Abstract
Qualitative and quantitative information are crucial to a detailed understanding of the function of protein phosphorylation. MS is now becoming a quantitative approach to analyze protein phosphorylation. All methods that have been described either require the elaborate/expensive use of stable isotopes to compare a limited number of samples or do not provide phosphorylation stoichiometries. Here, we present stable isotope-free MS strategies that allow relative and absolute quantitation of phosphorylation stoichiometries. By using the developed methods, we can normalize to robustly account for run-to-run variations and variations in amounts of starting material. This procedure monitors the unmodified proteolytic peptides derived from the protein of interest and identifies peptides that are suitable for normalization purposes. Also, we can determine changes in phosphorylation stoichiometry by monitoring the changes in the normalized ion currents of the phosphopeptide(s) of interest. Absolute phosphorylation stoichiometry are measured by monitoring the ion currents of a phosphopeptide and its unmodified cognate as the signal intensity changes of both peptide species are correlated. The method is applicable to multiply phosphorylated species (for which one more sample with varying phosphorylation stoichiometry than number of phosphorylation sites is required to correct for the differences in the ionization/detection efficiencies of the phosphopeptide, its partially phosphorylated and unphosphorylated cognates). Last, we can quantitate species with ragged ends resulting from incomplete proteolysis and measure phosphorylation stoichiometries of single samples by controlled dephosphorylation. These approaches were validated and subsequently applied to the phosphorylation of the yeast transcription factor Pho4.
One of the most common and important posttranslational protein modification (PTM) is protein phosphorylation (1). It is estimated that ≈30% of all proteins in mammalian cells are phosphorylated at any given time and ≈5% of a vertebrate genome encodes protein kinases and phosphatases (2), underscoring the importance of this PTM. The presence of various protein kinases and phosphatases permits the use of quickly reversible phosphorylation in a vast number of different, highly regulated pathways and functions, including signal transduction, cell division, and cell differentiation.
Knowledge of the phosphorylation site is crucial to a detailed understanding of regulatory processes in cells; this knowledge requires sensitive-analysis methods. Theoretically, the most sensitive methods for the detection of phosphorylation incorporate radioactive phosphorus isotopes before phosphopeptide mapping and/or Edman degradation (3). However, the incorporation of radioactive isotopes is not possible (e.g., in tissue samples) or is very inefficient in the case of cell culture because of the presence of endogenous unlabeled ATP. Also, high levels of radioactive phosphate incorporation cause cellular damage and, thereby, can alter phosphorylation. Alternative nonradioactive strategies use antibodies or MS. However, the former strategy cannot be used to discover phosphorylation sites. The latter strategy is currently the most commonly used nonradioactive method for the analysis of protein phosphorylation because of the sensitivity and speed provided by this technology as compared with traditional biochemical methods.
Although commonly applied, protein phosphorylation analysis by MS is still far from being routine. Whereas any set of peptides suffices for protein identification purposes, the modified peptides have to be observed and selected for sequencing during MS experiments for protein phosphorylation analysis. The problems associated with observing the modified species among a large excess of unmodified species resulted in the development of strategies for the selective enrichment and/or selective detection of phosphorylated species (4–9).
Identification of a phosphorylation site is only the first step. Two further aspects have to be addressed to understand the regulatory significance; i.e., (i) how the degree of phosphorylation of a particular site changes over time; and (ii) what the degree of phosphorylation is. Although the quantitation of protein phosphorylation by MS is in its infancy, several strategies have been introduced. Most of them address the issue of relative quantitation and follow two approaches. (i) The proteins or peptides are generally labeled with different isotopes by chemical or metabolic means (10–12); and (ii) phosphopeptide-specific derivatization methods for the incorporation of different isotopes are applied, enabling the selective enrichment and/or detection (13–15).
The issue of absolute phosphorylation stoichiometry is less commonly addressed. We are aware of two different methods that facilitate the quantitation of protein phosphorylation stoichiometry. In the first approach, the sample is split, both halves are differentially isotopically labeled, and one of the two fractions is dephosphorylated before pooling of the two fractions and MS analysis. The degree of phosphorylation can then be derived by comparing the signal intensity of the two differentially labeled unphosphorylated species, assuming that the increase in signal intensity of the species is due to the dephosphorylation of the singly phosphorylated species (16, 17). The other strategy is based on stable isotope dilution; i.e., the expected unphosphorylated and phosphorylated proteolytic peptides are synthesized as isotopologues and spiked into the samples in known quantities, such that the degree of phosphorylation can be inferred by monitoring the intensities of the two isotopologue pairs corresponding to the phosphorylated and unphosphorylated peptides (18).
