Abstract
Proliferation‐related gene signatures have been proposed to aid breast cancer management by providing reproducible prognostic and predictive information on a patient‐by‐patient basis. It is unclear however, whether a less demanding assessment of cell division rate (as determined in clinical setting by expression of Ki67) can function in place of gene profiling.
We investigated agreement between literature‐, distribution‐based, as well as signature‐derived values for Ki67, relative to the genomic grade index (GGI), 70‐gene signature, p53 signature, recurrence score (RS), and the molecular subtype models of Sorlie, Hu, and Parker in representative sets of 253 and 159 breast cancers with a median follow‐up of 13 and 14.5 years, respectively. The relevance for breast cancer specific survival was also addressed in uni‐ and bivariate Cox models.
Taking both cohorts into account, our broad approach identified ROC optimized Ki67 cutoffs in the range of 8–28%. With optimum signature‐reproducing cutoffs, similarity in classification of individual tumors was higher for binary signatures (72–85%), than multi‐level signatures (67–73%). Consistent with strong agreement, no prognostic superiority was noted for either Ki67 or the binary GGI, 70‐gene and p53 signatures in the Uppsala dataset by bivariate analyses. In contrast, Ki67‐independent prognostic capacity could be demonstrated for RS and molecular subtypes according to Sorlie, Hu and Parker in both datasets.
Our results show that the added prognostic value of binary proliferation‐related gene signatures is limited for Ki67‐assessed breast cancers. More complex, multi‐level descriptions have a greater potential in short‐ and long‐term prognostication for biologically relevant breast cancer subgroups.
Keywords: Ki67, Gene expression signatures, Breast cancer prognosis, Long‐term follow‐up, ROC cutoff
Highlights
Patient survival within gene expression signature subtypes can vary greatly over time.
Ki67 and binary gene expression signatures similarly classify breast cancer patients.
Multi‐level gene expression signatures outperform Ki67 for patient prognosis.
1. Introduction
Prognostic indicators and therapy predictive factors provide vital tools to assess and manage the long‐term survival of cancer patients. Microarray technology has provided scientists with a method to improve on traditional prognostic indicators and develop RNA gene‐expression signatures capable of dividing patients into biologically relevant subtypes (Sørlie et al., 2003). These signatures range from simpler binary tumor classifiers to multi‐level five‐group approaches, which provide prognostic (Hu et al., 2006; Miller et al., 2005; Paik et al., 2004; Parker et al., 2009; Perou et al., 2000; Sørlie et al., 2003; Sotiriou et al., 2006; Van't Veer et al., 2002; Van de Vijver et al., 2002) and therapy predictive (Chang et al., 2003; Chia et al., 2012; Gianni et al., 2005) information. Despite good inter‐observer reproducibility following normalization (Wirapati et al., 2008), widespread implementation of microarray analysis in a clinical setting has been hampered by the cost of the analysis, the requirement of good quality RNA derived from frozen tissue and concerns as to whether similar molecular signatures classify patients into the same breast cancer subtypes (Weigelt et al., 2010). Furthermore, as it is accepted that proliferative genes play a central role in gene expression classifiers (Sotiriou and Pusztai, 2009), it is unclear whether a simpler assessment of cell division rate can function in place of gene profiling.
The Ki67 protein, as analyzed by immunohistochemical staining, is widely used as a marker of cell proliferation. Meta‐analyses have highlighted the independent prognostic capabilities of Ki67 in primary breast cancer with reduced overall and disease‐free survival observed in patients with tumors displaying high expression of the protein (De Azambuja et al., 2007; Stuart‐Harris et al., 2008). Despite this evidence, and recommendation by the St. Gallen Consensus (Goldhirsch et al., 2009), Ki67 is still not widely employed in the clinical setting, in part due to question marks over its reproducibility between observers and between laboratories (Mengel et al., 2002; Nielsen et al., 2012; Polley et al., 2013; Varga et al., 2012).
Given the involvement of proliferative genes in gene expression signatures and the proposed prognostic role of Ki67, the aim of our study is to determine whether Ki67 alone can achieve analogous classification of tumors compared to current gene expression signatures. In order to assess this we determined the similarity in classification and prognostic capacity between Ki67 and binary (GGI, 70‐gene, p53) or multi‐level (recurrence score, Sorlie, Hu, and Parker) signatures in testing and validation breast cancer cohorts containing 253 and 159 patients respectively, by Kaplan–Meier, cross‐table, and bivariate survival analyses.
2. Materials and methods
2.1. Patients and specimens*
Our study population consists of two previously published breast cancer cohorts referred to as the Uppsala and Stockholm cohort, (Bergh et al., 1995; Miller et al., 2005; Pawitan et al., 2005). Briefly, Uppsala cohort: This cohort consists of 484 breast cancer patients who received primary therapy in the county of Uppsala between 1987 and 1989. Of these, enough frozen material remained, was available and provided sufficient quality expression profiles for 253 patients. Stockholm cohort: This cohort consists of 524 breast cancer patients who were identified through the Swedish regional cancer registry and received therapy at the Karolinska Hospital between 1994 and 1996. Of these, frozen material was available for 159 patients for which sufficient quality expression profiles could be obtained. Causes of death from 1987 to 2008 (Uppsala cohort) and 1994 to 2010 (Stockholm cohort) were obtained from the National Board of Health and Welfare (Socialstyrelsen). Both microarray studies were approved by the ethics committees at Karolinska Institutet and Karolinska university Hospital, respectively, Stockholm, Sweden.
2.2. Immunohistochemical assessment of Ki67 expression*
Briefly, staining was performed in the Dako Autostainer Plus automated slide processing system (Dako) with a 1:100 dilution of the monoclonal mouse anti‐human Ki67 antibody, clone MIB‐1 (DAKO, Glostrup, Denmark). Staining was evaluated by an experienced pathologist (JC) without prior knowledge of patient outcome or tumor characteristics. Cells with nuclear expression of Ki67 were considered positive and counts were performed at the invasive edge of the tumor as deemed important for prognostic evaluation of Ki67 (Dowsett et al., 2011). This manuscript was carried out and is reported in accordance with REMARK guidelines* (McShane et al., 2005).
