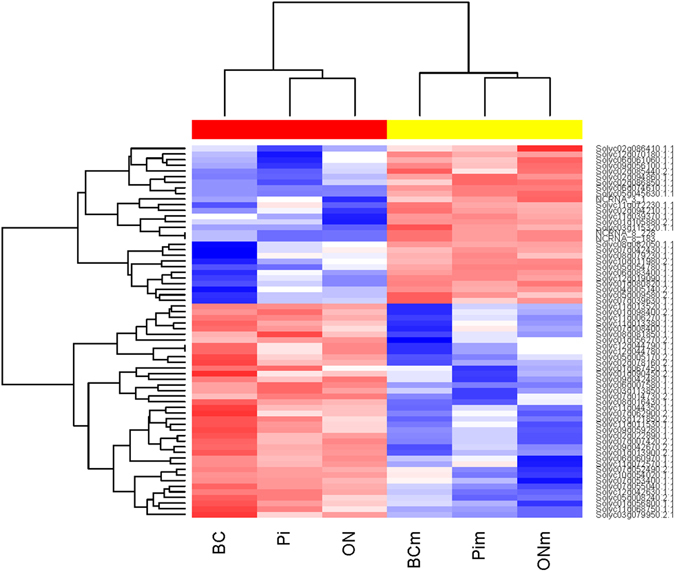
EBSeq pattern “healthy vs. diseased” ([PIm&BCm&ONm] vs. [PI&BC&ON]) identified DE genes from pathogen- and mock-inoculated tissues, comparing the overall similarity between the treatments. Of a total 63 genes were differentially expressed in the healthy and infected samples, independent of the inoculum source. Data is given for B. cinerea (BC), P. infestans (Pi), O. neolycopersici (ON), and their corresponding mock (m) inoculations. Blue represents up-regulated genes; red, down-regulated genes.
The most DE genes were found in B. cinerea inoculated tissues
Gene expression following infection with each disease and their corresponding mock inoculations were analysed by pairwise differential expression analysis using edgeR22. Pairwise comparison of B. cinerea and the corresponding mock infected samples at 24 hpi revealed 2476 up-regulated and 2264 down-regulated differentially expressed (DE) genes (Supplementary Table S3). At 24 hpi, 661 genes were up-regulated and 49 were down-regulated in P. infestans compared to the mock infections (Supplementary Table S3). Following O. neolycopersici leaf tissue infection, one gene was up-regulated and 22 were down-regulated compared to the water control (Supplementary Table S3). Comparing the differential expression between the diseases, we found 511 up-regulated genes in both the B. cinerea and the P. infestans infected tissues, and 24 and 3 down-regulated genes in B. cinerea-P. infestans and B. cinerea-O. neolycopersici infected leaf tissue, respectively (Supplementary Figure S4).
Grey mould and late blight-specific regulated genes were identified
In an attempt to identify disease-specific genes, the data were analysed with EBseq, and genes of informative patterns (i.e. sample combinations clustering for their gene expression against other samples which indicate whether they are “healthy” (BCm&PIm&ONm) or “diseased” (BC&PI&ON)) were extracted and compared to a pairwise comparison build with edgeR using a false discovery rate (FDR) < 1% and a logFC > 0.9. Genes belonging to a disease-specific pattern (only expressed for one disease and not for any other tested pathogen or mock inoculated samples) were extracted and analysed for their putative function using (1) Gene Ontology (GO) enrichment analysis, (2) an ortholog gene search of A. thaliana and Solanum tuberosum derived genes, and (3) protein database comparison.
EBseq resulted in 46 patterns for B. cinerea, 3 for P. infestans and 3 for O. neolycopersici, include gene candidates that are potentially disease-specifically regulated (Table 1). The 46 B. cinerea-specific patterns comprised 10009 genes, while the three patterns specific for P. infestans and O. neolycopersici included 178 and 80 genes, respectively (Table 1). These genes selected with EBseq were compared with the results of the DE of the pairwise comparison of the following pairs: BC vs. BCm, PI vs. PIm, ON vs. ONm, BC vs. PI, BC vs. ON and PI vs. ON. If the pattern-identified disease-specific genes were ‘DE’ for the specific disease-mock comparison and ‘not DE’ for the other pairs, they were selected for further characterization. This allowed 28 up- and 22 down-regulated genes to be identified that were specific for B. cinerea infections at 24 hpi, 18 up-regulated and no down-regulated genes with late blight specificity, and no DE genes for powdery mildew (Table 2). The B. cinerea infection specific up-regulated genes had a maximum logFC of 8.2 (Solyc04g028460.1.1, unknown protein) and the down-regulated genes a maximum of 5.3 (Solyc12g096490.1.1, GDU1). The strongest up-regulated gene during P. infestans infection was measured with a logFC of 4.4 (Solyc02g068670.1.1, Ankyrin repeat-containing protein).
Table 1.
Overview on informative patterns and their numbers of genes.
| Pattern associated genes | Pattern type | #Patterns |
|---|---|---|
| 10009 | [BC] vs. [all others] | 46 |
| 178 | [PI] vs. [all others] | 3 |
| 80 | [ON] vs. [all others] | 3 |
| 193 | [BCm] vs. [all others] | 4 |
| 23 | [PIm] vs. [all others] | 3 |
| 71 | [ONm] vs. [all others] | 2 |
| 609 | [BC] vs. [BCm] vs. [all others] | 4 |
| 668 | [PI] vs. [PIm] vs. [all others] | 9 |
| 337 | [ON] vs. [ONm] vs. [all others] | 1 |
| 89 | [BC&BCm] vs. [PI&PIm] vs. [ON&ONm] | 2 |
| 63 | [Disease] vs. [healthy] | 1 |
| 655 | [BC&PI] vs. [all others] | 3 |
| 26 | [PI&ON] vs. [all others] | 1 |
| 54 | [BCm&ONm] vs. [all others] | 1 |
| 19 | [BC&PI] vs. [ON&BCm] vs. [ONm&PIm] | 1 |
| 12 | [PI] vs. [BCm] vs. [all others] | 1 |
| 8 | [BC] vs. [PIm] vs. [all others] | 1 |
For pattern type, the single clade that is separated from the others (i.e. BC: BC vs “BCm, PI, PIm, ON, ONm”) is provided. BC, B. cinerea inoculated, BCm, inoculation with BC-corresponding mock solution; PI, P. infestans; ON, O. neolycopersici. Possibilities: BC, BCm, PI, PIm, ON, ONm, and their given combination.
