Abstract
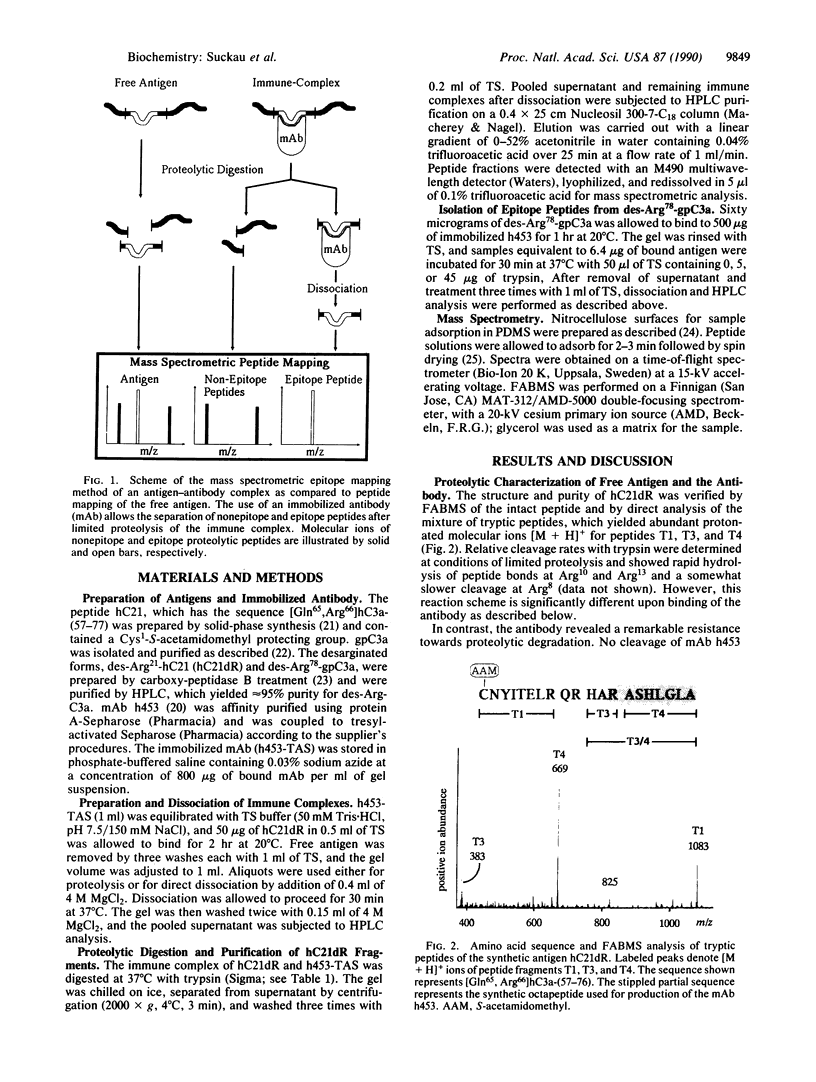
Sequences of antigenic determinants were identified by limited proteolysis of peptide antigens bound to an immobilized monoclonal antibody and direct molecular weight determination of the monoclonal antibody-bound peptide fragments by 252Cf plasma desorption mass spectrometry. The epitope peptides to the monoclonal antibody h453 [Burger, R., Zilow, G., Bader, A., Friedlein, A. & Naser, W. (1988) J. Immunol. 141, 553-558] were isolated from immobilized antigen-antibody complexes by partial trypsin digestion. A synthetic eicosapeptide comprised of the C-terminal sequence of the human complement component polypeptide des-Arg77-C3a as well as guinea pig des-Arg78-C3a was used as an antigen. Conditions were developed under which trypsin specifically degraded the antigens without inactivation of the immobilized antibody. After proteolysis, epitope peptides were dissociated from the antibody with 4 M MgCl2. The antigenic peptides were purified by HPLC and identified by 252Cf plasma desorption mass spectrometry. The epitope recognized by h453 resides on the C-terminal tryptic peptides of human (residues 70-76) and guinea pig (residues 70-77) C3a. As an estimation of accuracy this method is able to provide, trypsin digestion of immune complexes caused cleavage of the antigen within a distance of two amino acid residues upstream from the epitope.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambrosius D., Casaretto M., Gerardy-Schahn R., Saunders D., Brandenburg D., Zahn H. Peptide analogues of the anaphylatoxin C3a; syntheses and properties. Biol Chem Hoppe Seyler. 1989 Mar;370(3):217–227. doi: 10.1515/bchm3.1989.370.1.217. [DOI] [PubMed] [Google Scholar]
- Benjamin D. C., Berzofsky J. A., East I. J., Gurd F. R., Hannum C., Leach S. J., Margoliash E., Michael J. G., Miller A., Prager E. M. The antigenic structure of proteins: a reappraisal. Annu Rev Immunol. 1984;2:67–101. doi: 10.1146/annurev.iy.02.040184.000435. [DOI] [PubMed] [Google Scholar]
- Berzofsky J. A. Intrinsic and extrinsic factors in protein antigenic structure. Science. 1985 Sep 6;229(4717):932–940. doi: 10.1126/science.2410982. [DOI] [PubMed] [Google Scholar]
- Burger R., Zilow G., Bader A., Friedlein A., Naser W. The C terminus of the anaphylatoxin C3a generated upon complement activation represents a neoantigenic determinant with diagnostic potential. J Immunol. 1988 Jul 15;141(2):553–558. [PubMed] [Google Scholar]
- Chazin W. J., Hugli T. E., Wright P. E. 1H NMR studies of human C3a anaphylatoxin in solution: sequential resonance assignments, secondary structure, and global fold. Biochemistry. 1988 Dec 27;27(26):9139–9148. doi: 10.1021/bi00426a011. [DOI] [PubMed] [Google Scholar]
- Cotter R. J. Plasma desorption mass spectrometry: coming of age. Anal Chem. 1988 Jul 1;60(13):781A–793A. doi: 10.1021/ac00164a002. [DOI] [PubMed] [Google Scholar]
- Eisenberg R. J., Long D., Pereira L., Hampar B., Zweig M., Cohen G. H. Effect of monoclonal antibodies on limited proteolysis of native glycoprotein gD of herpes simplex virus type 1. J Virol. 1982 Feb;41(2):478–488. doi: 10.1128/jvi.41.2.478-488.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauss C., Klein J., Post K., Suckau D., Schneider K., Thomas H., Oesch F., Przybylski M. Mass spectrometric peptide mapping analysis and structural characterization of dihydrodiol dehydrogenase isoenzymes. Environ Health Perspect. 1990 Aug;88:57–62. doi: 10.1289/ehp.908857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerard N. P., Lively M. O., Gerard C. Amino acid sequence of guinea pig C3a anaphylatoxin. Protein Seq Data Anal. 1988;1(6):473–478. [PubMed] [Google Scholar]