2.3. KI67 cutoff selection*
Four approaches for finding relevant Ki67 cutoffs were used. Firstly, a MEDLINE search for Ki67 AND breast cancer AND 2010 or 2011 retrieved 164 publications of which 22 were directly relevant, of these 22, 10% (n = 7) and 20% (n = 8) were the most commonly used cutoffs. Secondly, simple medians of 11% and 12% respectively, were considered. Thirdly, we applied a model of two normal distributions, attempting to capture the visually apparent distribution of low‐expressing tumors versus the dispersed high‐expressing using the mixtools package (Benaglia et al., 2009) in R. With default settings, cutoffs of 16% (Uppsala) and 17% (Stockholm) contained 90% of the lower modeled distribution. Fourthly, receiver operating characteristic (ROC) analysis was used to identify optimal cutoffs for individual gene signatures, resulting in signature specific cutoffs ranging from 10–20% (Uppsala) and 8–28% (Stockholm).
2.4. Expression profiling
In brief, RNA was prepared from fresh frozen, homogenized tumors with RNeasy spin column kits (Qiagen). Evaluation of RNA quality was performed with an Agilent 2100 Bioanalyzer. Two to 5 μg of RNA was subjected to in vitro transcription; products were hybridized to HGU133A and B arrays (Affymetrix), with washing and scanning carried out according to the manufacturer's instructions. For a more detailed account, see here (Calza et al., 2006).
2.5. Prognostic gene signatures*
All tumors in both cohorts were classified according to seven prognostic gene signatures: Genomic Grade Index (Sotiriou et al., 2006) (GGI), 70‐gene signature (Van't Veer et al., 2002) (Mammaprint®), p53 signature (Miller et al., 2005), the recurrence score (Paik et al., 2004) (Oncotype DX®), intrinsic molecular subtype model (Perou et al., 2000; Sørlie et al., 2003) ‐referred to as Sorlie, and the closely related Hu (Hu et al., 2006) and Parker (Parker et al., 2009) (PAM50) classifications. For the Uppsala p53 signature we used the previously published tumor classifications from Miller et al. (2005), which were available for 251 of 253 tumors. Additional information can be found in the Supplementary Methods and code files.
2.6. Statistical analyses*
Differences in Kaplan–Meier estimates were assessed with the two‐sided log‐rank test in SPSS version 19.0. Univariate, bivariate and multivariate hazard ratios were calculated using the coxph function of the survival package in R. Analysis of Ki67 distribution and ROC curve plotting was performed using the R mixtools and Daim packages, respectively. Similarity in classification of a tumour is defined as a case where both a gene expression signature and Ki67 provide the same prognostic classification e.g. poor prognosis. In order to calculate this for a specific gene signature/Ki67 pairing, we simply add all cases where prognostic classification is identical by both classifiers and divide by the total number of patients. An example is shown in the Supplementary methods.
*Indicates extended in the Supplementary methods.
3. Results
3.1. Ki67 and gene expression signatures provide long‐term prognostic information in breast cancer
Patient characteristics for our previously described Uppsala cohort (Bergh et al., 1995; Miller et al., 2005) in relation to estrogen receptor (ER) status and Ki67 expression are shown in Supplementary Tables S1 and S2, respectively. In total, tumours from 253 patients were available for Ki67 staining and expression array profiling, exclusion criteria are displayed in Supplementary Figure S1. A general, literature based Ki67 cutoff of 10% divided the patients of the Uppsala cohort into two groups of low and high expression. We also applied binary and multi‐level gene expression signatures to microarray data from the same patients and used Kaplan–Meier analysis to determine the relationship between Ki67 or signature status and breast cancer specific survival (BCSS). Ki67 (p < 0.001, log‐rank test) and the GGI (p = 0.003), 70‐gene (p = 0.005) and p53 (p = 0.017) binary signatures successfully separated patients into two groups of longer and shorter survival (Figure 1A–D). The recurrence score (p = 0.004) separated patients into three groups‐ “low”, “intermediate” and “high”, where most notably, survival in the “intermediate” group (p = 0.018 vs. low) was similar to that of the “high” group (p = 0.001 vs. low) after twenty‐one years follow‐up (Figure 1E and Supplementary Table S3). The Sorlie (p = 0.024), Hu (p = 0.002) and Parker (p < 0.001) signatures separated patients into five subtypes with varying patient survival (Figure 1F–H). In long‐term follow up the normal‐like, luminal A and basal subtypes retained similar survival information across the five‐group signatures and patients with luminal B and HER2‐like subtype tumors had the worst breast cancer specific survival. One exception to this was the Sorlie HER2‐like group (p = 0.363 vs. normal‐like), which did not display significantly different survival to patients with normal‐like tumors. All log‐rank p‐values for multi‐group signatures are shown in Supplementary Table S3. These results demonstrate the ability of Ki67 and gene expression signatures to divide patients into subtypes with prognostic significance. Notably the basal subtype of Sorlie, Hu and Parker classifications, which contain triple‐negative tumors, did not show poorer outcome in long‐term survival analysis. This highlights the importance of relating gene signatures to long‐term survival data where the outcomes can be dramatically different to those with only five or ten years' follow‐up.
Figure 1.
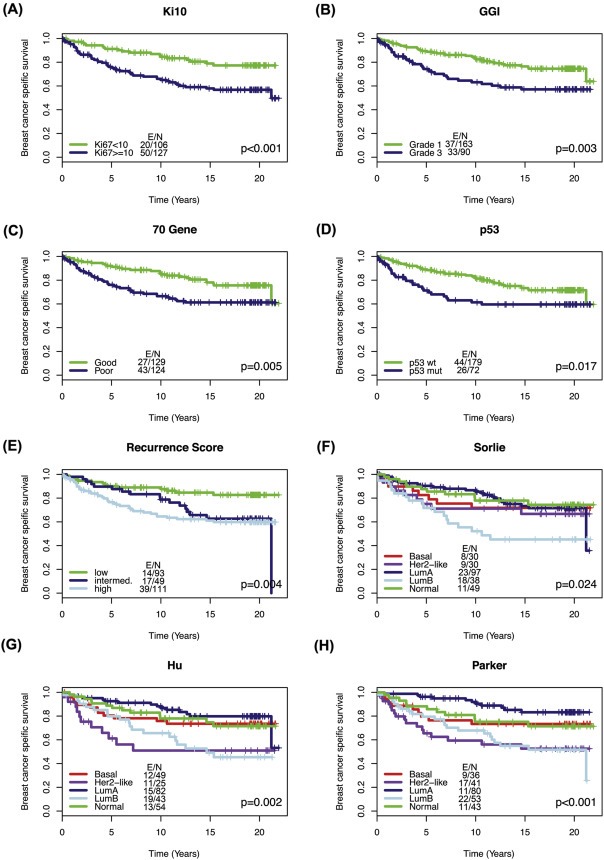
Kaplan–Meier analysis of Ki67 and gene expression signatures in the Uppsala dataset Tumors in the Uppsala cohort were classified according to Ki67 expression below or above/equal to 10% and gene expression signatures. This classification and patient data were subsequently used to produce the following Kaplan–Meier curves (A) Ki67, 10% cutoff (B) GGI (C) 70‐gene (D) p53 (E) Recurrence score (F) Sorlie (G) Hu (H) Parker. P‐value is based on log‐ranked test, E/N = Number of events/number of patients.