Table 2.
Disease-specific genes that are differentially regulated in tomato leaves at 24 hpi.
| Gene ID | Functional description | Putative disease specificity | DE compared to mock treatment | logFC | PValue | FDR | Gene size |
|---|---|---|---|---|---|---|---|
| Solyc04g028460.1.1 | Unknown Protein | BCspec | up | −8.2 | 0.000459735 | 0.00329139 | 336 |
| Solyc05g013650.2.1 | Lysine ketoglutarate reductase trans-splicing related 1 | BCspec | up | −7.7 | 4.07E-05 | 0.000417875 | 1215 |
| Solyc06g066040.1.1 | Unknown Protein | BCspec | up | −6.9 | 0.000356986 | 0.002655293 | 558 |
| Solyc09g074220.1.1 | Unknown Protein | BCspec | up | −6.7 | 0.000516971 | 0.003618668 | 666 |
| Solyc09g090390.1.1 | Unknown Protein | BCspec | up | −6.3 | 0.001655282 | 0.009693517 | 852 |
| Solyc01g016380.2.1 | Os06g0207500 protein (Fragment) | BCspec | up | −5.1 | 0.001600352 | 0.009431806 | 738 |
| Solyc02g088840.2.1 | Unknown Protein | BCspec | up | −5.1 | 0.000590923 | 0.004061215 | 1248 |
| Solyc04g074360.1.1 | UDP-glucuronosyltransferase | BCspec | up | −5.0 | 8.24E-05 | 0.000767071 | 1467 |
| Solyc09g056290.1.1 | RNA-dependent RNA polymerase | BCspec | up | −4.8 | 0.000872922 | 0.005660068 | 399 |
| Solyc10g079900.1.1 | Unknown Protein | BCspec | up | −4.6 | 0.000631817 | 0.004307987 | 1494 |
| Solyc03g113580.1.1 | Germin-like protein | BCspec | up | −4.5 | 0.000121579 | 0.001068314 | 654 |
| Solyc01g099210.2.1 | Lipoxygenase | BCspec | up | −4.4 | 0.000468489 | 0.003341085 | 2592 |
| Solyc10g005440.1.1 | Serine/threonine-protein kinase receptor | BCspec | up | −4.1 | 0.001589537 | 0.009380068 | 2505 |
| Solyc09g011350.1.1 | Plant-specific domain TIGR01570 family protein | BCspec | up | −3.9 | 0.001126102 | 0.007018453 | 747 |
| Solyc04g005240.1.1 | Unknown Protein | BCspec | up | −3.8 | 4.85E-05 | 0.000485408 | 276 |
| Solyc06g053440.2.1 | Unknown Protein | BCspec | up | −3.7 | 0.000586649 | 0.004035862 | 579 |
| Solyc07g008360.1.1 | p-coumarate CoA-ligase 2 | BCspec | up | −2.6 | 0.000875671 | 0.005673897 | 1707 |
| Solyc11g017390.1.1 | Unknown Protein | BCspec | up | −2.5 | 0.000248335 | 0.001958543 | 459 |
| Solyc08g063030.2.1 | ADP,ATP carrier protein 1, mitochondrial | BCspec | up | −2.2 | 0.001274919 | 0.007787885 | 750 |
| Solyc08g075230.1.1 | Genomic DNA chromosome 5 P1 clone MDA7 | BCspec | up | −2.2 | 0.00034842 | 0.002607255 | 597 |
| Solyc03g097230.1.1 | Protein containing AIG2-like domain | BCspec | up | −2.1 | 0.000771587 | 0.005104815 | 585 |
| Solyc10g080560.1.1 | DNA-3-methyladenine glycosylase | BCspec | up | −1.9 | 0.000871383 | 0.005652742 | 867 |
| Solyc02g071520.2.1 | RAG1-activating protein 1 homolog | BCspec | up | −1.5 | 0.000604591 | 0.004142788 | 708 |
| Solyc01g087570.2.1 | Unknown Protein | BCspec | up | −1.5 | 0.000438104 | 0.003162748 | 270 |
| Solyc01g007770.2.1 | Genomic DNA chromosome 5 P1 clone MHF15 | BCspec | up | −1.4 | 0.000594061 | 0.004078728 | 525 |
| Solyc06g068960.1.1 | Calmodulin | BCspec | up | −1.3 | 0.001619588 | 0.009518784 | 465 |
| Solyc07g048030.2.1 | Heterogeneous nuclear ribonucleoprotein A3 | BCspec | up | −1.3 | 0.001003225 | 0.006355831 | 1314 |
| Solyc12g095790.1.1 | Integral membrane protein like | BCspec | up | −1.2 | 0.001201251 | 0.007411601 | 1029 |
| Solyc05g006900.1.1 | Unknown Protein | BCspec | down | 1.4 | 0.001087962 | 0.006805303 | 990 |
| Solyc11g065180.1.1 | THUMP domain-containing protein | BCspec | down | 1.4 | 0.001054798 | 0.006623311 | 1152 |
| Solyc01g111600.2.1 | Metal ion binding protein | BCspec | down | 1.4 | 0.000815055 | 0.005341251 | 462 |
| Solyc05g048810.2.1 | tRNA-specific adenosine deaminase | BCspec | down | 1.4 | 0.00165971 | 0.00971739 | 1296 |
| Solyc06g076850.2.1 | Binding protein | BCspec | down | 1.5 | 0.00115087 | 0.007150384 | 1656 |
| Solyc06g063300.2.1 | Kelch-domain-containing protein | BCspec | down | 1.6 | 0.000487788 | 0.003460853 | 1845 |
| Solyc06g053840.2.1 | Auxin responsive protein | BCspec | down | 1.7 | 8.40E-05 | 0.000779433 | 573 |
| Solyc11g005640.1.1 | Ubiquitin | BCspec | down | 2.0 | 0.000607185 | 0.004158502 | 669 |
| Solyc09g005020.1.1 | Unknown Protein | BCspec | down | 2.2 | 0.000987737 | 0.00627497 | 753 |
| Solyc06g030540.2.1 | Unknown Protein | BCspec | down | 2.4 | 0.001068037 | 0.00669279 | 258 |
| Solyc11g007530.1.1 | Ring H2 finger protein | BCspec | down | 2.4 | 0.000724629 | 0.004842834 | 672 |
| Solyc08g007430.1.1 | Nitrate transporter | BCspec | down | 2.4 | 0.001056343 | 0.006627024 | 1773 |
| Solyc04g028470.1.1 | GDSL esterase/lipase 2 | BCspec | down | 2.5 | 0.000325126 | 0.002467012 | 963 |
