Abstract
The histone methyltransferase EZH2 is required for B and T cell development; however the molecular mechanisms underlying this requirement remain elusive. In a murine model of lymphoid specific EZH2-deficiency we found that EZH2 was required for proper development of adaptive, but not innate, lymphoid cells. In adaptive lymphoid cells EZH2 prevented the premature expression of Cdkn2a and the consequent stabilization of p53, an effector of the pre-antigen receptor checkpoints. Deletion of Cdkn2a in Ezh2-deficient lymphocytes prevented p53 stabilization, extended lymphocyte survival, and restored differentiation resulting in the generation of mature B and T lymphocytes. Our results uncover a crucial role for EZH2 in adaptive lymphocytes to control the developmental timing of effectors of the pre-antigen receptor checkpoints.
Introduction
The lymphoid arm of the immune system is divided into innate and adaptive components that can be distinguished by the mechanisms they use to detect and respond to invading pathogens(1). Innate lymphoid cells (ILCs) express an array of germ line-encoded receptors that rapidly induce cytokine production or cytotoxic functions when triggered by invading pathogens. In contrast, adaptive lymphoid cells detect pathogens using antigen-specific receptors that are generated through the recombinase activating gene (RAG)-dependent assembly of variable (V), diversity (D), and joining (J) gene segments. The process of V(D)J recombination creates unique receptors on each developing B or T lymphocyte, but it also produces many defective receptors and puts the cells at risk for oncogenic transformation. Therefore, developing B and T cells must pass through “checkpoints” to ensure that a functional receptor is produced before they progress to subsequent stages of differentiation and before they undergo further proliferation(2). The mechanisms that ensure appropriate activation of antigen-receptor checkpoints after initiation of V(D)J recombination are not well understood.
V(D)J recombination occurs in an ordered manner in which pro-B lymphocytes rearrange their immunoglobulin heavy chain gene (Igh) segments before they progress to the pre-B cell stage, where they rearrange the light chain (IgL) genes(3). Similarly, Tcrb rearrangements occur in double negative (DN) 3 thymocytes followed by Tcra rearrangements at the double positive (DP) stage of T cell development. The production of a functional IgH chain or TCRβ chain is ensured by a checkpoint in which these proteins, as part of a pre-BCR or pre-TCR, impart a signal to the cell that inhibits the p53-induced apoptosis pathway(4, 5). Destabilizing p53 is a critical requirement for progression beyond the pre-antigen receptor checkpoint as revealed in mice lacking the pre-TCR (i.e. Rag1−/− or Cd3γ−/− mice), which fail β-selection but can by-pass this checkpoint when Trp53 (p53) is deleted or mutated(6, 7). Stabilization of p53 prevents B and T cell development beyond the pre-antigen receptor checkpoints, as evidenced in mice lacking genes that directly regulate p53 at the transcriptional (WIP1(8, 9), MYSM1(10)), post-transcriptional (CNOT(11)), translational (RPL22(12, 13), MIZ1(14, 15)), or post-translational (YY1(16–18), BCL11A(19), NIR(20)) levels. Remarkably, B and T cell maturation are substantially restored in these mice upon deletion of p53.
p53 is exquisitely regulated in lymphocyte progenitors to ensure that it is only transiently expressed at the time of pre-antigen receptor selection. While Trp53 mRNA is constitutively transcribed in pro-B lymphocytes prior to the requirement for pre-BCR selection, p53 is not present due to its rapid degradation by the proteasome(21). Degradation of p53 is initiated by the ubiquitin ligase, MDM2(21). At the pre-BCR checkpoint p53 becomes stabilized when MDM2 is inhibited by p19ARF, which is one of two proteins produced from the Cdkn2a gene(22). Transcription of Cdkn2a is induced, in part, by the transcription factor BACH2, which is expressed in pro-B lymphocytes; but how transcription of Cdkn2a is restrained until the pre-BCR checkpoint is not well understood(5, 23). Signaling through the pre-BCR restores p53 degradation by inducing the activity of the transcriptional repressor BCL6, which displaces BACH2 from the Cdkn2a gene and silences Cdkn2a/p19 expression thereby allowing the survival of appropriately signaled pre-B cells. The proper activation and repression of the pre-antigen receptor checkpoints are essential for proper lymphocyte development and prevent lymphocyte transformation.
The developmental programs of innate and adaptive lymphocytes are established by lineage defining transcription factors in collaboration with chromatin modifying complexes that regulate genome accessibility. Polycomb repressive complex 2 (PRC2) is a protein complex that contains the histone methyltransferase EZH2, which catalyzes the methylation of histone 3 lysine 27 (H3K27)(24). H3K27me3 is found at the promoters and along the gene bodies of repressed genes, and at the promoters of genes that are poised for activation(25). Inducible deletion of the SET domain of EZH2 in all hematopoietic cells results in arrested lymphocyte development at the pro-B cell stage(26) and DN3 stage(27). In both cases the arrest is consistent with a failure of EZH2-deficient cells to pass through the pre-antigen receptor checkpoints; however, it is not known why these cells should fail these checkpoints. EZH2-deficient pro-B cells have proximal V(D)J rearrangements and a subset of cells express intracellular IgH suggesting that the arrested differentiation extends beyond the ability to create an IgH chain(26). Nonetheless, transgenic expression of a pre-arranged IgH chain partially restores B cell development in these mice, consistent with the hypothesis that EZH2-deficient B cell progenitors fail the pre-BCR checkpoint. EZH2-deficient DN3 cells have normal TCRβ and TCRγδ rearrangements and transgenic expression of a functional TCRβ chain fails to rescue T differentiation (27). Therefore, while EZH2 is clearly required for B and T lymphocyte development, the mechanisms underlying the developmental arrest and how EZH2 impacts the pre-antigen receptor checkpoints is not understood.
Here we demonstrate that lymphoid specific deletion of Ezh2 has little effect on innate lymphoid cell development but impacts adaptive lymphopoiesis due to the premature activation of critical effectors of the pre-antigen receptor checkpoint. We show that EZH2 is required in B and T lymphocytes to repress expression of Cdkn2a and prevent stabilization of p53. Deletion of Cdkn2a from EZH2-deficient lymphoid cells extended lymphocyte progenitor survival and allowed for the development of mature B and T lymphocytes. Our results reveal a critical requirement for EZH2 to maintain repression of Cdkn2a in adaptive lymphoid cells prior to its programmed activation during the process of pre-antigen receptor selection.
Materials and Methods
Mice
All mice were on a C57BL/6 background and were housed at the animal resource center at the University of Chicago. Animal protocols were carried out in accordance with guidelines set by The University of Chicago Institutional Animal Care and Use Committee. Ezh2fl/fl mice were from A. Tarakhovsky (Rockefeller University, New York)(26). Il7racre mice were from H.-R. Rodewald (Deutsches Krebsforschungszentrum, Heidelberg)(28). Cdkn2a−/− mice were obtained from the National Cancer Institute(29). Rosa26YFP and CD45.1 C57BL/6 mice were purchased from Jackson Labs.
Flow Cytometry
Antibodies were obtained from ebioscience, BD, Biolegend, and Cell Signaling and were conjugated to biotin, FITC, PE, APC, APC-EF780, PECy7, PerCP-Cy5.5, EF450, Pacific Blue, or Brilliant Violet 421. Specific antibody clones are available on request. To examine or sort common lymphoid progenitors (CLPs) and ILC2 progenitors (ILC2Ps), bone marrow was depleted with: B220, CD11b, Gr1, and Ter119. For ETP analysis and DN3 sorts, thymocytes were depleted with: B220, CD3e, CD8, and Ter119. Lineage for CLP, ILC2P, ETP was defined as: B220, CD11b, CD11c, CD3e, CD8, TCRb, TCRgd, NK11, Gr1, Ter119. Lineage for NK: CD19, CD3, CD8, CD4, Ter119. Propidium iodide (PI) was used for live/dead exclusion. Intracellular staining for EZH2 and H3K27me3 was performed with the FoxP3/Transcription Factor Staining Kit (ebioscience). H3K27me3 was detected with a secondary anti-rabbit IgG directly conjugated to PE. Samples were analyzed on an LSRII or Fortessa and cells were sorted on a FACS Aria running FACS Diva software. Analysis was carried out in FlowJo.