3.2. High similarity of classification between Ki67 and binary but not multi‐level gene expression signatures
Due to the considerable range of cutoffs used in the literature (Dowsett et al., 2011) (1%–28.6%), we used a broad approach to identify all reasonable levels above or equal to which staining was considered positive. This and our experimental work‐flow for the Uppsala cohort is depicted in Figure 2. In recent literature, 10% and 20% were most frequently used, and were therefore included (described in more detail in Supplementary methods). As the relevance of a certain cutoff may to some extent be laboratory‐specific, median and distribution‐based cutoffs of 11% and 16% were identified as potentially more relevant for the present data. Given our objective of determining whether Ki67 can achieve similar classifications compared to widely used gene expression signatures, cutoffs most relevant (producing the most similar separation of tumors) for the GGI (15%), 70‐gene (11%), and p53 (16%), were identified with receiver operating characteristics (ROC); Table 1. For the multi‐level signatures, binary collapses were used to avoid a signature disadvantage (for the recurrence score, low vs. intermediate or high; for Sorlie, Parker and Hu, normal‐like, luminal A, or basal vs. HER2‐like or luminal B), best reflecting the separation of survival estimates. The ROC cutoffs, and resulting classification similarity compared to gene signatures is displayed in Table 1.
Figure 2.
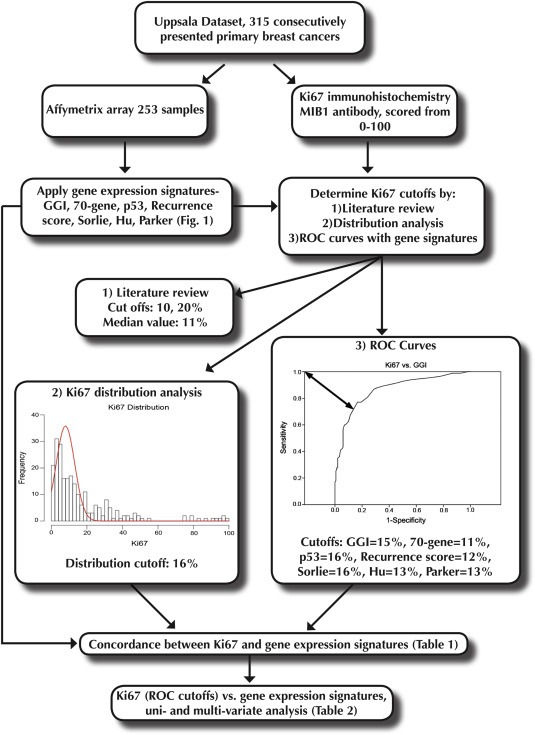
Analysis work‐flow diagram, Uppsala cohort.
Table 1.
Similarity in classification between Ki67 and prognostic gene signatures, Uppsala cohort.
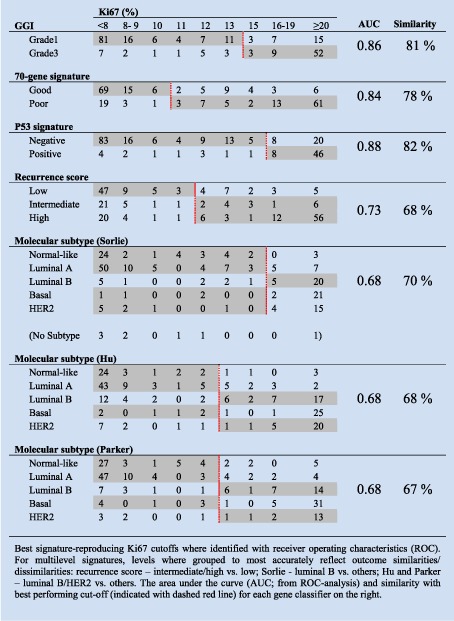
With Ki67, the separation of tumors in low‐ versus high‐proliferating groups demonstrated considerable similarity with the binary signatures (GGI 81%, 70‐gene signature 78%, p53 82%), whereas the similarity was lower for the multi‐level signatures (67–70%; Table 1). Similarly, using kappa statistics as a measurement of agreement, we noted higher kappa values for binary signatures compared to Ki67 (0.55–0.60) than those for multi‐level signatures (0.26–0.36, Supplementary Table S4). All signature and Ki67 classifications on a per patient basis are shown in Supplementary Table S5. Next, as the strongest prognostic capacity of the GGI, 70‐gene, and recurrence score signatures may lie in the ER‐positive subgroup of patients, we performed a subgroup analysis. Analyzing only ER‐positive patients the signature specific Ki67 cutoffs remained the same and the similarity in classification remained high between Ki67 and GGI (81%), and Ki67 and the 70‐gene signature (76%) and remained lower for recurrence score (65%, Supplementary Table 6).
Of note, better agreement could be achieved for the multi‐level classifications in all patients: similarity for the recurrence score increases to 73% when considering low and intermediate vs. high (rather than low vs. intermediate and high); for Sorlie, Parker and Hu similarity could be increased to 81%, 77% and 80%, when grouping basal tumors with luminal B and HER2‐like rather than with the normal‐like and luminal A tumors, illustrating that proliferation is a significant underlying driver in these signatures as well. These alternative groupings would however be a poorer reflection of the long‐term prognostic information in these signatures: the RS seems to identify a group of tumors (intermediate) that despite frequently low Ki67 expression are associated with poorer long‐term outcome; the Sorlie, Hu, Parker on the other hand appear to capture an outcome heterogeneity among highly proliferating tumors not captured by a simple proliferation‐based dichotomization with Ki67 or binary gene expression signatures.