Here, we present a stable isotope-free MS approach that leads to the relative and absolute quantitation of protein phosphorylation stoichiometry. This approach has three facets. (i) A robust normalization routine is established that accounts for run-to-run variations and variations in amount of starting material. This routine monitors the ion currents of numerous unmodified peptides derived from the protein of interest and identifies peptides suitable for normalization purposes. (ii) Relative quantitation of phosphorylation is accomplished by following variations in the normalized ion currents of the phosphopeptide(s) of interest, which mirror changes in phosphorylation stoichiometry. (iii) Absolute phosphorylation stoichiometry is determined by measuring the ion currents of a particular phosphopeptide and its unmodified cognate because the changes in the signal intensities of the phosphorylated and unphosphorylated form of a peptide are correlated.
This stable isotope-free quantitation method is easily extended to multiply phosphorylated peptides or peptides whose phosphorylation sites affect the cleavage efficiencies of the protease. Furthermore, single samples are also amenable to our absolute quantitation of phosphorylation stoichiometry method if it is combined with a protocol for controlled enzymatic dephosphorylation, which is independent of the amount of phosphoprotein/peptide. The strategies were validated on synthetic peptides before application to the quantitative phosphorylation analysis of the yeast transcription factor Pho4.
Materials and Methods
All chemicals were purchased from Sigma, unless specified otherwise. Solvents used for MS analysis and gel-band preparation were HPLC-grade from Burdick and Jackson. The synthetic peptides were kindly provided by Nick Morrice (Medical Research Council Protein Phosphorylation Unit, University of Dundee, Nethergate, Dundee, Scotland) and John Rush (Cell Signaling Technology, Beverly, MA). All synthetic peptides were purified and quantitated in duplicate by amino acid analysis. The phosphopeptide and its unphosphorylated counterpart were mixed in at least two different defined ratios (final concentrations, 30–150 nM). To minimize systematic errors due to imprecise pipetting, a freshly calibrated pipette was used for reversed pipetting. To avoid carry-over problems, the solutions of two different peptide pairs were analyzed by liquid chromatography (LC)/MS in an alternating fashion.
Controlled Dephosphorylation. Controlled dephosphorylation was performed in a buffer containing 25 mM ammonium bicarbonate and 0.5 mM phosphotyrosine, with a final alkaline phosphatase concentration of 100 milliunits/μl (Roche Diagnostics, Indianapolis). Robust dephosphorylation kinetics were observed taking aliquots after 0, 5, and 15 sec, and 2, 8, and 20 min of incubation at 37°C.
Pho4. Recombinant Pho4, Pho80, and Pho85 were kindly provided by M. Byrne and E. O'Shea (Department of Biochemistry and Biophysics, University of California, San Francisco). The in vitro phosphorylation was carried out as described (19) before isolation by SDS/PAGE (4–12% Novex NuPage Mops buffer, Invitrogen) and visualized with colloidal Coomassie blue staining (Invitrogen) and in-gel digestion according to protocols described in ref. 20. Subsequently, the digests were desalted by using in-house prepared stop-and-go extraction (STAGE) tips (21).
LC/MS Analysis. All experiments were performed by using a QSTAR XL mass spectrometer (AB/MDS Sciex, Concord, Canada) hyphenated with a microscale capillary HPLC (Famos autosampler, LC Packings, Sunnyvale, CA) and an Agilent 1100 HPLC pump (Agilent, Andover, MA). Columns were packed in-house by using Magic C18 beads (Michrom BioResources, Auburn, CA). Buffer A was 2.5% acetonitrile/0.2% formic acid; buffer B was 2.5% water/0.2% formic acid; and loading buffer was buffer A plus 5% formic acid). A 5-min gradient (5–35% buffer A, linear) was used with MS-acquisition times of 150 msec. The isotopes used for quantitation were chosen such that none exceeded the saturation limit of the instrument used. Quantitative information was obtained by using the algorithm provided with the analyst software package (AB/MDS Sciex).
Results and Discussion
Normalization. Most MS methods for the quantitation of protein phosphorylation assume that MS is not quantitatively reproducible. However, there is increasing evidence that MS can provide some quantitation of protein abundances (22–24) and protein modifications (25, 26) based on the ion signal intensities (ion current). These experiments show that appropriate normalization procedures are essential for deriving quantitative information without the use of stable isotopes. Unlike stable isotope labeling, normalization can also correct for varying amounts of starting materials.
Ruse et al. (25) suggested the use of a single peptide derived from the protein of interest for normalization purposes in samples like single protein digest. The idea of using peptides derived from the protein of interest has the advantages that they (i) are always present and (ii) reflect the amount of material present in the sample. However, the approach using only one peptide can be prone to error because it is not obvious whether the peptide chosen as the internal standard is affected by other time-dependent or random modifications. Such unaccounted variations in signal intensity of the normalization standard would result in erroneous normalization of the ion currents and incorrect quantitation.