Cell culture
Cells were maintained in OPTI-MEM media supplemented with 10% FBS, 80 mM 2-mercaptoethanol, 100 units/ml penicillin, 100 mg/ml streptomycin, and 29.2 mg/ml glutamine. Pro-B cultures were initiated with B220+ cells magnetically enriched from the bone marrow by MACS-based separation (Mitenyl) and were supplemented with IL-7 (produced from J558 cells stably transduced with an IL-7 expression vector)(30). NK cell cultures were initiated with DX5+ cells enriched from the spleen and were supplemented with IL2 (1000 IU/mL, NIH Reagents Program). GSK126 (10 uM) and DZnep (5 uM) were added to WT pro-B cultures after 3 days.
Apoptosis and proliferation assays
Apoptosis was measured by pan-caspase (FAM)-FLICA detection kit (Immunochemistry Technologies) according to the manufacturer’s protocol. As a positive control for in vitro apoptosis assays, cells were treated with etoposide (10 uM, Sigma) for 4h prior to caspase detection. For in vitro proliferation assays, cycling pro-B cells were incubated with BrdU for 45 min at 37 °C prior to surface staining and anti-BrdU detection with the FITC BrdU Flow Kit (BD Biosciences).
Chimeras
Chimeric mice were generated through retro-orbital injection of 5 × 106 total BM cells into lethally irradiated (1000 rad) recipient CD45.1 mice. Injected BM consisted of a 1:1 mix of CD45.1 or CD45.1/CD45.2 WT competitor BM with CD45.2 BM from Il7racre/+RosaYFP or Il7racre/+Ezh2fl/flRosaYFP mice. Chimeric mice were kept on Bactrim water and were analyzed 10–12 weeks post reconstitution.
Western Blot
Cultured pro-B cells or CD3-CD4-CD8-Ter119-depleted thymocytes were lysed in RIPA buffer. 10 ug total protein was run on a 10% acrylamide gel and transferred to PVDF by wet electrophoretic transfer. Membranes were probed with anti-p53 (clone 1C12, Cell Signaling Technologies) and anti-beta actin (product ab8227, Abcam) followed by secondary HRP-conjugated antibodies and detection by ECL according to the manufacturer’s protocol (SuperSignal West Dura Extended Duration Substrate, Thermoscientific).
ChIP
Cultured pro-B cells or NK cells from WT mice were crosslinked in 1% formaldehyde at room temperature for 10 minutes and then quenched with glycine (125 mM). 2 × 107 cells were lysed in 600 uL RIPA prior to sonication with the Diagenode Biorupter water bath sonicator at high intensity in cycles of 30s ON-30s OFF for 45 minutes. Protein-DNA complexes were precipitated with H3K27me3 antibody (clone ab6002, Abcam). Samples were decrosslinked and DNA was purified with a PCR purification kit (Qiagen). DNA was amplified by quantitative RT-PCR with SYBR green (Bio Rad Laboratories). Enrichment is reported relative to input DNA. Primer sequences are available on request.
RNAseq and Data Processing
pro-B, DN3, ILC2P, splenic NK, and CLPs were sorted from Il7racre/+ and Il7racre/+Ezh2fl/fl mice into RLT buffer. Total RNA was isolated using RNAeasy MicroKit (Qiagen) according to manufacturer’s recommendations. Libraries were constructed using Nugen’s Ovation Ultralow Library systems and were subsequently subjected to 76 cycles of NextSeq500 sequencing. Raw sequence reads were trimmed using Trimmomatic v 0.33 (TRAILING:30 MINLEN:20)(31) and then aligned to mouse genome assembly mm10 with Tophat v 2.1.0(32). Reads were assigned to genes using the htseq-count tool from HTSeq v 0.6.1(33) and gene annotations from Ensembl release 78(34). The R package EdgeR(35) was used to normalize the gene counts and to calculate differential expression statistics for each gene for each pairwise comparison of sample groups. Gene set enrichment analysis (GSEA) analysis was performed using gene sets from the Hallmark Pathways of the MSigDB(36). Genes were considered differentially expressed if the |fold change| >= 2 and FDR < 0.05. RNA-sequencing data can be accessed in the Gene Expression Omnibus (GSE97462).
qPCR
RNA was isolated from sorted or cultured cells by Trizol (Invitrogen) or the RNAeasy mini kit (Qiagen) and was reverse-transcribed with Superscript III (Invitrogen). Quantitative RT-PCR was performed in an iCycler (Bio Rad Laboratories) with SYBR green (Bio Rad Laboratories). Expression values were normalized to Hprt and were calculated by the ΔCT method. Primer sequences are available on request.
Statistics
Unless otherwise noted a Student’s T test was used to establish significant differences for in vitro and in vivo phenotypes. To compare between the three genotypes in Figure 6, the statistical test used was a repeated mean one-way anova followed by fisher’s least significant difference test uncorrected for multiple hypothesis testing. Statistical tests were calculated in Excel or with GraphPad Prism software. * p < 0.05, ** p < 0.01, *** p < 0.001.
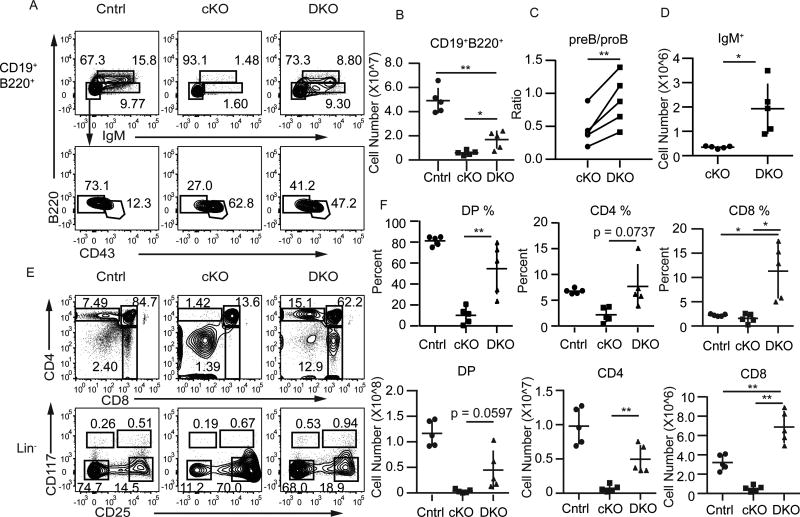
FIGURE 6.
Cdkn2a limits maturation of Ezh2Δ/Δ B and T cell progenitors. (A) Flow cytometry analysis showing IgM versus B220 on gated B220+CD19+ bone marrow cells (upper, Mature-Recirculating B220hiCD19+IgM+, Immature B220loCD19+IgM+) and CD43 versus B220 on B220+CD19+IgM cells (lower, pre-B B220loCD19+CD43, pro-B B220loCD19+CD43+) in Cdkn2a−/−Il7racre/+Ezh2fl/fl (DKO, right) mice compared to Il7racre/+Ezh2fl/fl (cKO, middle) or control (Cntrl, left). Control genotypes include Cdkn2a−/−Il7racre/+, Cdkn2a−/−, and Il7racre/+ mice. (B) Summary of the number of B220+CD19+ cells in the bone marrow of Cntrl, cKO, or DKO mice. (C) Summary of the ratio of pre-B to pro-B lymphocytes in cKO and DKO mice. (D) Summary of the number of IgM+ cells in the bone marrow of cKO and DKO mice. (E) Flow cytometry analysis showing CD4 and CD8 (top) or CD117 and CD25 on Lin cells in the thymus of Cntrl (left), cKO (middle), and DKO (right) mice. (F) Summary of the frequency (top) and number (bottom) of DP (left, CD4+CD8+), CD4 (middle), and CD8 (right) thymocytes. Data represent five independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001.