3.3. Binary, but not multi‐level gene expression signatures, give similar prognostic information to Ki67
As previously, we used the Ki67 ROC cutoffs and related signature groupings, examining each in a univariate Cox analysis (Table 2) with BCSS as endpoint. For comparison, uni‐ and multivariate hazard ratios for standard clinicopathological characteristics are shown in Supplementary Tables S7 and S8. All Ki67 and gene expression signature groups provided statistically significant prognostic information with hazard ratios (HR) ranging from 1.79 (95%CI 1.10–2.92) for the p53 signature to 2.75 (95%CI 1.72–4.42) for the Parker classifier (Table 2, “Separate models” column). Next, we compared Ki67 cutoffs head to head with gene signatures in bivariate analysis. Importantly, we found that neither Ki67 nor binary gene expression signatures (GGI, 70‐gene and p53) provided significant prognostic information when both where included in a bivariate model (see Table 2, “Combined model” column). In the case of multi‐level signatures, the recurrence score (HR 2.07, 95%CI 1.12–3.85), Sorlie (HR 2.02, 95%CI 1.15–3.55), Hu (HR 2.17, 95%CI 1.31–3.60) and Parker (HR 2.22, 95%CI 1.35–3.67) provided superior prognostic information to that given by Ki67 (Table 2).
Table 2.
Cox uni‐ and bivariate analyses, BCSS endpoint, hazard ratio (HR) and 95% conf. interval (CI), Uppsala.
| Covariate | Separate models HRs (CI) | Combined model HRs (CI) |
|---|---|---|
| Genomic grade index | 2.03 (1.27–3.25) | 1.57 (0.84–2.90) |
| Ki 67 ≥ 15 | 1.93 (1.21–3.09) | 1.46 (0.79–2.70) |
| 70‐gene signature | 1.96 (1.21–3.17) | 1.50 (0.84–2.68) |
| Ki 67 ≥ 11 | 2.04 (1.24–3.34) | 1.61 (0.89–2.94) |
| P53 signature | 1.79 (1.10–2.92) | 1.21 (0.64–2.28) |
| Ki 67 ≥ 16 | 1.84 (1.15–2.95) | 1.66 (0.90–3.08) |
| Recurrence scorea | 2.57 (1.43–4.62) | 2.07 (1.12–3.85) |
| Ki 67 ≥ 12 | 2.09 (1.28–3.41) | 1.66 (0.99–2.79) |
| Molecular subtype (Sorlie)b | 2.38 (1.39–4.08) | 2.02 (1.15–3.55) |
| Ki 67 ≥ 16 | 1.84 (1.15–2.95) | 1.46 (0.89–2.40) |
| Molecular subtype (Hu)c | 2.52 (1.57–4.06) | 2.17 (1.31–3.60) |
| Ki 67 ≥ 13 | 2.09 (1.29–3.38) | 1.66 (1.00–2.76) |
| Molecular subtype (Parker)c | 2.75 (1.72–4.42) | 2.22 (1.35–3.67) |
| Ki 67 ≥ 13 | 2.09 (1.29–3.38) | 1.61 (0.97–2.68) |
Hazard ratio for high or intermediate recurrence score; low as reference.
Hazard ratio for luminal B subtype; others as reference.
Hazard ratio for luminal B or HER2 subtype; others as reference.
We then extended our head to head bivariate analysis to include Ki67 cutoffs identified by all methods, and compare them to all gene expression signature groupings, again with BCSS as endpoint (Supplementary Table S9). In general, Ki67 cutoffs performed consistently well against binary but not multi‐level gene expression signatures. The exception to this was the Ki20 cutoff, which was out‐performed by all signatures (Supplementary Table S9, far right column).
Taken together, these results show that the tested binary gene expression signatures classify breast cancer patients into similar good/poor prognosis groups as Ki67 alone. However, the recurrence score, Sorlie, Hu and Parker signatures appear to provide prognostic information that is different to that given by Ki67 alone.
3.4. High similarity of classification between Ki67 and gene expression signatures in a validation cohort
Next, we sought to validate our findings in a second clinical cohort of 159 breast cancer patients (referred to as the Stockholm cohort (Pawitan et al., 2005)). Patient characteristics for this cohort in relation to ER and Ki67 = 8% are shown in Supplementary Tables 10 and 11 respectively.
Firstly, performing an identical analysis to that of the Uppsala cohort, we noted the ability of both Ki67 and gene expression signatures to separate breast cancer patients into prognostically significant subgroups using Kaplan–Meier analysis (Supplementary Figure S2 and Supplementary Table S12).
Secondly and similarly to the Uppsala cohort, tumors separated into low‐ versus high‐proliferating groups by Ki67 demonstrated an overlap with binary signatures that was higher than that for the multi‐level signatures (72–85% vs. 68–73% respectively, Table 3). Additionally, agreement between binary signatures and Ki67 was generally higher (kappa = 0.44 − 0.58) than for multi‐level signatures (kappa = 0.31 − 0.49). All signature and Ki67 classifications on a per patient basis for the Stockholm cohort are shown in Supplementary Table S13. The percentage similarity between Ki67 and gene signatures were slightly lower when considering ER‐positive patients only, 64% for GGI, 69% for 70‐gene and 64% for the recurrence score (Supplementary Table 14).
Table 3.
Similarity in classification between Ki67 and prognostic gene signatures, Stockholm cohort.