| Solyc06g075090.2.1 | Lysine decarboxylase-like protein | BCspec | down | 2.8 | 0.000124617 | 0.001090846 | 576 |
| Solyc08g066450.1.1 | Unknown Protein | BCspec | down | 2.8 | 0.000395385 | 0.002898268 | 564 |
| Solyc06g060830.2.1 | Homeobox-leucine zipper protein | BCspec | down | 3.0 | 5.86E-06 | 7.72E-05 | 897 |
| Solyc01g010970.2.1 | ARGONAUTE 1 | BCspec | down | 3.2 | 0.001442447 | 0.008661893 | 3003 |
| Solyc10g076790.1.1 | Auxin transporter-like protein 1 | BCspec | down | 3.3 | 0.001119952 | 0.006989584 | 1458 |
| Solyc01g066640.2.1 | Os04g0405500 protein | BCspec | down | 3.5 | 0.00011117 | 0.000990706 | 1152 |
| Solyc06g053210.2.1 | Ubiquitin | BCspec | down | 3.9 | 0.000567333 | 0.003921524 | 567 |
| Solyc11g010340.1.1 | BHLH transcription factor | BCspec | down | 5.0 | 0.000959904 | 0.00612349 | 897 |
| Solyc12g096490.1.1 | GDU1 | BCspec | down | 5.3 | 4.39E-06 | 5.97E-05 | 495 |
| Solyc02g068670.1.1 | Ankyrin repeat-containing protein At3g12360 | PIspec | up | −4.4 | 7.93E-18 | 1.54E-14 | 1704 |
| Solyc07g056210.2.1 | Unknown Protein | PIspec | up | −4.4 | 1.02E-17 | 1.64E-14 | 456 |
| Solyc08g062490.2.1 | WRKY transcription factor 16 | PIspec | up | −3.0 | 3.83E-10 | 1.13E-07 | 546 |
| Solyc01g079140.2.1 | Unknown Protein | PIspec | up | −2.7 | 1.82E-07 | 2.28E-05 | 423 |
| Solyc06g069740.1.1 | Calmodulin-like protein | PIspec | up | −2.3 | 0.000194837 | 0.007839798 | 558 |
| Solyc11g017280.1.1 | Receptor like kinase, RLK | PIspec | up | −2.3 | 3.00E-10 | 9.30E-08 | 2796 |
| Solyc01g009930.1.1 | LRR receptor-like serine/threonine-protein kinase, RLP | PIspec | up | −2.3 | 0.000192504 | 0.007768893 | 1758 |
| Solyc07g056200.2.1 | NBS-LRR class disease resistance protein | PIspec | up | −2.2 | 2.92E-07 | 3.44E-05 | 390 |
| Solyc02g077040.2.1 | Cathepsin B-like cysteine proteinase 5 | PIspec | up | −2.2 | 1.07E-07 | 1.42E-05 | 1038 |
| Solyc03g115930.1.1 | Calmodulin-like protein | PIspec | up | −2.2 | 1.65E-06 | 0.000152033 | 609 |
| Solyc03g122350.2.1 | Cytochrome P450 | PIspec | up | −2.1 | 1.20E-10 | 4.37E-08 | 1527 |
| Solyc03g095770.2.1 | WRKY transcription factor 6 | PIspec | up | −1.9 | 2.56E-06 | 0.000214804 | 822 |
| Solyc04g074000.2.1 | Receptor like kinase, RLK | PIspec | up | −1.9 | 8.80E-08 | 1.20E-05 | 3114 |
| Solyc03g033840.2.1 | 26S protease regulatory subunit 6B homolog | PIspec | up | −1.8 | 6.24E-07 | 6.59E-05 | 1518 |
| Solyc11g005630.1.1 | Receptor-like protein kinase | PIspec | up | −1.8 | 3.56E-06 | 0.000282714 | 2334 |
| Solyc02g081360.2.1 | Long-chain-fatty-acid–CoA ligase | PIspec | up | −1.8 | 8.06E-06 | 0.000574741 | 1728 |
| Solyc02g081350.2.1 | Acyl-CoA synthetase/AMP-acid ligase II | PIspec | up | −1.7 | 0.000135705 | 0.005812497 | 1782 |
| Solyc07g049660.2.1 | Acetyl coenzyme A cis-3-hexen-1-ol acetyl transferase | PIspec | up | −1.6 | 1.43E-05 | 0.000924682 | 1404 |
The genes have been selected using R package EBseq and pairwise comparison (edgeR). Full description of the selected genes is provided in Supplementary Table S4.
It was possible to identify several disease-related genes. Taking into account (1) the ITAG2.4 S. lycopersicum gene sequence list and characterization, (2) A. thaliana and S. tuberosum orthologs, (3) UniProt characterization and (4) Blast2GO annotation, the analysis revealed two of the B. cinerea specific (Solyc01g099210.2.1, lipoxygenase; Solyc10g005440.1.1, serine/threonine protein kinase receptor) and two of the P. infestans infection specific up-regulated genes (Solyc08g062490.2.1, WRKY transcription factor; and Solyc07g056200.2.1, NBS-LRR class disease resistance protein) known to be related to disease response.
18 of B. cinerea-up-regulated genes, 18 of the B. cinerea-down-regulated genes and 16 of the P. infestans-up-regulated genes were characterized by an ITAG functional description. Among these 52 genes, we identified one carrier protein, one glycosylase, one ion binding protein, three kinases, three ligases, one lipase, one oxygenase, one proteinase, one receptor protein, one reductase, one RNA polymerase, two transferases, one nitrate transporter, one leucine zipper and three transcriptions factors (Table 2).
According to KEGG pathway information generated using STRING24, two up-regulated genes are involved in plant-pathogen interaction (Solyc06g068960.1.1, calmodulin; Solyc06g069740.1.1, calmodulin-like protein) and two down-regulated genes are involved in plant hormone signal transduction (Solyc10g076790.1.1, ARGONAUTE; Solyc06g053840.2.1, auxin-responsive protein).