RESULTS
B and T cell progenitors required EZH2 prior to the pre-antigen receptor checkpoints
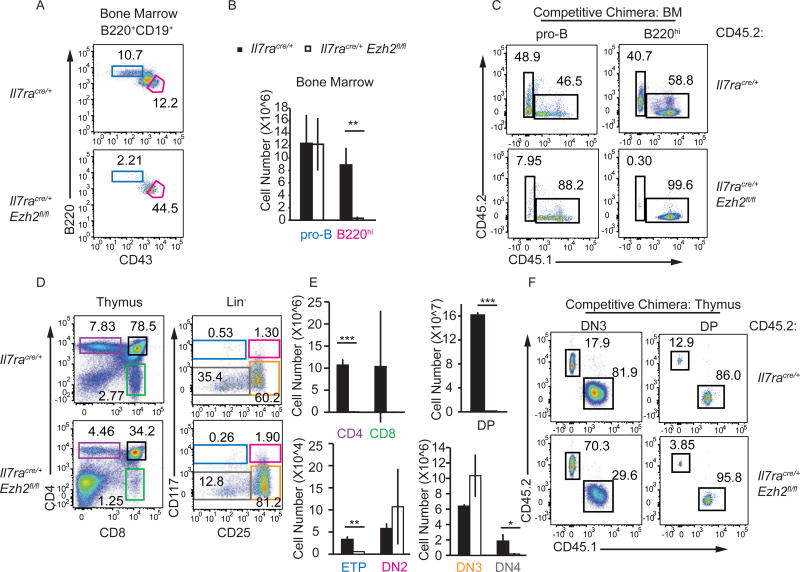
To gain insight into the requirement for EZH2 in B and T cell development we created Il7racre/+Ezh2fl/fl (Ezh2Δ/Δ) mice. Consistent with the phenotype observed in MxcreEzh2fl/fl mice(26, 27), there were few mature CD19+B220hi cells or CD19+B220loCD43− pre-B and immature B cells in the bone marrow of Ezh2Δ/Δ mice. However, CD19+B220loCD43+ pro-B cells and B-lymphoid biased Ly6D+ CLPs (also called B lymphocyte progenitors/BLP) were present at numbers similar to controls (Fig. 1A, 1B, Supplemental Fig. 1A). To examine B cell development under more stringent conditions we set up bone marrow chimeric mice in which Ezh2Δ/Δ cells developed in competition with congenic WT cells. In these chimeras, Ezh2Δ/Δ pro-B cells failed to develop, although they effectively reconstituted the BLP compartment (Fig. 1C, Supplemental Fig. 1B). Therefore, deletion of Ezh2 in lymphocyte progenitors compromised early B cell development revealing that EZH2 has important functions prior to the pre-BCR checkpoint.
FIGURE 1.
EZH2 is required for B and T cell development. (A) Flow cytometry analysis showing B220 versus CD43 on bone marrow B220+CD19+ cells from Il7racre/+ and Il7racre/+Ezh2fl/fl mice. The gated regions show pro-B cells (red, CD19+B220+CD43+) and mature B cells (blue, CD19+B220hi). (B) The bargraph shows the mean number of pro-B and B220hi mature B cells in the bone marrow of Il7racre/+ (black) and Il7racre/+Ezh2fl/fl (white) mice. (C) Flow cytometry analysis for CD45.1 and CD45.2 on pro-B cells and B220hi mature B cells from the bone marrow of competitive bone marrow chimeric mice 10 weeks post-reconstitution. Chimeras were established using a 1:1 ratio of bone marrow cells from CD45.1 WT B6 and CD45.2 Il7racre/+RosaYFP (top) or Il7racre/+Ezh2fl/flRosaYFP (bottom). (D) Flow cytometry analysis for CD4 versus CD8 on total thymocytes or CD117 versus CD25 on Lin thymocytes from Il7racre/+ or Il7racre/+Ezh2fl/fl mice. (E) The bargraphs show the mean number of CD4, CD8, DP (CD4+CD8+), ETP (LinCD117+CD25), DN2 (LinCD117+CD25+), DN3 (LinCD117CD25+) and DN4 (LinCD117CD25) cells in the thymus of Il7racre/+ (black) and Il7racre/+Ezh2fl/fl (white) mice. (F) Flow cytometry analysis showing CD45.1 versus CD45.2 on DN3 and DP thymocytes from competitive chimeric mice as in C. Data are representative of at least three independent experiments (mean ± SD). * p<0.05, ** p< 0.01, *** p< 0.001
As expected, Ezh2Δ/Δ mice had a reduced frequency and number of CD4, CD8, and DP thymocytes (Fig. 1D). ETP numbers were reduced in Ezh2Δ/Δ mice but there were normal or increased numbers of DN2 and DN3 thymocytes (Fig. 1E). DN4 cell numbers, however, were reduced (Fig. 1E). Interestingly, CD25 expression was increased on Ezh2Δ/Δ DN3 thymocytes suggesting that these cells may be arrested at an early DN3 stage (Fig. 1D). Competitive chimeras revealed that the failure to generate DP cells was intrinsic to the Ezh2Δ/Δ cells, although Ezh2Δ/Δ cells effectively competed for the DN3 compartment (Fig. 1F). These data suggest that EZH2 was required prior to, or at the time of, β-selection.
EZH2 was dispensable for innate lymphoid cell development
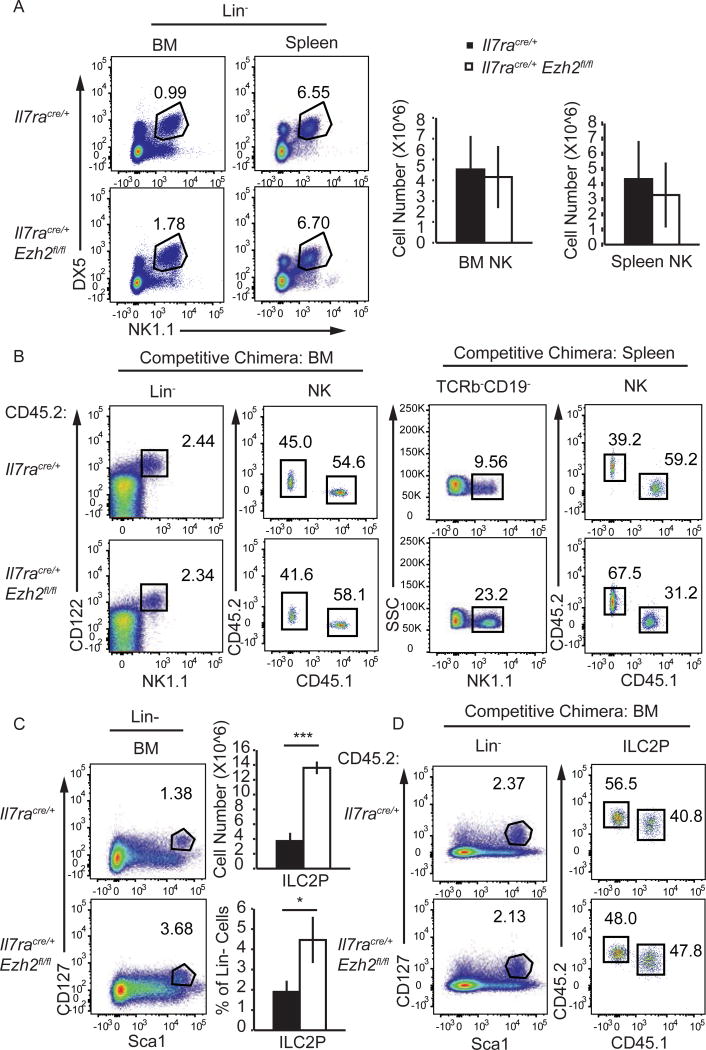
In contrast to adaptive lymphoid cells, NK cells were present in normal numbers in the BM and spleen of Ezh2Δ/Δ mice (Fig. 2A). Our results differed from a prior study that reported a modest increase in NK cell number in the spleen of Vav1creEzh2fl/fl mice(37). In competitive chimeras, Ezh2Δ/Δ bone marrow NK cells were present and splenic NK cell numbers were expanded when compared to controls, indicating that EZH2 limits NK cell expansion or survival (Fig. 2B). We also found that the frequency and number of bone marrow ILC2P was increased in Ezh2Δ/Δ mice (Fig. 2C). However, in mixed bone marrow chimeras Ezh2Δ/Δ ILC2P were present at a similar frequency as the control indicating that the expansion was not intrinsic to ILC2P (Fig. 2D). Overall, our data revealed that EZH2 was not required for NK cell or ILC2P development and that it limited NK cell expansion under competitive conditions.
FIGURE 2.