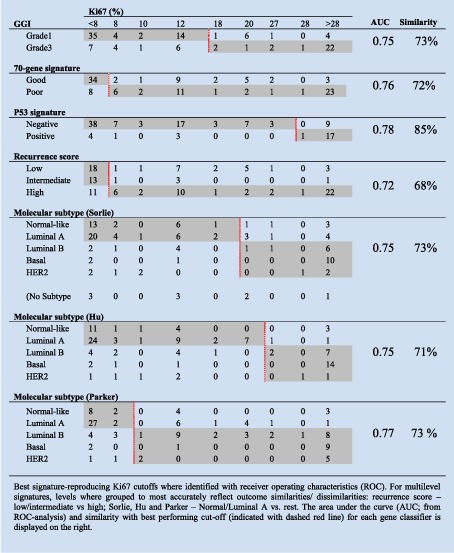
Thirdly, employing as before Ki67 ROC cutoffs and related signature groupings, we determined the prognostic capabilities of Ki67 and signatures in both uni‐ and bivariate analysis. For comparison, uni‐ and multivariate hazard ratios for standard clinicopathological characteristics are shown in Supplementary Tables S7 and S8. Whilst Ki67 was generally significant in univariate analysis (Table 4, see “Separate models HRs” column), it tended to be out‐performed by gene signatures in bivariate analysis (Table 4, “Combined model HRs” column). Of note the hazard ratios for multi‐level signatures increased when Ki67 was included in a bivariate model relative to their univariate values (Table 4, compare “Separate models HRs” column to “Combined model HRs” column).
Table 4.
Cox uni‐ and bivariate analyses, BCSS endpoint, hazard ratio (HR) and 95% conf. interval (CI), Stockholm.
| Covariate | Separate models HRs (CI) | Combined model HRs (CI) |
|---|---|---|
| Genomic grade index | 4.05 (2.06–8.00) | 3.43 (1.43–8.30) |
| Ki 67 ≥ 18 | 1.97 (0.92–4.18) | 1.17 (0.52–2.67) |
| 70‐gene signature | 4.61 (2.12–10.03) | 3.70 (1.36–10.05) |
| Ki 67 ≥ 8 | 3.20 (1.21–8.46) | 1.75 (0.60–5.07) |
| P53 signature | 1.88 (1.00–3.55) | 0.92 (0.33–2.58) |
| Ki 67 ≥ 28 | 2.45 (1.12–5.37) | 2.57 (0.97–6.81) |
| Recurrence scorea | 2.87 (1.43–5.75) | 4.62 (1.67–12.74) |
| Ki 67 ≥ 8 | 3.20 (1.21–8.46) | 1.92 (0.70–5.30) |
| Molecular subtype (Sorlie)b | 3.19 (1.65–6.14) | 3.80 (1.57–9.21) |
| Ki 67 ≥ 20 | 2.31 (1.08–4.92) | 1.16 (0.49–2.74) |
| Molecular subtype (Hu)b | 4.27 (2.12–8.58) | 4.58 (1.81–11.58) |
| Ki 67 ≥ 27 | 2.44 (1.13–5.27) | 1.11 (0.47–2.60) |
| Molecular subtype (Parker)b | 5.51 (2.43–12.49) | 6.78 (2.38–19.33) |
| Ki 67 ≥ 10 | 1.88 (0.84–4.19) | 0.87 (0.37–2.05) |
Hazard ratio for high recurrence score; others as reference.
Hazard ratio for luminal B/HER2 or basal subtypes; others as reference.
Fourthly, we compared all Ki67 cutoffs to all gene expression signatures in a head to head bivariate analysis. The most competitive Ki67 cutoff in the Stockholm cohort was 8, however in general, gene signatures performed consistently better than Ki67 with the exception of the p53 (Supplementary Table S15). Moreover, the highest hazard ratios were seen amongst the multi‐level gene expression signatures and lowest among the binary signatures.
Taken together these results provide a similar picture to that of the Uppsala cohort. Both Kaplan–Meier and similarity of classification analysis show that Ki67 and gene signatures can classify patients into comparable good/poor prognosis groups. In this instance however, the ability of Ki67 to compete with binary signatures in a bivariate analysis was lessened, potentially due to reduced patient numbers in comparison to the Uppsala cohort. Most notably, it was again readily apparent that Ki67 cannot provide similar prognostic information to that given by multi‐level gene expression signatures.
4. Discussion
In this study we have assessed whether Ki67 alone can achieve similar classifications of tumors compared to gene expression signatures, and whether Ki67‐based classification can compete with regards to prognostic capacity in cohorts of 253 and 159 tumors with a median follow‐up of 13 and 14.5 years, respectively. Our initial analysis in both cohorts showed similarity in tumor classification by Ki67 and binary gene expression signatures (GGI, 70‐gene and p53). Moreover, in the Uppsala cohort it was also not possible to demonstrate that these signatures or Ki67 had superior prognostic capacity, which is to be expected for highly correlated descriptions (Vittinghoff and Stephen Shiboski Eric, 2009). Less similarity in classification was observed between Ki67 and the molecular subtype signatures (Sorlie, Hu and Parker) in both cohorts, and these signatures provided independent prognostic information indicating their ability to delineate tumor groups with good/poor prognosis better than Ki67 can alone.
It has been suggested that widespread adoption of Ki67 as a prognostic indicator may have been hampered in part by poor inter‐observer or inter‐laboratory reproducibility (Mengel et al., 2002; Nielsen et al., 2012; Polley et al., 2013; Varga et al., 2012) and linked to that, the large range of reported cutoffs for assessing positivity of the protein (Dowsett et al., 2011) (1%–28.6%). We addressed this by employing a broad approach for determining relevant cutoffs, including literature‐ and distribution‐based as well as signature‐reproducing threshold values. Importantly, we did not include a Ki67 cutoff that maximized prognostic capacity, thereby precluding a potential over‐fit to the present data. Rather, and in consistency with the aim of investigating if Ki67 can function in place of individual gene expression signatures, we tested each signature against a Ki67 cutoff producing the most similar classification of tumors, thereby avoiding a potential advantage over gene signatures that have not been optimized for prognostic capacity in the present data. For the multi‐level signatures, cutoffs most accurately reflecting signature prognostic capacity were used: e.g. Uppsala cohort: Ki67 values were chosen to best reflect low vs. high and intermediate designations for the recurrence score; luminal B vs. all other for Sorlie; and luminal B or HER2‐like vs. all other for Hu and Parker. These groupings of levels were the ones that performed best in both univariate and bivariate analyses.