GO classification and enrichment analysis
The DE genes in the B. cinerea infection were classified with Blast2GO (v4.0.7) into 33 GO terms, and the ones related to the P. infestans infection into 14. The DE genes from the B. cinerea- and P. infestans-inoculated leaf samples shared three GO terms. GO enrichment analysis using the R package GOstat25, 26 identified ten GO terms related to biotic stress and defence that were overrepresented in the P. infestans inoculations at 24 hpi (P < 0.05; Supplementary Table S5). No such relation could be identified for the up-regulated genes from the B. infestans-inoculated samples (P < 0.05; Supplementary Table S6). The GO term “response to endogenous stimulus” was overrepresented in the down-regulated genes from the B. cinerea infection (P < 0.05; Supplementary Table S7).
Ortholog genes in A. thaliana and S. tuberosum
Comparing a total of 68 DE genes in tomato leaves infected with B. cinerea or P. infestans at 24 hpi with 41 different plant species, 38 ortholog A. thaliana genes and 43 ortholog S. tuberosum genes (Fig. 3) were identified. 26 DE genes were not represented by any ortholog A. thaliana genes and 11 were not represented by any S. tuberosum genes. Additionally, nine and 14 A. thaliana and S. tuberosum orthologues were described as “unknown protein”, “Conserved gene of unknown function” or “gene of unknown function” (Supplementary Table S4). For the Solyc04G028460.1.1, Solyc06G066040.1.1, Solyc09G074220.1.1 and Solyc04G005240.1.1 genes, all coding for unknown proteins and all being up-regulated after B. cinerea infection, no single ortholog could be identified. Orthologues of the Solyc01g079140.2.1 (P. infestans up-regulated), Solyc07g056210.2.1 (B. cinerea up-regulated), Solyc09g090390.1.1 (B. cinerea up-regulated) and Solyc10g079900.1.1 (B. cinerea up-regulated) genes–all coding for unknown proteins –were only found in S. tuberosum.
Figure 3.

Gene orthologues of B. cinerea- and P. infestans-specific DE genes. Ortholog search performed with g:profiler64.
Performing species co-occurrence analysis (Supplementary Figures S5–S7) using STRING, Solyc01g079140.2.1 (unknown protein), which was up-regulated during the P. infestans infection, was identified as being restricted to the Solanaceae family. Solyc04g005240.1.1 (unknown protein) and Solyc09g074220.1.1 (unknown protein), which were both up-regulated during B. cinerea infection, occurred only in S. lycopersicum, whereas Solyc06g066040.1.1, Solyc09g090390.1.1 and Solyc11g017390.1.1 (all coding for unknown proteins) were only found in the Solanaceae family. Since the orthologues have, to our knowledge, not yet been well characterized, the orthologue and co-occurrence analyses only provided information on the genes to be compared in future work.
Discussion
According to our knowledge, this paper represents the first differential gene expression study of S. lycopersicum affected by different diseases. Pairwise comparison of pathogen- and mock-treated samples at 24 hpi revealed that the greatest number of DE genes in the tomato specimens occurred when the plants were infected with B. cinerea. This number was less for P. infestans and even lower for O. neolycopersici. 75% of the DE genes induced by P. infestans were also found to be DE in the tomato-B. cinerea interaction. This is in contrast to a study into the compatible interaction of tomato with Cladosporium fulvum and Verticillium dahlia, which shared 454 DE genes out of a total of 4693 seven days after inoculation27. Furthermore, we identified genes that are specifically expressed at 24 hpi for both B. cinerea and P. infestans infected leaves. No such specific DE genes could be found for the infection with O. neolycopersici. Both the pairwise comparison and the disease-specific differential expression underline the limitations of the selected experimental setup, which used 24 hpi as a fixed time point for comparison, since each of the diseases has differing invasion and colonization strategies, as well as different development strategies over time and space. Due to its necrotrophic lifestyle B. cinerea is the fastest of the three pathogens in terms of invasion and destruction of host tissue, leading to visible symptoms within 24 hours4. An intermediate development speed was reported for P. infestans 7, and this was also visible in our experiments. Powdery mildew was the last of the three diseases to display evident symptoms12. This is also true for other Erysiphaceae, whose symptoms develop in a range from 5.1 to 8 days, as has been shown for powdery mildew species screening in grapevines28. Additionally, the water-based inoculation method used may not provide optimal infection conditions for O. neolicopersici, since powdery mildew does not need complete wetness during infection29. Spores were often transferred by touching healthy tissue with sporulating lesions12, but the water-based method allows control and standardisation of the experimental setup in terms of experiment replication comparability with O. neolycopersici and other pathogen inoculations. And liquid conidia suspensions were used successfully in experiments with O. neolycopersici 30, 31.
We identified 68 specific differentially expressed genes for grey mould and late blight at a relatively early disease progression state at 24 hpi. These genes were either up- or down-regulated and never lead to resistance, since the Heinz 1706 cultivar is highly susceptible to both diseases. Nevertheless, many of the up- and down-regulated candidate genes are related to environmental stress. All 68 identified gene accession numbers were checked using google scholar for citings in other publications. Referenced publications could only be found for eight genes. The disease-specific genes include six genes that are related to plant-pathogen interactions and two that are related to hormone signalling. One of these, the lipoxygenase (Solyc01g099210.2.1, up-regulated during B. cinerea infection) was reported to be up-regulated during root nematode infection32. SlWRKY80 (Solyc03g095770.2.1), which is up-regulated in P. infestans infected tissue, was also reported to be up-regulated during Pseudomonas syringae infiltration at 12 hpi33 and 6 days after infection with Xanthomonas perforans race T334. Solyc03g113580.1.1 (up-regulated during B. cinerea infection) coding for a Germin-like protein, was found to be expressed in tomato radicles grown under enhanced aluminium conditions35. Solyc06g053840.2.1, which is down-regulated during B. cinerea infection and coding for an auxin responsive protein (SlIAA4), is involved in hormone signal transduction and was up-regulated in young and old leaves and cotyledons compared to root36. Solyc06g075090.2.1 was down-regulated in B. cinerea infected leaf tissue and was reported to be involved in cytokinin related synthesis and signalling37. Solyc08g062490.2.1 (up-regulated in P. infestans infected leaf tissue) was annotated in the tomato genome (release ITAG2.4) as WRKY transcription factor 16, but is reported in literature as SlWRKY5038, a well-studied protein that mediates signalling of JA- and SA-pathways when the JA-pathway is repressed39.