EZH2 is not required for NK cell or ILC2 development. (A) Flow cytometry analysis showing NK1.1 versus DX5 on Lin cells in the bone marrow and spleen of Il7racre/+ and Il7racre/+Ezh2fl/fl mice. Mature NK cells (LinDX5+NK1.1+) are shown in the gated regions. The bargraphs show the mean number of mature NK cells in the bone marrow and spleen of Il7racre/+ (black) and Il7racre/+Ezh2fl/fl (white) mice. (B) Flow cytometry analysis showing NK1.1 versus CD122 on Lin cells and CD45.1 versus CD45.2 on the gated Lin or TCRβCD19 CD122+NK1.1+ NK cells in the bone marrow (left) and spleen (right) of competitive chimeric mice as in Fig. 1C. (C) Flow cytometry analysis showing Sca1 versus CD127 on Lin cells from the bone marrow of Il7racre/+ or Il7racre/+Ezh2fl/fl mice. The gated region shows ILC2P (LinScalhiCD127+). The bargraphs show the mean number (top) or frequency (bottom) of ILC2P among Lin cells in the bone marrow of Il7racre/+ (black) and Il7racre/+Ezh2fl/fl (white) mice. (D) Flow cytometry analysis for ILC2P in the bone marrow of competitive chimeric mice as in Fig. 1C. Data are representative of at least three (A, C), five (B), or six (D) independent experiments (mean ± SD). * p < 0.05, ** p < 0.01, *** p < 0.001.
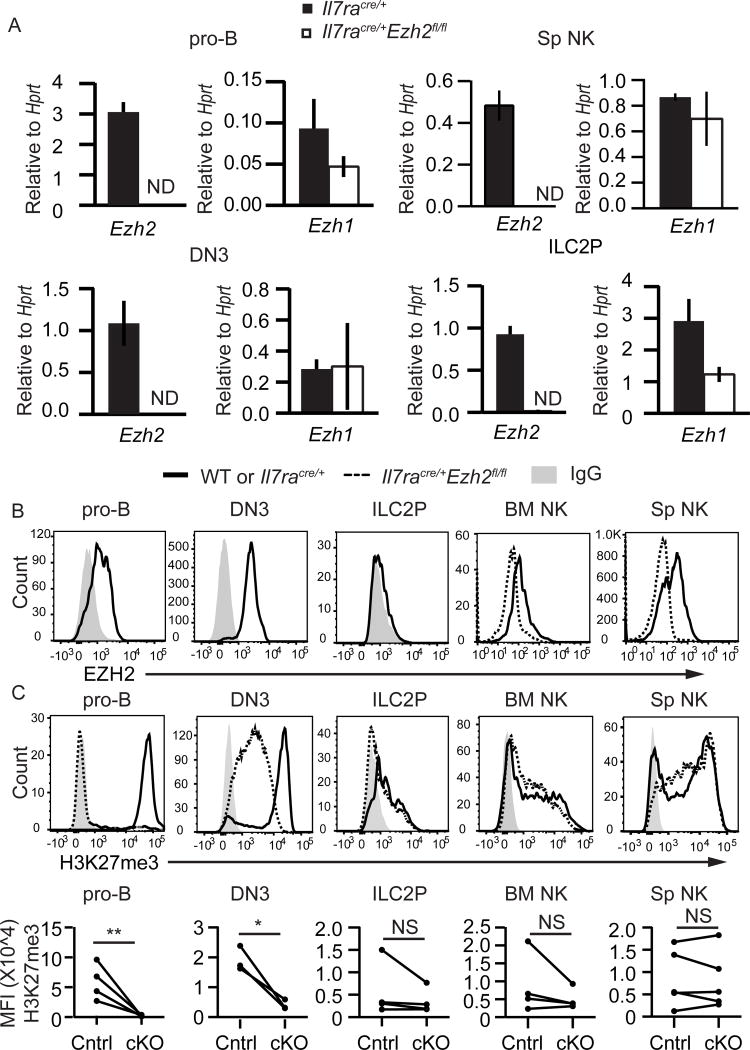
EZH2 was the primary H3K27me3 methyltransferase in B and T cell progenitors
To better understand how H3K27me3 is regulated in lymphocytes we examined the expression of known regulators of this histone modification. EZH2 is the primary enzyme to catalyze H3K27me3 modifications but there is an alternative PRC2 complex that contains a related enzyme, EZH1(25). EZH1 has lower methyltransferase activity in vitro than EZH2(38), however, it could maintain H3K27me3 marks that are already present at the time EZH2 is deleted (39). In both pro-B cells and DN3 cells, there was more Ezh2 mRNA than Ezh1 mRNA and deletion of Ezh2 did not result in increased Ezh1 mRNA (Fig. 3A). In contrast, NK cells and ILC2Ps expressed more Ezh1 mRNA than Ezh2 mRNA (Fig. 3A). EZH2 protein was expressed in pro-B cells and DN3 cells, and at low levels in NK cells but it was not detected in ILC2Ps (Fig. 3B). In Ezh2Δ/Δ pro-B cells H3K27me3 was reduced to background levels indicating that EZH2 is the major regulator of H3K27me3 in these cells (Fig. 3C). Ezh2Δ/Δ DN3 cells had reduced levels of H3K27me3 whereas the amount of H3K27me3 in Ezh2Δ/Δ NK cells and ILC2P was similar to controls (Fig. 3D). Taken together, these data indicate that EZH2 controlled the amount of H3K27me3 in adaptive lymphocyte progenitors but was not required for H3K27me3 in innate lymphoid cells, consistent with their development in Ezh2Δ/Δ mice.
FIGURE 3.
EZH2 is the primary H3K27me3 methyltransferase in B and T cell progenitors. (A) Quantitative RT-PCR analysis of Ezh2 and Ezh1 mRNA expression in sorted pro-B, DN3, splenic NK, and BM ILC2P cells from Il7racre/+ (black) or Il7racre/+Ezh2fl/fl (white) mice. Results are presented relative to Hprt. (B) Flow cytometry analysis showing EZH2 protein expression in pro-B (B220+CD19+CD43+), DN3 (LinCD25+CD117), NK (LinDX5+NK1.1+), and BM ILC2P (LinSca1hiCD127+) cells in WT or Il7racre/+ mice (solid) relative to expression in Il7racre/+Ezh2fl/fl mice (dashed) or IgG control (shaded histogram). (C) Flow cytometry analysis showing H3K27me3 in pro-B, DN3, NK, and BM ILC2P cells (top). Summary of H3K27me3 mean fluorescent intensity (bottom). Each circle represents one sample. Data are representative of at least three independent experiments. Bargraphs are mean ± SD. * p < 0.05, ** p < 0.01, *** p < 0.001.
EZH2 repressed Cdkn2a and the p53 pathway in B and T progenitors
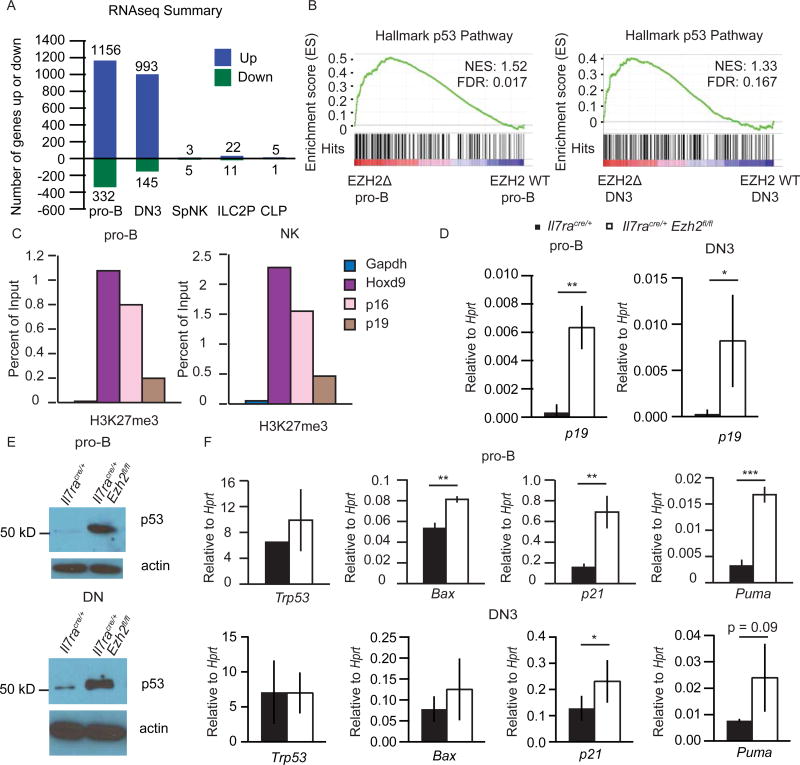
To better understand the role of EZH2 in lymphocyte development, we examined gene expression profiles in pro-B, DN3, splenic NK, ILC2P cells, and CLPs by RNA-sequencing. Notably, 1156 and 993 genes were increased in expression in Ezh2Δ/Δ pro-B and DN3 cells, respectively, as compared to controls. In contrast, very few genes were increased in expression in Ezh2Δ/Δ ILC2P or NK cells (Fig. 4A). In pro-B and DN3 cells only 332 and 145 genes decreased, respectively, in the absence of EZH2 and even fewer genes decreased in ILC2P or NK cells. GSEA revealed enrichment for genes associated with the Hallmark p53 pathway in both Ezh2Δ/Δ pro-B and DN3 cells (Fig. 4B). One of the genes in this pathway, Cdkn2a, is a known target of EZH2 in multiple cell types(24). Cdkn2a codes for p19ARF, which stabilizes p53, and p16INK4a, an inhibitor of CDK4(22), both of which could impact the survival and expansion of adaptive lymphoid cells.