Whilst others have employed alternative fixed Ki67 cutoffs including 14% or 20% (Goldhirsch et al., 2009; Naoi et al., 2011a; Toyoda et al., 2011), scrutinizing the particular choice of cutoff in our data seems less important: for the range of tested cutoffs in the Uppsala cohort (10%–20%), findings were largely consistent. GGI and the 70‐gene signature had significant HR estimates in only one of these tests (when run against Ki67 ≥ 20%), whereas the p53 signature HRs were never significantly different from Ki67 (Supplementary Table S9). In contrast, the recurrence score (low vs. high and intermediate) was associated with significant HRs regardless of cutoff, which was also the case for Sorlie (Luminal B vs. other), Hu and Parker (both Luminal B or HER2 vs. other; Supplementary Table S9).”
Regarding future cutoff selection by researchers undertaking Ki67 analysis (and in the interest of being explicit), it is worth detailing performance of the values obtained in the Uppsala dataset based on fitting a data distribution (16%), choosing a median value (11%) and by literature review (10%, 20%). In general, no binary gene expression signatures were significantly different to the 10, 11 and 16% cutoffs in a multivariate analysis. We therefore believe that in order to benefit from the implementation of Ki67 in a clinical setting, individual labs must conduct a study of how the protein is distributed in their samples, and choose a reasonable median value. Failing this, if the current lack of standardization in staining procedures (Colozza et al., 2010) can be improved upon, a cutoff of 10% would likely provide prognostically significant results. In our study, we found the much‐published cutoff of 20% (Naoi et al., 2011, 2011, 2011) to be a poor indicator of prognosis, and as such would recommend against its use unless it represented a median value of staining in a given laboratory.
It should be stressed that whilst we are comparing whether Ki67 can provide similar prognostic information to gene expression signatures, other routinely used prognostic markers could also be taken in account. The ability of existing biomarkers to identify the same subtypes of tumors as gene signatures is a much‐debated topic (Tang et al., 2009). Numerous studies have successfully recapitulated classification schemes that closely mimic gene expression subtyping using markers such as the estrogen and progesterone receptors, Her2, cytokeratins 5/6 and 18, and the epidermal growth factor (Abd El‐Rehim et al., 2005; Livasy et al., 2006; Nielsen et al., 2010). The addition of Ki67 to standard markers has previously enabled researchers to make the clinically important discrimination between the luminal A and B subtypes of breast cancer (Nielsen et al., 2010), and Ki67 is used in some clinical settings to identify high‐risk ER+ positive patients. However, we are only addressing the straight forward question of which prognosticator is better in a head to head comparison and as such have drawn our conclusions based on Ki67 and gene expression alone. It could be argued that inclusion of additional biomarkers would strengthen the performance of Ki67 as a prognostic tool against multi‐level gene signatures, but this was not our aim. Notably, when Chang et al. performed such an analysis against one gene signature, they found clinical factors and immunohistochemical stainings (including Ki67) gave less prognostic information than Parker (Cheang et al., 2009). Similarly, Dowsett et al. have recently found the PAM50 risk of recurrence score (ROR) to provide more prognostic information than IHC4 (a recurrence risk signature that utilizes ER/PR/HER2 and Ki67 protein expression) within a breast cancer subgroup of HER2‐negative/node negative patients (Dowsett et al., 2013).
Whilst we have focused on seven specific gene expression signatures here, other signatures with relevance to routine clinical practice could have been taken into account. Of particular note is the EndoPredict assay which is used to predict the risk of late metastasis in breast cancer patients with ER+, HER2‐ disease (Dubsky et al., 2012). With regard to the potential of Ki67 to compete well against this assay, Varga et al. have performed a direct comparison of the concordance between Ki67 as a continuous variable and the EndoPredict (EP) score in 34 hormone receptor positive invasive breast cancer patients (Varga et al., 2013). The author's noted a moderate statistically significant correlation between Ki67 and the EP score (Person correlation 0.55, p < 0.0001), but no correlation between Ki67 and the EP clin score (defined as the EP score combined with tumour size and nodal status) was found (Person correlation 0.24, p < 0.16). Using the St. Gallen Ki67 cutoff of 14% Dubsky et al. divided over 1000 patients into Luminal A and B subtypes and noted that the EP clin score can readily divide the patients of these subtypes into 2 additional groups of patients with better and worse prognosis (Dubsky et al., 2012). Similarly, Filipits et al. demonstrated the EP score provides independent prognostic information in a multivariate analysis with standard clinicopathological variables including Ki67 with a cutoff of 11% (Filipits et al., 2011). Taken together these data suggest the EP clin score will likely perform better than Ki67 alone for the prognosis of breast cancer patients, but the performance of the EP score vs. Ki67 is less clear.
Other groups have also examined the relationship between Ki67 and gene expression signatures, albeit in smaller cohorts, and found similar findings to ours. Niikurra et al. demonstrated a strong correlation between Ki67 expression as measured by immunohistochemistry (IHC) and the GGI and recurrence score signatures in 39 tumours, without adjusting for ER status (Niikura et al., 2012). Using Ki67 IHC as a continuous variable, Reyal et al. found a positive correlation between the protein and GGI (Reyal et al., 2012). In the same study 69/78 genomic grade 1 (GG1) tumours were classified as Ki67 low (cut‐off ≤20%) whilst 45/50 GG3 tumours were classified as Ki67 high (<20%). In contrast, others have shown the ability of GGI to outcompete Ki67 IHC (as a continuous variable) in a multivariate analysis including 204 patients with a disease‐free survival endpoint (Bertucci et al., 2012). Our study differs from those aforementioned not only in the aspect that we employ larger cohorts for our analysis, but also in that we implement relevant Ki67 cut‐offs and present a much broader analysis including comparisons to 7 gene expression signatures.
Our analysis has also highlighted a subset of patients where the same tumour sample is assigned to opposing prognostic subgroups by gene expression signatures and Ki67. This is most clearly demonstrated by the data in Supplementary Tables 5 and 13 for the Uppsala and Stockholm cohorts respectively. One plausible explanation for opposing classifications of the same sample, is that as only a small fraction of a tumour sample is sectioned for immunohistochemistry (as little as 4–5 μm for formalin fixed, paraffin‐embedded material), high numbers of Ki67 positive cells present only at the invasive tumour edge could easily be missed. Similarly, samples with high levels of Ki67 expression at the invasive tumour front but lower levels in bulk of the tumour body could be classified as good prognosis by gene expression signatures as the proliferative signal is drowned‐out by shear number of low/slowly proliferating cells.