To avoid this problem, we opted for monitoring numerous peptide ion signals derived from the protein of interest. First, for each peptide, the mean and relative deviations from this mean are calculated from signal intensities in the different MS experiments. This procedure converts absolute error to relative error. Subsequently, for each experiment, the median relative deviation is calculated from all measured peptides. “Good” peptides are identified based on comparing their fit with the median relative deviation from each experiment. This comparison can often be done by eye; however, with a view toward future automation, the normalized square deviation from the median curve χ2 can be calculated as it has been used in protein correlation profiling (27). If the curve of a particular peptide is significantly different from the median curve, it is considered a “bad” peptide and excluded from further calculations. Last, the mean relative standard deviation of the remaining peptides is calculated and used for the normalization (for an example of this normalization procedure, see Fig. 4, which is published as supporting information on the PNAS web site). Although not suitable for normalization purposes, the bad peptides might correlate with peptides that are biologically modified, and the method described above is possibly a modification identification tool. Standard deviations of <10% are observed for our normalization approach (see below). Similar deviations have been reported for other MS methods for the quantitation of protein phosphorylation based on stable-isotope labeling (12, 16, 25).
Relative Quantitation of Protein Phosphorylation. The strategy described above was subsequently used to monitor the changes in phosphorylation stoichiometry of the yeast transcription factor Pho4 that was phosphorylated to different degrees by the cyclin–cyclin-dependent kinase complex Pho80/85, the in vivo kinase of Pho4 (19). Pho4 was isolated by SDS/PAGE from different time points during a kinase assay. After in-gel tryptic digestion, the samples were analyzed in replicate by LC/MS.
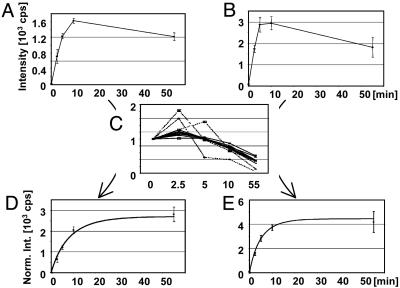
The results for the integrated signal intensities of the tryptic phosphopeptide ion SNSpSPYLNKR at m/z 623.28 ([MH2]2+) and the tryptic phosphopeptide TSSSAEGVVVASEpSPVIAPHGSTHAR at m/z 871.75 ([MH3]3+) before normalization are shown in Fig. 1 A and B (p denotes phosphorylation). It appears from the signal intensities that the degree of phosphorylation first increases and then decreases during the time course. However, when the data displayed in Fig. 1 A and B are normalized as described above (see Fig. 1C; the mean is shown in bold, whereas the curves for the four bad peptides are dotted), a steady increase of phosphorylation is observed (Fig. 1 D and E). The normalization accounted for a decrease of >50% in the amount of Pho4 from the first (0 min) to the last (55 min) time point. The experimental data were compared with a computational model derived from quantitative studies using 32P-labeling approaches (Fig. 1 E and D, curves) (19). This excellent agreement with the previous quantitation data underscores that stable isotope-free LC/MS experiments combined with appropriate normalization procedures are a cheap, fast, and accurate method for the relative quantitation of protein phosphorylation.
Fig. 1.
The ion currents of phosphopeptides as a function of the incubation time with Pho80/Pho85 before normalization. (A) SNSpSPYLNKR ([MH2]2+ at m/z 623.28). (B) TSSSAEGVVVASEpSPVIAPHGSTHAR ([MH3]3+ at m/z 871.75). Error bar indicates one standard deviation. (C) Plot of the relative deviations from the mean signal intensities of proteolytic peptides derived from Pho4 to determine good peptides (solid lines) and outliers (broken lines). The normalized signal intensities for the phosphopeptides SNSpSsPYLNKR ([MH2]2+ at m/z 623.28) (D) and TSSSAEGVVVASEpSPVIAPHGSTHAR ([MH3]3+ at m/z 871.75) (E) are shown. The curves represent the modeled phosphorylation kinetics based on previous 32P-labeling and phosphopeptide-mapping data (19).
Absolute Quantitation of the Phosphorylation Stoichiometry. The examples described above illustrate that the ions currents allow for relative quantitation of phosphorylation without stable-isotope labeling. However, the absolute phosphorylation stoichiometry is often of greater interest. Determination of the stoichiometry would be an easy task if peptides and their phosphorylated cognates had identical ionization/detection efficiencies (here also called “flyability”) because it would be sufficient to monitor the peptide and the phosphorylated complement and calculate the intensity ratio between these two species. Although it is tempting to assume equivalent flyabilities for the absolute quantitation of the degree of phosphorylation this assumption is invalid because peptides and their phosphorylated counterparts can have significantly different flyabilities, sometimes varying by several orders of magnitude (e.g., see Fig. 5, which is published as supporting information on the PNAS web site, and H.S., J.A.J., J. Rush, N. Morrice, M.W.K., unpublished data).