FIGURE 4.
EZH2 represses the Cdkn2a–p53 pathway in B and T cell progenitors. (A) Summary of RNA-seq data from sorted pro-B (B220+CD19+CD43+), DN3 (LinCD25+cKit), splenic NK (LinDX5+NK1.1+), BM ILC2P (LinSca1hiCD127+) and CLPs (LinCD117intCD127+CD135+). The number of genes that increased or decreased >= 2-fold averaged across 2–3 samples per genotype with FDR < 0.05 in Ezh2Δ/Δ vs. Il7racre/+ control is shown. (B) Gene set enrichment analysis of p53 pathway enriched in Ezh2Δ/Δ pro-B and DN3 cells relative to Il7racre/+ control. (C) ChIP analysis with anti-H3K27me3 antibody followed by quantitative RT-PCR using primers specific for the promoters of p16 and p19 in WT pro-B or NK cells from culture as in Fig. 5A. (D) Quantitative RT-PCR analysis of p19 mRNA expression in sorted pro-B and DN3 cells from Il7racre/+ (black) or Il7racre/+Ezh2fl/fl (white) mice. Results are presented relative to Hprt. (E) Western blot for p53 in pro-B cells from culture (top) and in thymocytes magnetically depleted for CD4, CD3, CD8 and Ter119 (DN, bottom). (F) Quantitative RT-PCR analysis of Trp53, Bax, p21, and Puma mRNA expression in sorted pro-B (top) and DN3 (bottom) cells from Il7racre/+ (black) or Il7racre/+Ezh2fl/fl (white) mice. Results are presented relative to Hprt. ChIP data are representative of four (C, pro-B) or two (C, NK) experiments. Data in D–F are representative of at least three independent experiments. Mean ± SD (D and F). * p < 0.05, ** p < 0.01, *** p < 0.001.
We next set out to rigorously address whether Cdkn2a and the p53 pathway were regulated by EZH2 in adaptive lymphocyte progenitors. We found that Cdkn2a was marked by H3K27me3 in pro-B and DN3 cells using publically available ChIP-seq data sets(40, 41) (Supplemental Fig. 2A), and we confirmed this in WT pro-B cells using ChIP-qPCR (Fig. 4C). The p16 and p19 promoters were also marked by H3K27me3 in WT NK cells but p16 and p19 mRNAs were not expressed in Ezh2Δ/Δ NK cells cultured in vitro (Fig. 4C, Supplemental Fig. 2B). Quantitative PCR analysis revealed that p19 mRNA was increased in both pro-B and DN3 cells from Ezh2Δ/Δ mice (Fig. 4D). As p19ARF stabilizes p53, we found that p53 was abundant in Ezh2Δ/Δ pro-B cells and DN thymocytes whereas it was barely detectable in control cells (Fig. 4E). Several canonical p53 target genes were also increased in Ezh2Δ/Δ pro-B and DN3 cells by qPCR and RNA sequencing (Fig. 4F). Trp53 mRNA was not altered consistent with a post-transcriptional regulation of this protein(21) (Fig. 4F). In addition to H3K27me3, the Cdkn2a locus was also marked by H3K4me2 in WT pro-B, DN3 and NK cells(41–43) suggesting that this locus was primed for expression in these cells (Supplemental Fig. 2A).
EZH2 was required for the proliferation and survival of B lymphocyte progenitors
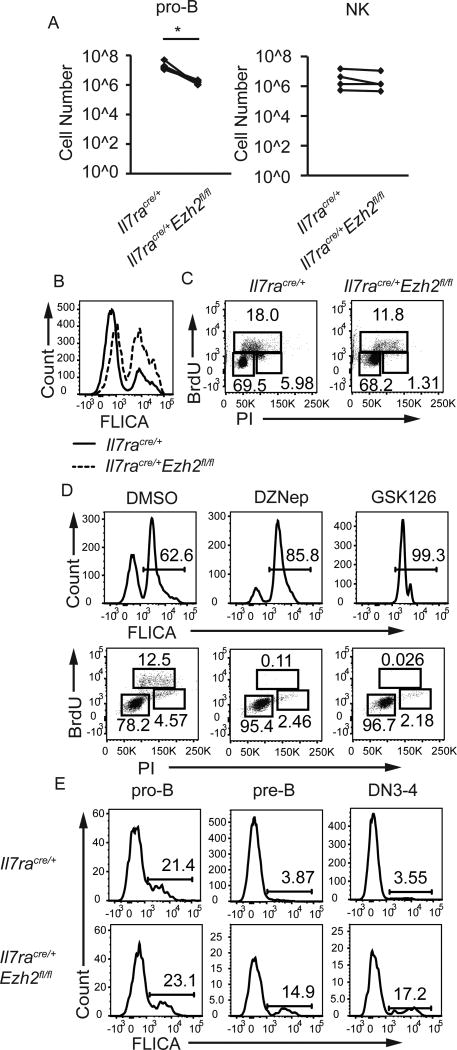
Cdkn2a gene products regulate proliferation and survival so we examined these properties in Ezh2Δ/Δ pro-B cells. In vitro Ezh2Δ/Δ pro-B cells generated fewer CD19+ cells than EZH2-sufficient cells (Fig. 5A). In contrast, NK cells from Ezh2Δ/Δ mice or control mice expanded equivalently in vitro (Fig. 5A). Cultured Ezh2Δ/Δ pro-B cells had an increased frequency of apoptotic cells as estimated by staining for activated caspases (FLICA) and a decreased frequency of S-phase and G2/M-phase cells compared to control pro-B cells, indicating both an induction of cell death and a possible block in the cell cycle at the G1-S transition (Fig. 5B, 5C). Increased apoptosis and cell cycle alterations were also observed when WT pro-B cells were cultured in the presence of the EZH2 inhibitors, DZnep (5 uM)(44) or GSK126 (10 uM)(45) (Fig. 5D). In vivo Ezh2Δ/Δ pre-B cells, and Lin−CD117−CD25lo/− (DN4) cells, were more apoptotic than control cells (Fig. 5E). Taken together, these data indicate that EZH2 is required for the proper proliferation and survival of pro-B and pre-B lymphocytes and for the survival of Lin−CD117−CD25lo/− (DN4) cells.
FIGURE 5.