One potential limitation of this study is that all of the signatures were applied to gene expression data from a single platform (Affymetrix). Whilst this makes the different signature classifications highly comparable within our datasets, they are not routinely run on this platform in a clinical setting. Take for example the recurrence score, commercially known as Oncotype DX® (Geomic Health, Redwood City, CA). This test is based on a 21 gene (qRT)‐PCR assay where five genes are used for normalization and another sixteen cancer‐related genes are used for tumour classification. Despite the strong correlation between microarray and qRT‐PCR data (Morey et al., 2006), it is plausible that had all tumour samples been run on the commercial Oncotype DX® assay, 100% agreement with classifications derived from the microarray data may not have been achieved. In general though, we are confident that the trends seen in the RS, GGI, 70‐gene and PAM50 classifications within our cohorts are highly reproducible and are on the whole representative of their commercial counterparts.
The future of individual patient prognostication may encompass a combination of established (ER, PR, HER2/neu and Ki67) and new biomarkers, or mRNA derived signatures describing similar tumor subtypes. Quantification possibilities, reproducibility, and user‐independence probably favors the latter, whereas pathologists and clinicians may prefer established markers until very substantial data demonstrating at least non‐inferiority for gene expression profiles are presented. The drawback to current pathological approaches is their reliance on techniques requiring ocular interpretation, thereby introducing problems with inter‐observer stability. Whilst the current trend towards digital pathology and automated image analysis systems (Klimowicz et al., 2012; Tobin et al., 2012) may alleviate these issues, gene expression markers appear to be more technically reliable and reproducible, particularly after normalization protocols. Regardless of technology, the biological heterogeneity of breast cancer seems to warrant a degree of complexity in these descriptions. Simple binary classifications designating tumors as either good or bad appear overly simplistic and offer limited improvement over a simple proliferation‐based marker such as Ki67.
In conclusion, we have found minimal added prognostic value of binary proliferation‐related gene signatures for Ki67‐assessed breast cancers. In contrast, multi‐level descriptions display a greater ability to pinpoint the short‐ and long term outcome heterogeneity in breast cancer, adding further credibility to their use as a routine clinical diagnostic tool.
Funding
This work was supported by grants from the Swedish Research Council, Gösta Miltons Donations fond, Karolinska Institutet Foundations, Swedish Cancer Society, Cancer Society in Stockholm, The King Gustaf V Jubilee Fund, Swedish Breast Cancer Association, Märit and Hans Rausing's Initiative Against Breast Cancer and BRECT to JB and by the Swedish Research Council [grant no: 524‐2011‐6857] and Gösta Miltons Donations fond to LL. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure of potential conflicts of interest
To err on the side of caution, we wish to disclose the following potential conflict of interests; Professor Bergh receives research funding from Merck, paid to Karolinska Institutet and from Amgen, Roche, Sanofi‐Aventis and Bayer, paid to Karolinska University Hospital. All remaining authors have declared no conflicts of interest.
Supporting information
The following are the supplementary data related to this article:
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary Figure S1Exclusion criteria and treatment information for the Uppsala and Stockholm cohorts.
Supplementary Figure S2Kaplan–Meier analysis of Ki67 and gene expression signatures in the Stockholm dataset.
Acknowledgements
We wish to thank Anna‐Lena Borg for her help and technical expertise in sample preparation and staining.
Supplementary data 1.
1.1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2014.02.007
Tobin Nicholas P., Lindström Linda S., Carlson Joseph W., Bjöhle Judith, Bergh Jonas and Wennmalm Kristian, (2014), Multi‐level gene expression signatures, but not binary, outperform Ki67 for the long term prognostication of breast cancer patients, Molecular Oncology, 8, doi: 10.1016/j.molonc.2014.02.007.
References
- Abd El-Rehim, D.M. , Ball, G. , Pinder, S.E. , Rakha, E. , Paish, C. , Robertson, J.F.R. , Macmillan, D. , Blamey, R.W. , Ellis, I.O. , 2005. High-throughput protein expression analysis using tissue microarray technology of a large well-characterised series identifies biologically distinct classes of breast cancer confirming recent cDNA expression analyses. Int. J. Cancer. 116, 340–350. [DOI] [PubMed] [Google Scholar]
- Benaglia, T. , Chauveau, D. , Hunter, D.R. , Young, D.S. , 2009. Mixtools: an R package for analyzing finite mixture models. J. Stat. Softw.. 32, 1–29. [Google Scholar]
- Bergh, J. , Norberg, T. , Sjögren, S. , Lindgren, A. , Holmberg, L. , 1995. Complete sequencing of the p53 gene provides prognostic information in breast cancer patients, particularly in relation to adjuvant systemic therapy and radiotherapy. Nat. Med.. 1, 1029–1034. [DOI] [PubMed] [Google Scholar]
- Bertucci, F. , Finetti, P. , Roche, H. , Doussal, J.M.L. , Marisa, L. , Martin, A.L. , Lacroix-Triki, M. , Blanc-Fournier, C. , Jacquemier, J. , Peyro-Saint-Paul, H. , Viens, P. , Sotiriou, C. , Birnbaum, D. , Penault-Llorca, F. , 2012. Comparison of the prognostic value of genomic grade index, Ki67 expression and mitotic activity index in early node-positive breast cancer patients. Ann. Oncol.. [DOI] [PubMed] [Google Scholar]