When taking into account the gene description provided in the ITAG2.4 release, we could identify the following genes that potentially belonging to the major resistance (R) gene classes40 from the 68 candidates: (1) three genes coding for receptor like kinases (RLKs; Solyc04g074000.2.1, Solyc11g017280.1.1, Solyc11g005630.1.1), (2) two genes coding for lysine rich repeat proteins ((LRR; Solyc07g056200.2.1, Solyc01g009930.1.1), (1) one for serine/threonine-protein kinase receptor (Solyc10g005440.1.1), and (4) additionally three for transcriptions factors (TF, Solyc11g010340.1.1, Solyc03g095770.2.1, Solyc08g062490.2.1). RLKs can be involved in multiple processes including biotic and abiotic stress41. The RLKs, LRRs and two of the TFs were up-regulated during P. infestans infection, indicating a response to the pathogen invasion in the susceptible tomato tissue. However, this response did not lead to resistance. Therefore, this may suggest a disruption to downstream processes in disease resistance response. The serine/threonine-protein kinase was up-regulated during B. cinerea infection, which also failed to lead to a resistance reaction. Solyc08g075230.1.1 includes a harpin-induced 1 interpro domain. The bacterial harpins induce disease resistance through the systemically acquired resistance pathways42.
Additionally, the B. cinerea inoculation down-regulated the Argonaute 1 gene (AGO1, Solyc01g010970.2.1), which may also be involved in biotic plant-environment interaction since an interaction of AGO1 with AGO2 in response to virus infection was demonstrated in A. thaliana 43. The argonaute Piwi subfamily to which AGO1 belongs supports the silencing of mobile genetic elements44 and antiviral RNA45. The down-regulated Solyc11g010340.1.1 gene is one of 152 bHLH transcription factors identified in tomato. One of these TFs (SlybHLH131, Solyc10g008270.2.1) was reported to be involved in the tomato reaction to tomato yellow leaf curl virus infection46. Solyc02g068670.1.1 was characterized to code for an “Ankyrin repeat-containing protein”. These proteins were reported to be related to resistance as a potential negative regulator of pathogen-induced protein PR1 and antioxidation metabolism47. Furthermore, a number of DE genes may be involved in abiotic interaction: Solyc06g060830.2.1 down-regulated during B. cinerea infection codes for a putative homeobox-leucine zipper protein which is known to be involved in response to abiotic stresses48. The two genes coding for the homeobox-leucine zipper proteins ATHB12 and ATHB7 were up-regulated during drought stress49. The homologue gene AtRNP1 of Solyc07g048030.2.1 (Heterogeneous nuclear ribonucleoprotein A3) was reported to be involved in the abiotic stress response in A. thaliana 50. In our experimental setup, the only remarkable abiotic difference of the disease infection setup of the B. cinerea infections was the media that was used for inoculation (half strength grape juice), which might have exerted some stress due to its higher sugar level and osmotic potential but Solyc07g048030.2.1 did not match the media-specific expression pattern. Solyc06g068960.1.1, Solyc03g115930.1.1 and Solyc06g069740.1.1 potentially code for calmodulin and calmodulin-like proteins which are Ca2+ sensor proteins known to be involve in environmental stress responses51, 52.
In summary, the genes mentioned in the previous sections are related to biotic and abiotic stresses. The Heinz 1706 tomato cultivar we used for our experiments is obviously highly susceptible to all three diseases, hence no single related stress pathway which might be induced by these genes leads to a resistant phenotype. Nevertheless, some of the 68 genes we identified as late blight and grey mould-specific could be of use as indicators for pre-symptomatic disease identification if they are expressed systemically.
The presented multi-disease comparison at 24 hpi revealed several major findings: First, we identified genes that are differentially regulated in tomato leaves both during B. cinerea and P. infestans infections by comparing grey mould, late blight and powdery mildew leaf diseases with their mock infections. The identified candidate genes may be of use in identifying one of these two diseases before symptoms development. Therefore, the regulation of candidates will be evaluated in future works for their temporal and spatial expression patterns. During O. neolycopersici infection, no disease-specific differentially expressed genes were identified when compared to late blight and grey mould disease. Nevertheless, a pairwise comparison of O. neolycopersici inoculated leaf tissue with corresponding mock-inoculated tissues identified some DE genes. Second, most of the late blight and grey mould specific DE tomato genes are apparently not directly related to plant-pathogen interactions. Third, the results suggest that inoculation location (abaxial and adaxial) and inoculum solution solvent (water and half-strength grape juice) both have an impact on gene expression.
In future studies we will analyse the expression of identified disease-specific candidate genes over time and space within the whole plant and assess the potential use of one or a combination of these genes for early pre-symptomatic disease detection.
Methods
Plant material
Tomato plants (S. lycopersicum, Heinz 1706 cultivar) were grown in standard soil in a semi-regulated greenhouse with open windows. The temperature was set to 20–26 °C with maxima during sunny summer days of up to 40 °C. On cloudy days, artificial light was used to achieve minimal constant lighting of 80 kW per square meter for 16 h per day. Once a week, cuttings were produced from the tomato plants, which were treated once with sulphur (Stulln WG, Andermatt Biocontrol, Grossdietwil, Switzerland) and then placed in approximately 100% rel. humidity for one week. Afterwards the cuttings were acclimatised to the same greenhouse conditions mentioned above. Young and fully unfolded leaflets were harvested from two-week old cuttings for inoculation trials. 15 leaflets were placed in miniature grow boxes (30 × 60 cm and 14 cm in height with a clear plastic cover) on paper towels wetted with distilled sterile water. A separate box was used for each pathogen and mock inoculation. All inoculations were repeated three times.
Inoculum preparation, inoculation and sampling for transcriptome analysis
A P. infestans strain K5276 that was isolated in Switzerland and kindly provided by Syngenta (Basel, Switzerland) was grown for 3–8 weeks on V8 medium (200 ml V8 Jus de Legume, Globus, Switzerland; 30 mM CaCO3, 1.5% (w/v) Agar, Sigma-Aldrich, Buchs, Switzerland). Sporangia were collected with 10 ml tap water, diluted to 4 × 105 sporangia per ml and stored in darkness for 2 h at 5 °C before inoculation. Slight shaking of the inoculum solution hindered sporangia and zoospore sedimentation. 10 μl of the suspension was applied to the abaxial leaf surface for inoculation. The inoculated leaves were stored in darkness at 16 °C for 24 h, followed by a 16/8 hour day/night regime. Mock inoculations were performed under the same conditions using tap water for inoculation.