We hypothesized that by determining the flyability ratio for a particular peptide/phosphopeptide pair and using this ratio to correct the signal intensities of the corresponding species, the absolute phosphorylation stoichiometry could be calculated. The key to calculating the f lyability ratios for peptide/phosphopeptide pairs without the use of internal standards or stable-isotope labeling is the simple assumption that the decrease in the amount of a peptide must result in an increase in the amount of its phosphorylated cognate and vice versa. Correspondingly, the ion signal intensities of the species of interest must be correlated. It is assumed that in the cases of single phosphorylation the total amount of the phosphopeptide and unmodified complement is constant. However, the concentrations of the peptide ([P]) and the phosphopeptide ([pP]) and correspondingly the total concentration ([Po]) are unknown and not easily measurable by MS. Instead, the signal intensities I are measurable entities that are the product of peptide concentration [P] and the flyability of the peptide, which we call μ (IP = μP[P]). This approach can be formalized for a sample A as follows:
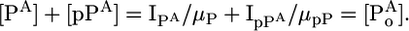 |
[1] |
Equivalent formulas can be written for a sample B. The experiments described above have shown that it is possible to account for different amounts of starting material and run-to-run variations by applying normalization procedures. Therefore, the measured ion intensities I can be normalized such that [Po] of two different samples A and B can be set equal. Thus, the flyability ratio can be calculated as follows:
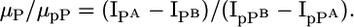 |
[2] |
By using this flyability ratio, the degree of phosphorylation can be determined without any isotope labeling or without the use of synthetic peptides as internal standards or for calibration curves. For a singly phosphorylated peptide, only two samples with different degrees of phosphorylation are necessary. The samples can be completely unrelated as long as complete digestion is ensured and the sample complexity of the samples is not vastly different. Similarly, the formulation of Eq. 1 is easily extended to multiply phosphorylated peptides, because the following equations hold true (e.g., for doubly phosphorylated peptides):
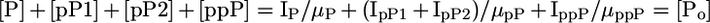 |
[3] |
Assuming close-to-identical flyabilities for the two different singly phosphorylated isoforms (pP1 and pP2), it can be rewritten in matrix form for three different samples:
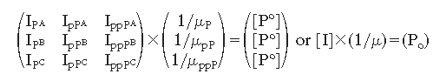 |
[4] |
Three samples with different degrees of phosphorylation are sufficient to calculate the flyability ratios for the unphosphorylated, singly phosphorylated, and doubly phosphorylated species and to calculate the stoichiometry of these species. In general, the number of samples needed is one more than the number of phosphorylation sites. If more samples than necessary are available, methods such as least-square-fit methods can be applied to estimate the error range of the determined flyability ratios. Requirement for the absolute quantitation of phosphorylation stoichiometry without stable isotopes is that differences in signal intensity can be measured reliably, i.e., that the changes in phosphorylation stoichiometry in the different samples is >10% (see above).
To test this approach, several synthetic peptides and their phosphorylated cognates were purified to homogeneity and quantitated by amino acid analysis. These synthetic standards were mixed in varying but defined ratios and analyzed in replicate by LC/MS. The flyability ratios for these synthetic peptide pairs were calculated by using the defined mixing ratio and measured ion currents; the values are summarized in Table 1, which is published as supporting information on the PNAS web site (listed as Method I). These values were compared with the flyability ratios calculated by following the approach described above, which requires samples with undefined but varying degrees of phosphorylation. To generate these samples, the same set of singly and doubly phosphorylated peptides were mixed with similar amounts of several unrelated unphosphorylated peptides, which were used for normalization. As such, this mixture mimicked a digest of a phosphoprotein. Subsequently, the sample was treated with phosphatase under controlled conditions to obtain a sufficient number of samples with varying degrees of phosphorylation. The major problem of controlled enzymatic dephosphorylation is adjusting the phosphatase concentration to the amount of sample present, which is normally ill-defined. The problem is avoided by adding a defined large excess of a phosphoester to the sample (mM vs. μM), thereby making kinetics of the enzymatic dephosphorylation essentially independent of the amount of phosphopeptides of interest. With biological samples, it is easy to ensure that the added phosphoester will be in excess.
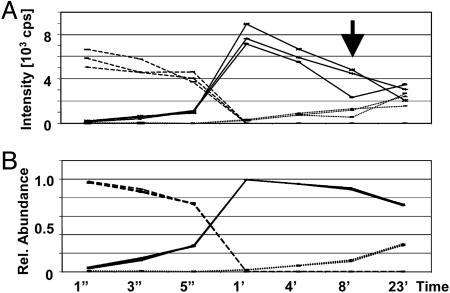
As an example, the time-dependent ion current changes of the doubly phosphorylated peptide RYPRPVpSVPPpSPSLSR (broken lines), the singly phosphorylated species RYPRPVSVPPpSPSLSR/RYPRPVpSVPPSPSLSR (solid lines; assuming near identical flyabilities for both species), and the unphosphorylated peptide (dashed lines) before normalization are shown in Fig. 2A. After normalization, the flyability ratios were calculated for the different species by following our approach described above, which in turn allowed us to calculate the relative abundances (see Fig. 2B). The benefits of the normalization procedure are obvious: before normalization, relative standard deviations in the range of 10–41% (see arrow) are observed, whereas after normalization, relative standard deviations are in the 1–8% range. The calculated flyability ratios for numerous tested peptides is given in Table 1 (Method II). These values agree very well with the flyability ratios determined by using the purified and quantitated synthetic peptide standards (deviations in the 1–16% range). The very good agreement between the two data sets further validates this stable isotope-free approach. Furthermore, this quantitation approach combined with controlled dephosphorylation provides a cheap and easy means to determine the flyability ratios of peptide/phosphopeptide species irrespective of the number of phosphorylation sites and phosphorylation stoichiometries as long as the latter is >10%.