EZH2 promotes proliferation and survival in lymphocyte progenitors. (A) Cell counts from pro-B cells (B220 enriched bone marrow cells in IL7 without stroma) cultured in vitro for 4 days (left) or NK cells (DX5 enriched splenocytes in IL2 without stroma) cultured for 4 days (right) from Il7racre/+ and Il7racre/+Ezh2fl/fl mice. Cultures were seeded with 8–50 × 105 cells. (B) Flow cytometry analysis for intracellular caspase activation (FLICA) to measure apoptosis in pro-B cell cultures from Il7racre/+ (solid) and Il7racre/+Ezh2fl/fl (dashed) mice after 3 days. (C) Flow cytometry analysis for PI and BrdU to measure cell cycle in pro-B cell cultures from Il7racre/+ (left) and Il7racre/+Ezh2fl/fl (right) mice. G0/G1 cells are BrdUPIlo, S-phase cells are BrdU+, and G2/M cells are BrdUPIhi. (D) Flow cytometry analysis for FLICA (top) and PI versus BrdU (bottom) in EZH2 inhibitor treated WT pro-B cell cultures. (E) Flow cytometry analysis for FLICA in pro-B (left, B220+CD19+CD43+), pre-B (middle, B220loCD19+CD43) and LinCD117CD25lo/− (DN4) thymocytes (right) from Il7racre/+ and Il7racre/+Ezh2fl/fl mice. Data are representative of at least three independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001.
Cdkn2a limited the maturation of Ezh2Δ/Δ B and T lymphocytes
To test the hypothesis that deregulation of the Cdkn2a locus contributed to the B and T cell developmental defects in Ezh2Δ/Δ mice, we generated Ezh2Δ/Δ mice that also had an inactivating mutation in Cdkn2a (DKO). All DKO mice had improved B cell development compared to Ezh2Δ/Δ mice (Fig. 6A). DKO mice had an increased number of bone marrow B220+CD19+ B lymphocytes, an increased ratio of pre-B to pro-B cells and an increased number of IgM+ cells in the bone marrow compared to Ezh2Δ/Δ mice, indicating that B cell maturation was at least partially restored (Fig. 6B, 6C, 6D). These results support the hypothesis that deletion of Cdkn2a improved differentiation beyond the pro-B cell stage. However, the DKO mice continued to have a reduced number of B cells in the BM when compared to EZH2-sufficient control mice (Fig. 6B).
T cell maturation was also restored in DKO mice when compared to Ezh2Δ/Δ mice with an increased frequency and number of DP, CD4 and CD8 thymocytes (Fig. 6E, 6F). Maturation from the DN3 to the DN4 stage was rescued and CD25 was restored to WT levels in the DKO mice (Fig. 6E). DKO mice had increased CD8 T cells but this was not a consequence of a block at the immature CD8 single positive (ISP) stage because immature single positive CD8 T cells (CD8+CD24hiTCRb−) were not increased in DKO mice (Fig. 6E, 6F, data not shown).
Deregulation of Cdkn2a in Ezh2Δ/Δ mice promoted p53 stabilization
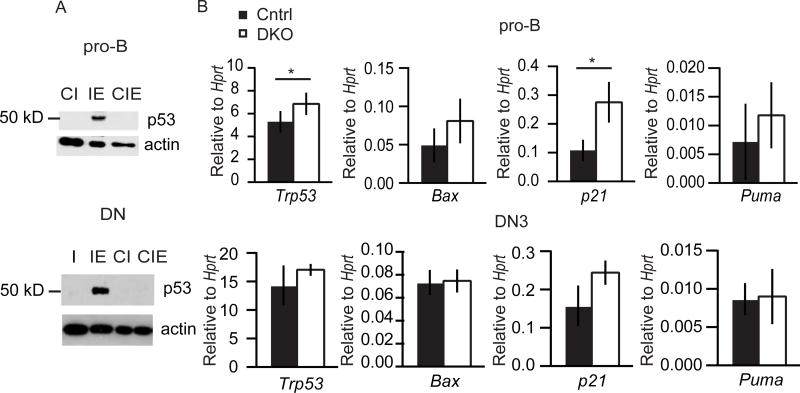
We next examined the possibility that Cdkn2a contributed to the activation of the p53-apoptosis pathway in Ezh2Δ/Δ cells. Indeed, p53 protein was reduced in pro-B cells and DN thymocytes from DKO mice when compared to Ezh2Δ/Δ cells and was indistinguishable from controls (Fig. 7A). Consistent with the reduced p53 stabilization, the p53 target genes Puma and Bax were not significantly elevated in DKO pro-B or DN3 cells compared to control cells (Fig. 7B). Our data demonstrate that the deregulation of Cdkn2a in Ezh2Δ/Δ pro-B and DN thymocytes was responsible for the premature stabilization of p53 and execution of apoptosis prior to the pre-antigen receptor checkpoints.
FIGURE 7.
Cdkn2a promotes p53 stabilization in Ezh2Δ/Δ pro-B and DN3 cells. (A) Western blot for p53 in pro-B cells from culture (top) and in depleted thymocytes as in Fig. 4e (bottom). (I - Il7racre/+, IE- Il7racre/+Ezh2fl/fl, CI - Cdkn2a−/−Il7racre/+, CIE - Cdkn2a−/−Il7racre/+Ezh2fl/fl) (B) Quantitative RT-PCR analysis of Trp53, Bax, p21, and Puma mRNA expression in sorted pro-B (top) and DN3 (bottom) cells from control (black) and DKO (white) mice. Results are presented as mean ± SD, relative to Hprt. Data represent at least three independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001.
DISCUSSION
Developing B and T cells must transition through checkpoints that ensure the appropriate recombination, expression, and function of their antigen receptors. How these checkpoints are regulated and restricted to stages where antigen receptors are expressed is a key question in lymphocyte development. In pro-B cells, BACH2 induces the transcription of the V(D)J recombinases, Rag1 and Rag2, as well as the pre-BCR checkpoint effectors Cdkn2a and Trp53(5). Cdkn2a, through p19ARF, promotes the stabilization of p53 and enforces the pre-BCR checkpoint by inducing apoptosis in cells that lack the pre-BCR. To ensure that p53 stabilization does not occur until the cell has had time to create a functional pre-BCR, p19ARF induction by BACH2 must be delayed relative to Rag1 and Rag2. Here we demonstrated that Cdkn2a was marked by H3K27me3 and was not expressed in pro-B cells. We propose that EZH2, through deposition of H3K27me3, impedes the induction of Cdkn2a by BACH2 in pro-B cells. This transcriptional hurdle delays the stabilization of p53 allowing the cells time to attempt recombination at the heavy chain locus and to express a functional pre-BCR. Therefore, we propose that EZH2 allows the induction of Cdkn2a to act as a molecular timer to induce apoptosis in cells that fail to generate a functional pre-BCR after initiation of Rag1/Rag2 gene transcription.
In accordance with this model, Ezh2Δ/Δ pro-B lymphocytes and DN3 thymocytes showed premature activation of the Cdkn2a/p53 pathway causing the cells to become apoptotic prior to the stage of the pre-antigen receptor checkpoints. Deletion of Cdkn2a in Ezh2Δ/Δ mice was sufficient to prevent the stabilization of p53 and partially restored B and T cell development. Our model is also consistent with a prior study showing that transgenic expression of the pre-BCR could partially rescue B cell development in EZH2-deficient hematopoietic cells since this transgene is expressed in all pro-B lymphocytes and could signal the down regulation of Cdkn2a in these cells(26). The model is also consistent with prior studies showing that augmented expression of the p19ARF/p53 pathway arrests T cell development at the DN3 to DN4 transition(6). Taken together, our data demonstrate that a major function of the PRC2 protein EZH2 in adaptive lymphocyte development is to prevent the premature activation of the pre-antigen receptor checkpoint.
The Cdkn2a gene was marked by H3K27me3 in both innate and adaptive lymphocytes but deletion of Ezh2 led to significant Cdkn2a expression only in B and T lymphocyte progenitors. Unlike B and T cell progenitors, developing NK and ILC2P cells do not create antigen receptors through the process of DNA recombination and therefore they do not need to have a checkpoint for functional receptor expression. Thus innate and adaptive lymphocytes may have evolved distinct mechanisms to regulate Cdkn2a and p53 during their development. Similar to long-term hematopoietic stem cells, innate lymphocytes may require EZH1 instead of EZH2 to maintain H3K27me3 at Cdkn2a(46). Indeed, NK and ILC2P expressed more Ezh1 than Ezh2 mRNA; and in the absence of EZH2, ILC2P and NK cells showed only a marginal decrease in global H3K27me3, indicating that EZH2 is not required in these cells for the maintenance of this histone modification. In contrast, we found that global H3K27me3 was entirely dependent on EZH2 in pro-B cells. Consistent with this finding, Ezh2 expression spikes at the pro-B cell stage while Ezh1 is downregulated(47). Thus it may be adaptive lymphoid cells that evolved to selectively utilize EZH2-containing PRC2 complexes to repress Cdkn2a at critical stages.