A B. cinerea strain T4 that was kindly provided by Philippe Nicot, INRA Centre de Recherche PACA, Montfavet, France, was grown on 15 g/l malt agar (Fluka, Sigma-Aldrich, Buchs, Switzerland) plates for 3–8 weeks. Spores were harvested with 20 ml half-strength grape juice (Farmer, Landi, Dotzingen, Switzerland) and diluted to 1.2 × 106 spores per ml. The spore suspension was used directly for inoculation, with 10 μl drops placed on the adaxial leaf surface. The inoculated leaves were stored without light at 18 °C. Mock inoculations were performed under the same conditions using half-strength grape juice for inoculation.
O. neolycopersici that was isolated on tomato plants in the greenhouse at our institute was maintained on S. lycopersicum cv. Heinz 1706. Spores were harvested with a wet paint brush and diluted in water to a concentration of 4 × 104 spores per ml, which was used to directly inoculate the adaxial leaf surface with 10 μl drops. The inoculated leaves were stored at 22 °C with a 16/8 hours day/night regime. Mock inoculations were performed under the same conditions using tap water for inoculation.
Inoculation and Sampling: Each inoculum was applied as approximately eight 10-μl-drops to the abaxial or adaxial surface of 15 leaflets (folioles53). Leaf disks (LD) of the inoculated sites were cut at 24 hours past inoculation (hpi) with a 5 mm-diameter cork borer for RNA-seq. LDs of eight leaflets were pooled, the remaining inoculum removed with a paper towel and frozen in liquid nitrogen. Samples were stored at −80 °C until further processing.
RNA extraction, transcriptome sequencing and quantitative real time PCR
Total RNA was extracted using NucleoSpin® RNA Plant (Macherey-Nagel, Düren, Germany) following the manufacturer’s instructions, including DNase treatment. The RNA quality and quantity was estimated using the Standard Sensitivity RNA Analysis Kit in a Fragment Analyzer (Advanced Analytical Technologies, Ames, USA) and analysed with the associated PROSize® 2.0 v.1.3 software. Total RNA with an RNA quality number (RQN) >= 6 was sequenced with Illumina Highseq2500 v4 chemistry by GATC (Constance, Germany) using a strand-specific cDNA library from purified poly-A containing mRNA molecules.
Validating the RNA-seq-derived differential expressed genes primers for 11 DE genes (Supplementary Table S1) were designed using Primer354. For qPCR total RNA was transcribed into first strand cDNA using an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA). Primers LSM7 and SlCBL1 that amplify genes that code for U6 snRNA-associated Sm-like protein LSm7 and Calcineurin B-like protein, respectively, were used as reference genes55, 56. The qPCR was conducted on a LifeCycler 480 (Roche, Basel, Switzerland) with a Fast EvaGreen® qPCR Master Mix (Biotium, Haywardm, CA, USA). For all primer pairs used in this study (Supplementary Table S1) the fast amplification protocol suggested by the master-mix provider was used, consisting of initial denaturation at 95 °C for 2’ followed by 40 cycles at 95 °C for 5” and 60 °C for 30”. A melting curve analysis was performed from 60–100 °C in 0.1 °C steps at the end of the run. All samples were analysed in duplicates. Primer efficiency was estimated by 1:10 dilution series run in triplicates (Fahrentrapp et al. under review). The same primers were applied to an additional time-course experiment (Fahrentrapp et al., unpublished, Supplementary Figure S2) conducted under the same conditions as described above. Leaf disc samples were taken at 3 hours intervals from 0–24 hpi and then at 48 hpi.
Data processing and statistics
To estimate the percentage of RNA-seq reads coming from the infection, the reads from all samples were aligned to the ENSEMBL57 (release 30) genome of S. lycopersicum and to the pathogen genomes. The B. cinerea T458 and P. infestans genomes59 were downloaded from the Broad Institute’s website. Since no reference genome was available for O. neolycopersici, reads from the infected and corresponding mock samples were assembled using Oases60 with a k-mer of 31. Resulting contigs were blasted against GenBank nt database using only contigs that matched fungal species for further analysis. The reads from the O. neolycopersici infected and mock-inoculated samples were mapped to the fungal contigs. For each sample, the percentage of reads that mapped the pathogen genome/contigs was calculated.
Differential gene expression analysis
Expression levels for each sample were estimated using the RSEM package (version 1.2.25)61 in paired-end and strand-specific mode with bowtie262 to the ENSEMBL (release 30) annotation of the S. lycopersicum genome. The RSEM “rsem-run-ebseq” tool was used to calculate the expression pattern based on the estimated expression levels for each gene based on the EBSeq method23. Significant patterns were assigned with a PPDE (posterior probability of being DE) >= 99%, which corresponds to a False Discovery Rate (FDR) of 1%. Additionally a pairwise comparison of all samples was performed using the R package edgeR22.
The resulting genes of interest were linked to their GO terms with Blast2GO (v4.0.7) and further analysed using g:Profiler63, 64, STRING24, 65 GOstat25 for GO enrichment analysis.
Availability of materials and data
RNA-seq data are available at European Nucleotide Archive http://www.ebi.ac.uk/ena/data/view/PRJEB21223.
Electronic supplementary material
Acknowledgements
This work was funded by the Swiss State Secretariat for Education, Research and Innovation SERI grant number C14.0094, and supported by COST Action FA1208. We would like to acknowledge technical assistance provided by the Bioinformatics Core Facility/Professorship of Systems Biology at JLU Giessen and access to their resources which are financially supported by the BMBF grant FKZ 031A533 and the BiGi center within the de.NBI network. We would also like to thank Syngenta, Basel, Switzerland, for providing the P. infestans isolate K5276 and INRA Centre de Recherche PACA, Montfavet, France, who provided the T4 B. cinerea strain.
Author Contributions
J.F. developed the research concept, conducted all experiments and interpreted the data. O.R. conducted the bioinformatics work. All authors wrote and reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-04792-5
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bergougnoux V. The history of tomato: From domestication to biopharming. Biotechnol. Adv. 2014;32:170–189. doi: 10.1016/j.biotechadv.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Elad, Y., Pertot, I., Prado, A. M. C. & Stewart, A. In Botrytis–the Fungus, the Pathogen and its Management in Agricultural Systems 413–486 (Springer, 2016).