Fig. 2.
Raw vs. processed data. (A) The unprocessed signal intensities of the doubly phosphorylated (RYPRPVpSVPPpSPSLSR; broken lines), singly phosphorylated (RYPRPVSVPPpSPSLSR/RYPRPVpSVPPSPSLSR; solid lines) and unphosphorylated peptide (RYPRPVSVPPSPSLSR; dashed lines) as a function of incubation time with phosphatase. (B) Relative abundances of the different species after normalization and calculation of the flyability ratios.
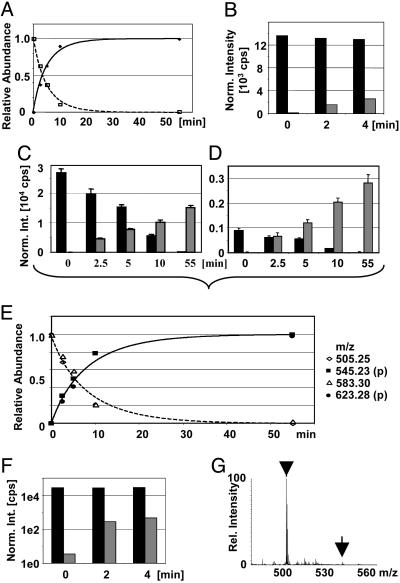
We then applied this method to a protein digest to quantitate the phosphorylation stoichiometry. In vitro phosphorylated Pho4 samples described above were chosen as independent quantitation data were available. The time-dependent changes in relative abundance of the peptide/phosphopeptide TSSSAEGVVVASE(pS)PVIAPHGSTHAR based on the ion currents of the triply charged species at m/z 845.09/875.75 are shown in Fig. 3A, together with the expected kinetics based on published 32P-labeling and phosphopeptide mapping data. For the triply charged peptide ion signals, a flyability ratio μP/μpP of 0.47 (±0.02) was calculated. As soon as the flyability ratio has been calculated with ease for the high ATP conditions, it could be applied to conditions in which it would be difficult to measure the ratio (i.e., in which the phosphorylation stoichiometry is <10%). For example, the degree of Ser-223 phosphorylation of Pho4 under low ATP conditions is <10% and, as such, not amenable to the reliable calculation of the flyability ratios and the degree of phosphorylation (see Fig. 3B). By using the calculated flyability ratio of 0.47, the degree of phosphorylation was found to be 4% after 2 min of incubation with Pho80/85 under low ATP conditions and 8% after 4 min, in good agreement with predicted results (19).
Fig. 3.
Pho4 phosphorylation stoichiometries. (A) Changes of the relative abundance of the peptide (▪) and phosphopeptide (•) TSSSAEGVVVASEpSPVIAPHGSTHAR based on the ion currents of the triply charged species at m/z 845.09/871.75 after normalization and calculation of the flyability ratio. The curves represent the modeled phosphorylation kinetics based on previous 32P-labeling data (19). (B) Normalized temporal changes of the signal intensities of the same peptide (black bars) and phosphopeptide (gray bars) as shown in A after incubation of Pho4 under low-ATP conditions. Normalized changes of the signal intensities of the peptide (black bars) and phosphopeptide (gray bars) are shown for SNSpSPYLNK ([MH2]2+at m/z 505.25/545.23) (C) and SNSpSPYLNKR ([MH2]2+at m/z 583.30/623.28) (D). Error bars indicate one standard deviation. (E) Kinetics for Ser-152. Changes of the relative abundance of the peptides and phosphorylated cognates by using the calculated flyability ratios (open symbols, unphosphorylated peptides; filled symbols, phosphopeptides) are shown. The curves represent the modeled phosphorylation kinetics based on previous 32P-labeling data (19). (F) Normalized temporal changes of the signal intensities of the peptide SNSpSPYLNK (black bars) and its phosphorylated complement (gray bars) after incubation under low-ATP conditions (note: logarithmic y scale). (G) Mass spectrum of the “low ATP/4 min”-sample averaged over the complete elution of the peptide SNSpSPYLNK (arrowhead) and its phosphorylated cognate (arrow).