Our data reveal that EZH2 is required to suppress Cdnk2a in developing B and T lymphocytes prior to the pre-antigen receptor checkpoint. While the Cdkn2a/p53 pathway needs to be repressed until the point of pre-antigen receptor selection it also needs to be accessible to protect developing B and T cells from oncogenic transformation(4). Indeed, many early B and T lineage acute leukemias harbor inactivating mutations at the Cdkn2a locus(48). Surprisingly, inactivating mutations in Ezh2 have been detected in T acute lymphoblastic leukemia (T-ALL) and these mutations are associated with increased Cdkn2a(49, 50), which should be detrimental to these cells. How these leukemias overcome the tumor suppressor activity of Cdkn2a remains to be determined but could involve alterations in the p53 pathway or cell cycle regulation. In contrast to T-ALL, germinal center B cell lymphomas have been shown to have activating Ezh2 mutations(51) and in these cells EZH2 may contribute to tumorigenesis, in part, through repression of Cdkn2a(52, 53). Therefore, EZH2-dependent repression of Cdkn2a is a critical control point for pre-antigen receptor selection that could be manipulated by lymphoid cells to promote transformation.
Supplementary Material
Acknowledgments
We thank members of the Kee Lab for helpful discussions, Dr. Alexander Tarakhovsky for Ezh2-floxed mice and comments on the manuscript, and the Cytometry and Antibody Technology Core facility for expert technical assistance.
Funding Sources: This work was supported by grants from the NIH (R21 AI109233, R01 AI106352) and a pilot project award from the University of Chicago Comprehensive Cancer Center (P30 CA014599). J.A.J. was supported by the Medical Scientist Training Grant (T32 GM007281), J.W. was supported by a fellowship from the Leukemia and Lymphoma Society (LLS), B.L.K was an LLS Scholar.
Abbreviations used
- CLP
common lymphoid progenitor
- ILC
innate lymphoid cell
- ETP
early thymic progenitor
- DN
double negative thymocytes
- DP
double positive thymocytes
- BM
bone marrow
- Sp
spleen
- DKO
double knock out
- cKO
conditional knockout
- WT
wild type
- PRC2
polycomb repressive complex 2
- H3K27
histone 3 lysine 27
- ChIP
chromatin immunoprecipitation
Footnotes
RNA-sequencing data can be accessed in the Gene Expression Omnibus (GSE97462).
AUTHOR CONTRIBUTIONS
J.A.J. performed experiments, analyzed and interpreted data, and wrote the manuscript. J.W. M.S. performed experiments. E.B. analyzed data, M.M. and M.R.C. provided advice and ChIP-sequencing data, B.L.K. directed the studies, analyzed and interpreted data, and wrote the paper.
References
- 1.De Obaldia ME, Bhandoola A. Transcriptional regulation of innate and adaptive lymphocyte lineages. Annu Rev Immunol. 2015;33:607–642. doi: 10.1146/annurev-immunol-032414-112032. [DOI] [PubMed] [Google Scholar]
- 2.Clark MR, Mandal M, Ochiai K, Singh H. Orchestrating B cell lymphopoiesis through interplay of IL-7 receptor and pre-B cell receptor signalling. Nat Rev Immunol. 2014;14:69–80. doi: 10.1038/nri3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109(Suppl):S45–55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- 4.Swaminathan S, Duy C, Müschen M. BACH2-BCL6 balance regulates selection at the pre-B cell receptor checkpoint. Trends Immunol. 2014;35:131–137. doi: 10.1016/j.it.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swaminathan S, Huang C, Geng H, Chen Z, Harvey R, Kang H, Ng C, Titz B, Hurtz C, Sadiyah MF, Nowak D, Thoennissen GB, Rand V, Graeber TG, Koeffler HP, Carroll WL, Willman CL, Hall AG, Igarashi K, Melnick A, Müschen M. BACH2 mediates negative selection and p53-dependent tumor suppression at the pre-B cell receptor checkpoint. Nat Med. 2013;19:1014–1022. doi: 10.1038/nm.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyazaki M, Miyazaki K, Itoi M, Katoh Y, Guo Y, Kanno R, Katoh-Fukui Y, Honda H, Amagai T, van Lohuizen M, Kawamoto H, Kanno M. Thymocyte proliferation induced by pre-T cell receptor signaling is maintained through polycomb gene product Bmi-1-mediated Cdkn2a repression. Immunity. 2008;28:231–245. doi: 10.1016/j.immuni.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 7.Jiang D, Lenardo MJ, Zúñiga-Pflücker JC. p53 prevents maturation to the CD4+CD8+ stage of thymocyte differentiation in the absence of T cell receptor rearrangement. J Exp Med. 1996;183:1923–1928. doi: 10.1084/jem.183.4.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schito ML, Demidov ON, Saito S, Ashwell JD, Appella E. Wip1 phosphatase-deficient mice exhibit defective T cell maturation due to sustained p53 activation. J Immunol. 2006;176:4818–4825. doi: 10.4049/jimmunol.176.8.4818. [DOI] [PubMed] [Google Scholar]
- 9.Yi W, Hu X, Chen Z, Liu L, Tian Y, Chen H, Cong YS, Yang F, Zhang L, Rudolph KL, Zhang Z, Zhao Y, Ju Z. Phosphatase Wip1 controls antigen-independent B-cell development in a p53-dependent manner. Blood. 2015;126:620–628. doi: 10.1182/blood-2015-02-624114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gatzka M, Tasdogan A, Hainzl A, Allies G, Maity P, Wilms C, Wlaschek M, Scharffetter-Kochanek K. Interplay of H2A deubiquitinase 2A-DUB/Mysm1 and the p19(ARF)/p53 axis in hematopoiesis, early T-cell development and tissue differentiation. Cell Death Differ. 2015;22:1451–1462. doi: 10.1038/cdd.2014.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoue T, Morita M, Hijikata A, Fukuda-Yuzawa Y, Adachi S, Isono K, Ikawa T, Kawamoto H, Koseki H, Natsume T, Fukao T, Ohara O, Yamamoto T, Kurosaki T. CNOT3 contributes to early B cell development by controlling Igh rearrangement and p53 mRNA stability. J Exp Med. 2015;212:1465–1479. doi: 10.1084/jem.20150384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson SJ, Lauritsen JP, Hartman MG, Foushee AM, Lefebvre JM, Shinton SA, Gerhardt B, Hardy RR, Oravecz T, Wiest DL. Ablation of ribosomal protein L22 selectively impairs alphabeta T cell development by activation of a p53-dependent checkpoint. Immunity. 2007;26:759–772. doi: 10.1016/j.immuni.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 13.Fahl SP, Harris B, Coffey F, Wiest DL. Rpl22 Loss Impairs the Development of B Lymphocytes by Activating a p53-Dependent Checkpoint. J Immunol. 2015;194:200–209. doi: 10.4049/jimmunol.1402242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rashkovan M, Vadnais C, Ross J, Gigoux M, Suh WK, Gu W, Kosan C, Möröy T. Miz-1 regulates translation of Trp53 via ribosomal protein L22 in cells undergoing V(D)J recombination. Proc Natl Acad Sci U S A. 2014;111:E5411–5419. doi: 10.1073/pnas.1412107111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saba I, Kosan C, Vassen L, Klein-Hitpass L, Möröy T. Miz-1 is required to coordinate the expression of TCRbeta and p53 effector genes at the pre-TCR “beta-selection” checkpoint. J Immunol. 2011;187:2982–2992. doi: 10.4049/jimmunol.1101451. [DOI] [PubMed] [Google Scholar]