- Gerardy-Schahn R., Ambrosius D., Casaretto M., Grötzinger J., Saunders D., Wollmer A., Brandenburg D., Bitter-Suermann D. Design and biological activity of a new generation of synthetic C3a analogues by combination of peptidic and non-peptidic elements. Biochem J. 1988 Oct 1;255(1):209–216. [PMC free article] [PubMed] [Google Scholar]
- Gerardy-Schahn R., Ambrosius D., Saunders D., Casaretto M., Mittler C., Karwarth G., Görgen S., Bitter-Suermann D. Characterization of C3a receptor-proteins on guinea pig platelets and human polymorphonuclear leukocytes. Eur J Immunol. 1989 Jun;19(6):1095–1102. doi: 10.1002/eji.1830190620. [DOI] [PubMed] [Google Scholar]
- Hoffmann T., Böttger E. C., Baum H. P., Messner M., Hadding U., Bitter-Suermann D. In vivo effects of C3a on neutrophils and its contribution to inflammatory lung processes in a guinea-pig model. Clin Exp Immunol. 1988 Mar;71(3):486–492. [PMC free article] [PubMed] [Google Scholar]
- Huber R., Scholze H., Pâques E. P., Deisenhofer J. Crystal structure analysis and molecular model of human C3a anaphylatoxin. Hoppe Seylers Z Physiol Chem. 1980 Sep;361(9):1389–1399. doi: 10.1515/bchm2.1980.361.2.1389. [DOI] [PubMed] [Google Scholar]
- Hugli T. E. Biochemistry and biology of anaphylatoxins. Complement. 1986;3(3):111–127. doi: 10.1159/000467889. [DOI] [PubMed] [Google Scholar]
- Hugli T. E. Human anaphylatoxin (C3a) from the third component of complement. Primary structure. J Biol Chem. 1975 Nov 10;250(21):8293–8301. [PubMed] [Google Scholar]
- Jemmerson R., Paterson Y. Mapping epitopes on a protein antigen by the proteolysis of antigen-antibody complexes. Science. 1986 May 23;232(4753):1001–1004. doi: 10.1126/science.2422757. [DOI] [PubMed] [Google Scholar]
- Kessler S. W. Rapid isolation of antigens from cells with a staphylococcal protein A-antibody adsorbent: parameters of the interaction of antibody-antigen complexes with protein A. J Immunol. 1975 Dec;115(6):1617–1624. [PubMed] [Google Scholar]
- Köhl J., Casaretto M., Gier M., Karwath G., Gietz C., Bautsch W., Saunders D., Bitter-Suermann D. Reevaluation of the C3a active site using short synthetic C3a analogues. Eur J Immunol. 1990 Jul;20(7):1463–1468. doi: 10.1002/eji.1830200709. [DOI] [PubMed] [Google Scholar]
- Moelling K., Scott A., Dittmar K. E., Owada M. Effect of p15-associated protease from an avian RNA tumor virus on avian virus-specific polyprotein precursors. J Virol. 1980 Feb;33(2):680–688. doi: 10.1128/jvi.33.2.680-688.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwyzer M., Weil R., Frank G., Zuber H. Amino acid sequence analysis of fragments generated by partial proteolysis from large simian virus 40 tumor antigen. J Biol Chem. 1980 Jun 25;255(12):5627–5634. [PubMed] [Google Scholar]
- Sheshberadaran H., Payne L. G. Protein antigen-monoclonal antibody contact sites investigated by limited proteolysis of monoclonal antibody-bound antigen: protein "footprinting". Proc Natl Acad Sci U S A. 1988 Jan;85(1):1–5. doi: 10.1073/pnas.85.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J., Bode W. The X-ray crystal structure analysis of the refined complex formed by bovine trypsin and p-amidinophenylpyruvate at 1.4 A resolution. Hoppe Seylers Z Physiol Chem. 1983 Aug;364(8):949–959. doi: 10.1515/bchm2.1983.364.2.949. [DOI] [PubMed] [Google Scholar]