The quantitation of the phosphorylation stoichiometry of Ser-152 is not as simple because the trypsin cleavage site C-terminal to Ser-152 is a KRK motif leading to miscleavages with ragged C termini. Because the phospho moiety can affect the cleavage efficiency, the ratio cleaved peptide vs. miscleaved peptide can be different for the phosphorylated and the unphosphorylated species. Miscleavages and ragged ends are a problem for all absolute quantitation methods using stable-isotope labeling because they do not account for differential cleavage efficiencies due to phosphorylation. Even isotopically labeled internal standards with short overhangs on the N and C termini to account for potential miscleavages can only be applied to this problem if the protease shows similar digestion efficiencies at the KK, RK, KR, or RR motif for the whole protein and the synthetic peptide with short overhangs. However, this issue of miscleavages does not matter for relative quantitation purposes. To merely observe the changes in Ser-152 phosphorylation, it is sufficient to monitor only one species because the signal intensity of each phosphorylated proteolysis product will increase (e.g., 2-fold) if the degree of phosphorylation increases 2-fold. However, to determine the stoichiometry, our method requires that the sum of a peptide and its phosphorylated cognate remains constant. This assumption is not valid when phosphorylation affects cleavage. Hence, it is impossible to calculate the different real flyability ratios for each species, even if an appropriate number of samples with various degrees of phosphorylation were available.
For the quantitation of the phosphorylation stoichiometry, instead of calculating the real flyability ratio by using the previously described approach, one can calculate the “apparent flyability ratio,” which is a function of the real flyability ratio and the cleavage efficiencies. Despite the fact that these values cannot be separated easily, this apparent flyability ratio can still be used to calculate the phosphorylation stoichiometry for a given set of samples. If this apparent flyability ratio is to be used for other sample sets, identical digestion conditions have to be ensured to achieve identical ratios for the completely cleaved vs. miscleaved species. If identical digestion conditions cannot be ensured but absolute quantitation of the phosphorylation stoichiometry is desired, the stoichiometry can still be obtained by monitoring the decrease of the signal of the unphosphorylated peptide in (partially) phosphorylated samples as compared with a completely unphosphorylated sample. Such a completely dephosphorylated sample can be generated by dephosphorylation before proteolysis if necessary.
In this particular case, after extensive searching, the following four different peptide species that contain the particular Ser-152 residue could be identified: SNSSPYLNK, SNSpSPYLNK, SNSSPYLNKR, and SNSpSPYLNKR. Peptides comprising the complete KRK motif at the C terminus were not observed. The various intensities for the phosphorylated and unphosphorylated peptides are shown in Fig. 3 C and D. It is apparent that completely cleaved species and the miscleaved species will give very different apparent flyability ratios. Subsequent calculations provided an apparent flyability ratio of 2.0 (± 0.2) for the completely cleaved species (vs. 0.3 ± 0.02 for the miscleaved species). These ratios enabled us to calculate the temporal changes in the relative abundances of the peptide/phosphopeptide pairs (see Fig. 3E), which is in good agreement with the expected phosphorylation kinetics based on published data represented by the curve (19). The flyability ratios also enabled us to determine the degree of phosphorylation for Ser-152 to be 3% after 4 min of incubation with Pho80/85 under low ATP concentrations (see Fig. 3 F and G), which is 2.7-fold lower than the degree of phosphorylation of Ser-223. This number is in good agreement with a 2.5-fold difference predicted from simulation after 32P-labeling experiments (19), validating the use of apparent flyability ratios in the case of differential miscleavages due to phosphorylation.
The approach involving the calculation of apparent flyability ratios does not apply to artificial modifications such as methionine oxidation, which occur to a various extent in different samples during the preparation, separation and digestion steps. In this case, the number of samples must be equal to or larger than the number of different species; i.e., four different species have to be expected for peptides containing one methionine residue and one phosphorylation site such that at least four different samples with various degrees of phosphorylation are required to calculate the degree of phosphorylation. To avoid these types of complications, we recommend completely oxidizing the samples with hydrogen peroxide or sodium periodate.
One issue of pivotal interest for phosphorylation analysis is the detection limit because endogenous phosphoproteins are often of low abundance. However, the sensitivity in this case is not limited by the method itself but by the general detection limits; i.e., ionization/detection efficiencies of the different peptide/phosphopeptide species, and hence, it is difficult to generalize. To examine further the issue of sensitivity, we tried to detect all of the expected tryptic phosphopeptides derived from Pho4. Whereas the phosphopeptide TSSSAEGVVVASEpSPVIAPHGSTHAR gave a very intense ion signal, the tryptic peptide/phosphopeptide AFELVEGMDMDWMMPSHAHHpSPATTATIKPR was hardly observable even when 6 pmol of digest was loaded onto the column. This problem persisted despite the fact that all Met residues were oxidized under mild condition (10 mM NaIO4 for 30 min at room temperature) to ensure minimal heterogeneity of the peptide species, and that the LC and the instrumental parameters were optimized for larger peptide ion species. If an ion signal is reliably observable, then quantitation is possible. For example, when the Pho4 sample was diluted and 50 fmol was injected onto the column, signals for the peptides with the best flyabilities were still observable, which allowed the changes in phosphorylation to be accurately monitored. Similarly, controlled dephosphorylation by using the conditions described in Materials and Methods performed on <400 fmol was still sufficient for two time points and four separate LC/MS experiments for each time point. In summary, the quantitation approach depends only on the detectability of the ion signals and on changes of >10%.