- 16.Sui G, Affar eB, Shi Y, Brignone C, Wall NR, Yin P, Donohoe M, Luke MP, Calvo D, Grossman SR. Yin Yang 1 is a negative regulator of p53. Cell. 2004;117:859–872. doi: 10.1016/j.cell.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 17.Liu H, Schmidt-Supprian M, Shi Y, Hobeika E, Barteneva N, Jumaa H, Pelanda R, Reth M, Skok J, Rajewsky K. Yin Yang 1 is a critical regulator of B-cell development. Genes Dev. 2007;21:1179–1189. doi: 10.1101/gad.1529307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L, Foreman DP, Sant’Angelo DB, Krangel MS. Yin Yang 1 Promotes Thymocyte Survival by Downregulating p53. J Immunol. 2016;196:2572–2582. doi: 10.4049/jimmunol.1501916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu Y, Wang J, Khaled W, Burke S, Li P, Chen X, Yang W, Jenkins NA, Copeland NG, Zhang S, Liu P. Bcl11a is essential for lymphoid development and negatively regulates p53. J Exp Med. 2012;209:2467–2483. doi: 10.1084/jem.20121846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma CA, Pusso A, Wu L, Zhao Y, Hoffmann V, Notarangelo LD, Fowlkes BJ, Jain A. Novel INHAT repressor (NIR) is required for early lymphocyte development. Proc Natl Acad Sci U S A. 2014;111:13930–13935. doi: 10.1073/pnas.1310118111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polager S, Ginsberg D. p53 and E2f: partners in life and death. Nat Rev Cancer. 2009;9:738–748. doi: 10.1038/nrc2718. [DOI] [PubMed] [Google Scholar]
- 22.Gil J, Peters G. Regulation of the INK4b–ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- 23.Buchner M, Swaminathan S, Chen Z, Müschen M. Mechanisms of pre-B-cell receptor checkpoint control and its oncogenic subversion in acute lymphoblastic leukemia. Immunol Rev. 2015;263:192–209. doi: 10.1111/imr.12235. [DOI] [PubMed] [Google Scholar]
- 24.Aloia L, Di Stefano B, Di Croce L. Polycomb complexes in stem cells and embryonic development. Development. 2013;140:2525–2534. doi: 10.1242/dev.091553. [DOI] [PubMed] [Google Scholar]
- 25.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su IH, Basavaraj A, Krutchinsky AN, Hobert O, Ullrich A, Chait BT, Tarakhovsky A. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat Immunol. 2003;4:124–131. doi: 10.1038/ni876. [DOI] [PubMed] [Google Scholar]
- 27.Su IH, Dobenecker MW, Dickinson E, Oser M, Basavaraj A, Marqueron R, Viale A, Reinberg D, Wülfing C, Tarakhovsky A. Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell. 2005;121:425–436. doi: 10.1016/j.cell.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 28.Schlenner SM, Madan V, Busch K, Tietz A, Läufle C, Costa C, Blum C, Fehling HJ, Rodewald HR. Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity. 2010;32:426–436. doi: 10.1016/j.immuni.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 29.Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 30.Kee BL, Rivera RR, Murre C. Id3 inhibits B lymphocyte progenitor growth and survival in response to TGF-beta. Nat Immunol. 2001;2:242–247. doi: 10.1038/85303. [DOI] [PubMed] [Google Scholar]
- 31.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kinsella RJ, Kähäri A, Haider S, Zamora J, Proctor G, Spudich G, Almeida-King J, Staines D, Derwent P, Kerhornou A, Kersey P, Flicek P. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database (Oxford) 2011;2011:bar030. doi: 10.1093/database/bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin J, Leavenworth JW, Li Y, Luo Q, Xie H, Liu X, Huang S, Yan H, Fu Z, Zhang LY, Zhang L, Hao J, Wu X, Deng X, Roberts CW, Orkin SH, Cantor H, Wang X. Ezh2 regulates differentiation and function of natural killer cells through histone methyltransferase activity. Proc Natl Acad Sci U S A. 2015;112:15988–15993. doi: 10.1073/pnas.1521740112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32:503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Son J, Shen SS, Margueron R, Reinberg D. Nucleosome-binding activities within JARID2 and EZH1 regulate the function of PRC2 on chromatin. Genes Dev. 2013;27:2663–2677. doi: 10.1101/gad.225888.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mandal M, Powers SE, Maienschein-Cline M, Bartom ET, Hamel KM, Kee BL, Dinner AR, Clark MR. Epigenetic repression of the Igk locus by STAT5-mediated recruitment of the histone methyltransferase Ezh2. Nat Immunol. 2011;12:1212–1220. doi: 10.1038/ni.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang JA, Mortazavi A, Williams BA, Wold BJ, Rothenberg EV. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell. 2012;149:467–482. doi: 10.1016/j.cell.2012.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, Ideker T, Glass CK, Murre C. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. 2010;11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lara-Astiaso D, Weiner A, Lorenzo-Vivas E, Zaretsky I, Jaitin DA, David E, Keren-Shaul H, Mildner A, Winter D, Jung S, Friedman N, Amit I. Immunogenetics. Chromatin state dynamics during blood formation. Science. 2014;345:943–949. doi: 10.1126/science.1256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miranda TB, Cortez CC, Yoo CB, Liang G, Abe M, Kelly TK, Marquez VE, Jones PA. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol Cancer Ther. 2009;8:1579–1588. doi: 10.1158/1535-7163.MCT-09-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A, Diaz E, LaFrance LV, Mellinger M, Duquenne C, Tian X, Kruger RG, McHugh CF, Brandt M, Miller WH, Dhanak D, Verma SK, Tummino PJ, Creasy CL. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 46.Hidalgo I, Herrera-Merchan A, Ligos JM, Carramolino L, Nuñez J, Martinez F, Dominguez O, Torres M, Gonzalez S. Ezh1 is required for hematopoietic stem cell maintenance and prevents senescence-like cell cycle arrest. Cell Stem Cell. 2012;11:649–662. doi: 10.1016/j.stem.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 47.Heng TS, Painter MW, Consortium IGP. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 48.Sulong S, Moorman AV, Irving JA, Strefford JC, Konn ZJ, Case MC, Minto L, Barber KE, Parker H, Wright SL, Stewart AR, Bailey S, Bown NP, Hall AG, Harrison CJ. A comprehensive analysis of the CDKN2A gene in childhood acute lymphoblastic leukemia reveals genomic deletion, copy number neutral loss of heterozygosity, and association with specific cytogenetic subgroups. Blood. 2009;113:100–107. doi: 10.1182/blood-2008-07-166801. [DOI] [PubMed] [Google Scholar]
- 49.Simon C, Chagraoui J, Krosl J, Gendron P, Wilhelm B, Lemieux S, Boucher G, Chagnon P, Drouin S, Lambert R, Rondeau C, Bilodeau A, Lavallée S, Sauvageau M, Hébert J, Sauvageau G. A key role for EZH2 and associated genes in mouse and human adult T-cell acute leukemia. Genes Dev. 2012;26:651–656. doi: 10.1101/gad.186411.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ntziachristos P, Tsirigos A, Van Vlierberghe P, Nedjic J, Trimarchi T, Flaherty MS, Ferres-Marco D, da Ros V, Tang Z, Siegle J, Asp P, Hadler M, Rigo I, De Keersmaecker K, Patel J, Huynh T, Utro F, Poglio S, Samon JB, Paietta E, Racevskis J, Rowe JM, Rabadan R, Levine RL, Brown S, Pflumio F, Dominguez M, Ferrando A, Aifantis I. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat Med. 2012;18:298–301. doi: 10.1038/nm.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, Yap D, Humphries RK, Griffith OL, Shah S, Zhu H, Kimbara M, Shashkin P, Charlot JF, Tcherpakov M, Corbett R, Tam A, Varhol R, Smailus D, Moksa M, Zhao Y, Delaney A, Qian H, Birol I, Schein J, Moore R, Holt R, Horsman DE, Connors JM, Jones S, Aparicio S, Hirst M, Gascoyne RD, Marra MA. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Velichutina I, Shaknovich R, Geng H, Johnson NA, Gascoyne RD, Melnick AM, Elemento O. EZH2-mediated epigenetic silencing in germinal center B cells contributes to proliferation and lymphomagenesis. Blood. 2010;116:5247–5255. doi: 10.1182/blood-2010-04-280149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Caganova M, Carrisi C, Varano G, Mainoldi F, Zanardi F, Germain PL, George L, Alberghini F, Ferrarini L, Talukder AK, Ponzoni M, Testa G, Nojima T, Doglioni C, Kitamura D, Toellner KM, Su IH, Casola S. Germinal center dysregulation by histone methyltransferase EZH2 promotes lymphomagenesis. J Clin Invest. 2013;123:5009–5022. doi: 10.1172/JCI70626. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.