- 3.van Kan JA. Licensed to kill: the lifestyle of a necrotrophic plant pathogen. Trends Plant Sci. 2006;11:247–253. doi: 10.1016/j.tplants.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 4.Asselbergh B, et al. Resistance to Botrytis cinerea in sitiens, an abscisic acid-deficient tomato mutant, involves timely production of hydrogen peroxide and cell wall modifications in the epidermis. Plant Physiol. 2007;144:1863–1877. doi: 10.1104/pp.107.099226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamoun S, Smart CD. Late blight of potato and tomato in the genomics era. Plant Dis. 2005;89:692–699. doi: 10.1094/PD-89-0692. [DOI] [PubMed] [Google Scholar]
- 6.Hardham A, Blackman L. Molecular cytology of Phytophthora-plant interactions. Australas. Plant Pathol. 2010;39:29–35. doi: 10.1071/AP09062. [DOI] [Google Scholar]
- 7.Nowicki M, Foolad MR, Nowakowska M, Kozik EU. Potato and tomato late blight caused by Phytophthora infestans: An overview of pathology and resistance breeding. Plant Dis. 2011;96:4–17. doi: 10.1094/PDIS-05-11-0458. [DOI] [PubMed] [Google Scholar]
- 8.Zuluaga AP, et al. Analysis of the tomato leaf transcriptome during successive hemibiotrophic stages of a compatible interaction with the oomycete pathogen Phytophthora infestans. Mol. Plant Pathol. 2016;17:42–54. doi: 10.1111/mpp.12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Llorente B, et al. A quantitative real-time PCR method for in planta monitoring of Phytophthora infestans growth. Lett. Appl. Microbiol. 2010;51:603–610. doi: 10.1111/j.1472-765X.2010.02942.x. [DOI] [PubMed] [Google Scholar]
- 10.Nonomura T, et al. Polymorphic change of appressoria by the tomato powdery mildew Oidium neolycopersici on host tomato leaves reflects multiple unsuccessful penetration attempts. Fungal Biology. 2010;114:917–928. doi: 10.1016/j.funbio.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 11.Jones H, Whipps JM, Gurr SJ. The tomato powdery mildew fungus Oidium neolycopersici. Mol. Plant Pathol. 2001;2:303–309. doi: 10.1046/j.1464-6722.2001.00084.x. [DOI] [PubMed] [Google Scholar]
- 12.Jacob D, David DR, Sztjenberg A, Elad Y. Conditions for development of powdery mildew of tomato caused by Oidium neolycopersici. Phytopathology. 2008;98:270–281. doi: 10.1094/PHYTO-98-3-0270. [DOI] [PubMed] [Google Scholar]
- 13.Zheng, Z. et al. Genome-Wide Study of the Tomato SlMLO Gene Family and Its Functional Characterization in Response to the Powdery Mildew Fungus Oidium neolycopersici. Frontiers in Plant Science7, doi:10.3389/fpls.2016.00380 (2016). [DOI] [PMC free article] [PubMed]
- 14.Kunkel BN, Brooks DM. Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 2002;5:325–331. doi: 10.1016/S1369-5266(02)00275-3. [DOI] [PubMed] [Google Scholar]
- 15.Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- 16.van Loon LC, Geraats BPJ, Linthorst HJM. Ethylene as a modulator of disease resistance in plants. Trends Plant Sci. 2006;11:184–191. doi: 10.1016/j.tplants.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Mulema JK, Denby K. Spatial and temporal transcriptomic analysis of the Arabidopsis thaliana–Botrytis cinerea interaction. Mol. Biol. Rep. 2012;39:4039–4049. doi: 10.1007/s11033-011-1185-4. [DOI] [PubMed] [Google Scholar]
- 18.Windram O, et al. Arabidopsis defense against Botrytis cinerea: chronology and regulation deciphered by high-resolution temporal transcriptomic analysis. Plant Cell. 2012;24:3530–3557. doi: 10.1105/tpc.112.102046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Birch PRJ, et al. The potato–Phytophthora infestans interaction transcriptome. Can. J. Plant Pathol. 2003;25:226–231. doi: 10.1080/07060660309507074. [DOI] [Google Scholar]
- 20.Gyetvai G, et al. The transcriptome of compatible and incompatible interactions of Potato (Solanum tuberosum) with Phytophthora infestans revealed by DeepSAGE analysis. PLoS ONE. 2012;7:e31526. doi: 10.1371/journal.pone.0031526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seifi A, et al. Genetics and molecular mechanisms of resistance to powdery mildews in tomato (Solanum lycopersicum) and its wild relatives. Eur. J. Plant Pathol. 2014;138:641–665. doi: 10.1007/s10658-013-0314-4. [DOI] [Google Scholar]
- 22.Robinson MD, McCarthy DJ, Smyth G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leng N, et al. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013;29:1035–1043. doi: 10.1093/bioinformatics/btt087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szklarczyk D, et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falcon S. & Gentleman, R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 26.Beißbarth T, Speed TP. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- 27.van Esse HP, Fradin EF, de Groot PJ, de Wit PJ, Thomma BP. Tomato transcriptional responses to a foliar and a vascular fungal pathogen are distinct. Mol. Plant-Microbe Interact. 2009;22:245–258. doi: 10.1094/MPMI-22-3-0245. [DOI] [PubMed] [Google Scholar]
- 28.Frenkel O, Brewer MT, Milgroom MG. Variation in pathogenicity and aggressiveness of Erysiphe necator from different Vitis spp. and geographic origins in the eastern United States. Phytopathology. 2010;100:1185–1193. doi: 10.1094/PHYTO-01-10-0023. [DOI] [PubMed] [Google Scholar]
- 29.Gadoury DM, et al. Grapevine powdery mildew (Erysiphe necator): a fascinating system for the study of the biology, ecology and epidemiology of an obligate biotroph. Mol. Plant Pathol. 2012;13:1–16. doi: 10.1111/j.1364-3703.2011.00728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huibers RP, et al. Powdery mildew resistance in tomato by impairment of SlPMR4 and SlDMR1. PLoS ONE. 2013;8:e67467. doi: 10.1371/journal.pone.0067467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao D, et al. Down-regulation of acetolactate synthase compromises Ol-1- mediated resistance to powdery mildew in tomato. BMC Plant Biol. 2014;14:32. doi: 10.1186/1471-2229-14-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iberkleid I, Sela N, Brown Miyara S. Meloidogyne javanica fatty acid- and retinol-binding protein (Mj-FAR-1) regulates expression of lipid-, cell wall-, stress- and phenylpropanoid-related genes during nematode infection of tomato. BMC Genomics. 2015;16:272. doi: 10.1186/s12864-015-1426-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang S, et al. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol. Genet. Genomics. 2012;287:495–513. doi: 10.1007/s00438-012-0696-6. [DOI] [PubMed] [Google Scholar]
- 34.Du H, Wang Y, Yang J, Yang W. Comparative transcriptome analysis of resistant and susceptible tomato lines in response to infection by Xanthomonas perforans race T3. Frontiers in Plant Science. 2015;6:1173. doi: 10.3389/fpls.2015.01173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okekeogbu I, et al. Effect of aluminum treatment on proteomes of radicles of seeds derived from Al-treated tomato plants. Proteomes. 2014;2:169. doi: 10.3390/proteomes2020169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Audran-Delalande C, et al. Genome-wide identification, functional analysis and expression profiling of the Aux/IAA gene family in tomato. Plant Cell Physiol. 2012;53:659–672. doi: 10.1093/pcp/pcs022. [DOI] [PubMed] [Google Scholar]
- 37.Yan F, Hu G, Ren Z, Deng W, Li Z. Ectopic expression a tomato KNOX Gene Tkn4 affects the formation and the differentiation of meristems and vasculature. Plant Mol. Biol. 2015;89:589–605. doi: 10.1007/s11103-015-0387-x. [DOI] [PubMed] [Google Scholar]
- 38.Etalo, D. W., Tameling, W. I., De Vos, R. C., Bouwmeester, H. J. & Joosten, M. H. Genome-wide functional analysis of WRKY transcription factors in resistance of tomato to Cladosporium fulvum Ph.D. thesis, Wageningen University (2014).