One of the major advantages of using flyability ratios of peptides and their phosphorylated cognate to measure protein phosphorylation quantitatively is that this ratio is constant when complete digestion, identical spray conditions, and identical instrumental parameter settings are ensured. As soon as such a flyability ratio has been determined for a given peptide/phosphopeptide pair, the degree of phosphorylation for this particular phosphorylation site can be calculated easily for single samples at a later stage by simply comparing the ratio of phosphorylated vs. unphosphorylated peptide. Constant flyability ratios are of particular interest for many biological applications because biologists often initially investigate function and regulation of proteins by using overexpressed, tagged proteins before studying the endogenous, normally much less abundant protein. Thus, the flyability ratios for phosphopeptides and their unmodified complements can be readily determined by using the larger quantities available of the overexpressed protein, and this calculated ratio can then be applied to any sample at a later stage when the endogenous protein is studied.
Conclusions and Perspectives
We show that microscale capillary LC/MS is sufficiently reproducible such that quantitative analysis of protein phosphorylation can be derived from LC/MS data based on the ion currents from the different ion species of interest without any stable-isotope labeling. Run-to-run variations and variations in the amount of starting material are accounted for by using a normalization procedure, which uses numerous unmodified peptides derived from the phosphoprotein of interest. These unmodified peptides from the protein of interest have the advantage that they reflect the exact amount of protein present. The approach to choose good peptides suitable for normalization purposes and to identify outliers, which might be interesting peptides themselves, is straightforward and lends itself to automation by using statistical “goodness-of-fit” methods similar to χ2 methods. By using this normalization procedure, changes in phosphorylation of a site (i.e., relative quantitation) can easily be monitored based on the ion currents of the phosphorylated species. By also monitoring the signal intensities of the unphosphorylated cognate, the flyability ratio of a peptide and its phosphorylated complement can be calculated. This approach is easily extended to quantitating the phosphorylation stoichiometry of a given site by merely comparing the ion currents of the phosphopeptide and its unphosphorylated cognate and correcting them for the differences in their flyabilities. Stable-isotope labeling and/or isotopically labeled internal standards are necessary neither for relative nor for absolute quantitiation. This method is not limited to singly phosphorylated species and can also be applied to multiply phosphorylated species if additional samples are available. Otherwise, additional samples with different degrees of phosphorylation can be generated by using a controlled enzymatic dephosphorylation protocol described here. These conditions are generally applicable and independent of the amount of phosphopeptide(s) present. Because these flyability ratios are constant for a given set of LC and instrumental parameters, they can be applied to additional samples at a later stage of the investigation of a particular protein. Furthermore, it is expected that we will be able to estimate the flyability ratios of unknown peptides and their phosphorylated cognates when sufficient empirical information about the determinants of the ionization/detection efficiencies of peptides have been derived.
The sensitivity of this approach depends on the absolute flyabilities of the phosphopeptides of interest; i.e., if a signal can be observed reliably, then the quantitation procedures can be applied. All of the described experiments were performed on a quadrupole–TOF-type mass spectrometer with a limited dynamic range. However, the quantitation methods lend themselves to the implementation on other instruments with larger dynamic ranges, such as the triple-quadrupole mass spectrometer, with which the use of appropriate multiple-reaction monitoring (MRM) strategies are feasible; by using MRM approaches, apparent flyability ratios, which are a function of ionization, detection, and fragmentation efficiencies, are amenable. Using MRM would further improve the sensitivity and compensate for the lower resolution/accuracy of triple-quadrupole instruments as compared with quadrupole–TOF-type instruments. Furthermore, the methods for the relative and absolute quantitation of modification stoichiometries are applicable not only to protein phosphorylation but to other protein modifications in general.
Supplementary Material
Acknowledgments
We thank N. Morrice for the synthetic peptides/phosphopeptides; J. Rush for the purification and quantitation of the synthetic peptides; and S. Gerber, W. Haas, and S. Gygi (Department of Cell Biology, Harvard Medical School) for fruitful discussions, instrument time, and instrument support. We also thank M. Byrne and E. O'Shea for the recombinant Pho4, Pho80, and Pho85. This work was funded by the National Institute of General Medical Sciences Biochemical Studies of Mitosis Grant GM02675.
Author contributions: H.S., J.A.J., M.S., and M.W.K. designed research; H.S. and J.A.J. performed research; H.S. analyzed data; and H.S., J.A.J., M.S., and M.W.K. wrote the paper.
Abbreviation: LC, liquid chromatography.
References
- 1.Yan, J., Packer, N., Gooley, A. & Williams, K. (1998) J. Chromatogr. 808, 23–41. [DOI] [PubMed] [Google Scholar]
- 2.Hunter, T. (1998) Philos. Trans. R. Soc. London B 353, 583–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell, D. & Morrice, N. (2002) J. Biomol. Tech. 13, 119–130. [PMC free article] [PubMed] [Google Scholar]
- 4.Posewitz, M. C. & Tempst, P. (1999) Anal. Chem. 71, 2883–2892. [DOI] [PubMed] [Google Scholar]
- 5.Nuhse, T. S., Stensballe, A., Jensen, O. N. & Peck, S. C. (2003) Mol. Cell. Proteomics 2, 1234–1243. [DOI] [PubMed] [Google Scholar]
- 6.Beausoleil, S. A., Jedrychowski, M., Schwartz, D., Elias, J. E., Villen, J., Li, J., Cohn, M. A., Cantley, L. C. & Gygi, S. P. (2004) Proc. Natl. Acad. Sci. USA 101, 12130–12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carr, S. A., Huddleston, M. J. & Annan, R. S. (1996) Anal. Biochem. 239, 180–192. [DOI] [PubMed] [Google Scholar]
- 8.Neubauer, G. & Mann, M. (1997) J. Mass Spectrom. 32, 94–98. [Google Scholar]
- 9.Steen, H., Kuster, B., Fernandez, M., Pandey, A. & Mann, M. (2001) Anal. Chem. 73, 1440–1448. [DOI] [PubMed] [Google Scholar]
- 10.Oda, Y., Huang, K., Cross, F. R., Cowburn, D. & Chait, B. T. (1999) Proc. Natl. Acad. Sci. USA 96, 6591–6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ibarrola, N., Kalume, D. E., M., G., Iwahori, A. & Pandey, A. (2003) Anal. Chem. 75, 6043–6049. [DOI] [PubMed] [Google Scholar]
- 12.Bonenfant, D., Schmelzle, T., Jacinto, E., Crespo, J. L., Mini, T., Hall, M. N. & Jenoe, P. (2003) Proc. Natl. Acad. Sci. USA 100, 880–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weckwerth, W., Willmitzer, L. & Fiehn, O. (2000) Rapid Commun. Mass Spectrom. 14, 1677–1681. [DOI] [PubMed] [Google Scholar]
- 14.Goshe, M. B., Conrads, T. P., Panisko, E. A., Angell, N. H., Veenstra, T. D. & Smith, R. D. (2001) Anal. Chem. 73, 2578–2586. [DOI] [PubMed] [Google Scholar]
- 15.Qian, W. J., Goshe, M. B., Camp, D. G., 2nd, Yu, L. R., Tang, K. & Smith, R. D. (2003) Anal. Chem. 75, 5441–5450. [DOI] [PubMed] [Google Scholar]
- 16.Zhang, X., Jin, Q. K., Carr, S. A. & Annan, R. S. (2002) Rapid Commun. Mass Spectrom. 16, 2325–2332. [DOI] [PubMed] [Google Scholar]
- 17.Hegeman, A. D., Harms, A. C., Sussman, M. R., Bunner, A. E. & Harper, J. F. (2004) J. Am. Soc. Mass Spectrom. 15, 647–653. [DOI] [PubMed] [Google Scholar]
- 18.Gerber, S. A., Rush, J., Stemman, O., Kirschner, M. W. & Gygi, S. P. (2003) Proc. Natl. Acad. Sci. USA 100, 6940–6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeffery, D. A., Springer, M., King, D. S. & O'Shea, E. K. (2001) J. Mol. Biol. 306, 997–1010. [DOI] [PubMed] [Google Scholar]
- 20.Shevchenko, A., Wilm, M., Vorm, O. & Mann, M. (1996) Anal. Chem. 68, 850–858. [DOI] [PubMed] [Google Scholar]
- 21.Rappsilber, J., Ishihama, Y. & Mann, M. (2003) Anal. Chem. 75, 663–670. [DOI] [PubMed] [Google Scholar]
- 22.Bondarenko, P. V., Chelius, D. & Shaler, T. A. (2002) Anal. Chem. 74, 4741–4749. [DOI] [PubMed] [Google Scholar]
- 23.Wang, W., Zhou, H., Lin, H., Roy, S., Shaler, T. A., Hill, L. R., Norton, S., Kumar, P., Anderle, M. & Becker, C. H. (2003) Anal. Chem. 75, 4818–4826. [DOI] [PubMed] [Google Scholar]
- 24.Chelius, D., Zhang, T., Wang, G. & Shen, R. F. (2003) Anal. Chem. 75, 6658–6665. [DOI] [PubMed] [Google Scholar]
- 25.Ruse, C. I., Willard, B., Jin, J. P., Haas, T., Kinter, M. & Bond, M. (2002) Anal. Chem. 74, 1658–1664. [DOI] [PubMed] [Google Scholar]
- 26.Willard, B. B., Ruse, C. I., Keightley, J. A., Bond, M. & Kinter, M. (2003) Anal. Chem. 75, 2370–2376. [DOI] [PubMed] [Google Scholar]
- 27.Andersen, J. S., Wilkinson, C. J., Mayor, T., Mortensen, P., Nigg, E. A. & Mann, M. (2003) Nature 426, 570–574. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.