- 39.Gao Q-M, Venugopal S, Navarre D, Kachroo A. Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiol. 2011;155:464–476. doi: 10.1104/pp.110.166876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.KRUIJT MARCO, DE KOCK MAARTEN J. D., DE WIT PIERRE J. G. M. Receptor-like proteins involved in plant disease resistance. Molecular Plant Pathology. 2005;6:85–97. doi: 10.1111/j.1364-3703.2004.00264.x. [DOI] [PubMed] [Google Scholar]
- 41.Wu J, et al. Transcriptional regulation of receptor-like protein genes by environmental stresses and hormones and their overexpression activities in Arabidopsis thaliana. J. Exp. Bot. 2016;67:3339–3351. doi: 10.1093/jxb/erw152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong H, Delaney TP, Bauer DW, Beer SV. Harpin induces disease resistance in Arabidopsis through the systemic acquired resistance pathway mediated by salicylic acid and the NIM1 gene. The Plant Journal. 1999;20:207–215. doi: 10.1046/j.1365-313x.1999.00595.x. [DOI] [PubMed] [Google Scholar]
- 43.Harvey JJW, et al. An Antiviral Defense Role of AGO2 in Plants. PLoS ONE. 2011;6:e14639. doi: 10.1371/journal.pone.0014639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Höck J, Meister G. The Argonaute protein family. Genome Biology. 2008;9:210–210. doi: 10.1186/gb-2008-9-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X, et al. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 2006;20:3255–3268. doi: 10.1101/gad.1495506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J, et al. Genome-wide analysis of bHLH transcription factor and involvement in the infection by yellow leaf curl virus in tomato (Solanum lycopersicum) BMC Genomics. 2015;16:39. doi: 10.1186/s12864-015-1249-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan J, Wang J, Zhang H. An ankyrin repeat-containing protein plays a role in both disease resistance and antioxidation metabolism. The Plant Journal. 2002;29:193–202. doi: 10.1046/j.0960-7412.2001.01205.x. [DOI] [PubMed] [Google Scholar]
- 48.Elhiti M, Stasolla C. Structure and function of homodomain-leucine zipper (HD-Zip) proteins. Plant Signaling & Behavior. 2009;4:86–88. doi: 10.4161/psb.4.2.7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olsson A, Engström P, Söderman E. The homeobox genes ATHB12 and ATHB7encode potential regulators of growth in response to water deficit in Arabidopsis. Plant Mol. Biol. 2004;55:663–677. doi: 10.1007/s11103-004-1581-4. [DOI] [PubMed] [Google Scholar]
- 50.Wang Z, et al. Overexpression of an Arabidopsis heterogeneous nuclear ribonucleoprotein gene, AtRNP1, affects plant growth and reduces plant tolerance to drought and salt stresses. Biochem. Biophys. Res. Commun. 2016;472:353–359. doi: 10.1016/j.bbrc.2016.02.081. [DOI] [PubMed] [Google Scholar]
- 51.Luan S. The CBL–CIPK network in plant calcium signaling. Trends Plant Sci. 2009;14:37–42. doi: 10.1016/j.tplants.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 52.Zheng X-y, et al. Spatial and temporal regulation of biosynthesis of the plant immune signal salicylic acid. Proceedings of the National Academy of Sciences. 2015;112:9166–9173. doi: 10.1073/pnas.1511182112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiménez-Saa H. Revised nomenclature of compound leaves as an aid in field identification of tropical trees and other woody plants. Vulpia. 2011;9:1–11. [Google Scholar]
- 54.Untergasser A, et al. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115–e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Müller OA, et al. Genome-wide identification and validation of reference genes in infected tomato leaves for quantitative RT-PCR analyses. PLoS ONE. 2015;10:e0136499. doi: 10.1371/journal.pone.0136499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pombo M, et al. Transcriptomic analysis reveals tomato genes whose expression is induced specifically during effector-triggered immunity and identifies the Epk1 protein kinase which is required for the host response to three bacterial effector proteins. Genome Biology. 2014;15:492. doi: 10.1186/s13059-014-0492-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bolser, D. M., Staines, D. M., Perry, E. & Kersey, P. J. In Plant Genomics Databases: Methods and Protocols (ed. Aalt D. J. van Dijk) 1-31 (Springer New York, 2017).
- 58.Amselem J, et al. Genomic analysis of the aecrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 2011;7:e1002230. doi: 10.1371/journal.pgen.1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haas BJ, et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461:393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- 60.Schulz MH, Zerbino DR, Vingron M, Birney E. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics. 2012;28:1086–1092. doi: 10.1093/bioinformatics/bts094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Meth. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reimand J, Kull M, Peterson H, Hansen J, Vilo J. g:Profiler—a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35:W193–W200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reimand, J. et al. g: Profiler—a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res., gkw199 (2016). [DOI] [PMC free article] [PubMed]
- 65.von Mering C, et al. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