- Calza, S. , Hall, P. , Auer, G. , Bjöhle, J. , Klaar, S. , Kronenwett, U. , Liu, E.T. , Miller, L. , Ploner, A. , Smeds, J. , Bergh, J. , Pawitan, Y. , 2006. Intrinsic molecular signature of breast cancer in a population-based cohort of 412 patients. Breast Cancer Res.. 8, R34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, J.C. , Wooten, E.C. , Tsimelzon, A. , Hilsenbeck, S.G. , Gutierrez, M.C. , Elledge, R. , Mohsin, S. , Osborne, C.K. , Chamness, G.C. , Allred, D.C. , 2003. Gene expression profiling for the prediction of therapeutic response to docetaxel in patients with breast cancer. The Lancet. 362, 361 [DOI] [PubMed] [Google Scholar]
- Cheang, M.C.U. , Chia, S.K. , Voduc, D. , Gao, D. , Leung, S. , Snider, J. , Watson, M. , Davies, S. , Bernard, P.S. , Parker, J.S. , Perou, C.M. , Ellis, M.J. , Nielsen, T.O. , 2009. Ki67 Index, HER2 status, and prognosis of patients with luminal B breast cancer. JNCI J. Natl. Cancer Inst.. 101, 736–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia, S.K. , Bramwell, V.H. , Tu, D. , Shepherd, L.E. , Jiang, S. , Vickery, T. , Mardis, E. , Leung, S. , Ung, K. , Pritchard, K.I. , Parker, J.S. , Bernard, P.S. , Perou, C.M. , Ellis, M.J. , Nielsen, T.O. , 2012. A 50-gene intrinsic subtype classifier for prognosis and prediction of benefit from adjuvant tamoxifen. Clin. Cancer Res.. 18, 4465–4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colozza, M. , Sidoni, A. , Piccart-Gebhart, M. , 2010. Value of Ki67 in breast cancer: the debate is still open. The Lancet Oncol.. 11, 414–415. [DOI] [PubMed] [Google Scholar]
- De Azambuja, E. , Cardoso, F. , de Castro, G. , Colozza, M. , Mano, M.S. , Durbecq, V. , Sotiriou, C. , Larsimont, D. , Piccart-Gebhart, M.J. , Paesmans, M. , 2007. Ki-67 as prognostic marker in early breast cancer: a meta-analysis of published studies involving 12,155 patients. Br. J. Cancer. 96, 1504–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowsett, M. , Nielsen, T.O. , A'Hern, R. , Bartlett, J. , Coombes, R.C. , Cuzick, J. , Ellis, M. , Henry, N.L. , Hugh, J.C. , Lively, T. , McShane, L. , Paik, S. , Penault-Llorca, F. , Prudkin, L. , Regan, M. , Salter, J. , Sotiriou, C. , Smith, I.E. , Viale, G. , Zujewski, J.A. , Hayes, D.F. , 2011. Assessment of Ki67 in breast cancer: recommendations from the International Ki67 in Breast Cancer working group. J. Natl. Cancer Inst.. 103, 1656–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowsett, M. , Sestak, I. , Lopez-Knowles, E. , Sidhu, K. , Dunbier, A.K. , Cowens, J.W. , Ferree, S. , Storhoff, J. , Schaper, C. , Cuzick, J. , 2013. Comparison of PAM50 risk of recurrence score with oncotype DX and IHC4 for predicting risk of distant recurrence after endocrine therapy. J. Clin. Oncol.. 31, 2783–2790. [DOI] [PubMed] [Google Scholar]
- Dubsky, P. , Filipits, M. , Jakesz, R. , Rudas, M. , Singer, C.F. , Greil, R. , Dietze, O. , Luisser, I. , Klug, E. , Sedivy, R. , Bachner, M. , Mayr, D. , Schmidt, M. , Gehrmann, M.C. , Petry, C. , Weber, K.E. , Kronenwett, R. , Brase, J.C. , Gnant, M. , 2012. EndoPredict improves the prognostic classification derived from common clinical guidelines in ER-positive, HER2-negative early breast cancer. Ann. Oncol. Mds. 334, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipits, M. , Rudas, M. , Jakesz, R. , Dubsky, P. , Fitzal, F. , Singer, C.F. , Dietze, O. , Greil, R. , Jelen, A. , Sevelda, P. , Freibauer, C. , Müller, V. , Jänicke, F. , Schmidt, M. , Kölbl, H. , Rody, A. , Kaufmann, M. , Schroth, W. , Brauch, H. , Schwab, M. , Fritz, P. , Weber, K.E. , Feder, I.S. , Hennig, G. , Kronenwett, R. , Gehrmann, M. , Gnant, M. , 2011. A new molecular predictor of distant recurrence in ER-positive, HER2-negative breast cancer adds independent information to conventional clinical risk factors. Clin. Cancer Res.. 17, 6012–6020. [DOI] [PubMed] [Google Scholar]
- Gianni, L. , Zambetti, M. , Clark, K. , Baker, J. , Cronin, M. , Wu, J. , Mariani, G. , Rodriguez, J. , Carcangiu, M. , Watson, D. , 2005. Gene expression profiles in paraffin-embedded core biopsy tissue predict response to chemotherapy in women with locally advanced breast cancer. J. Clin. Oncol.. 23, 7265–7277. [DOI] [PubMed] [Google Scholar]
- Goldhirsch, A. , Ingle, J.N. , Gelber, R.D. , Coates, A.S. , Thürlimann, B. , Senn, H.-J. , 2009. Thresholds for therapies: highlights of the st gallen international expert consensus on the primary therapy of early breast cancer 2009. Ann. Oncol.. 20, 1319–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Z. , Fan, C. , Oh, D.S. , Marron, J.S. , He, X. , Qaqish, B.F. , Livasy, C. , Carey, L.A. , Reynolds, E. , Dressler, L. , Nobel, A. , Parker, J. , Ewend, M.G. , Sawyer, L.R. , Wu, J. , Liu, Y. , Nanda, R. , Tretiakova, M. , Ruiz Orrico, A. , Dreher, D. , Palazzo, J.P. , Perreard, L. , Nelson, E. , Mone, M. , Hansen, H. , Mullins, M. , Quackenbush, J.F. , Ellis, M.J. , Olopade, O.I. , Bernard, P.S. , Perou, C.M. , 2006. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 7, 96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimowicz, A.C. , Bose, P. , Nakoneshny, S.C. , Dean, M. , Huang, L. , Chandarana, S. , Magliocco, A.M. , Wayne Matthews, T. , Brockton, N.T. , Dort, J.C. , 2012. Basal Ki67 expression measured by digital image analysis is optimal for prognostication in oral squamous cell carcinoma. Eur. J. Cancer. 48, 2166–2174. [DOI] [PubMed] [Google Scholar]
- Livasy, C.A. , Karaca, G. , Nanda, R. , Tretiakova, M.S. , Olopade, O.I. , Moore, D.T. , Perou, C.M. , 2006. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod. Pathol.. 19, 264–271. [DOI] [PubMed] [Google Scholar]
- McShane, L.M. , Altman, D.G. , Sauerbrei, W. , Taube, S.E. , Gion, M. , Clark, G.M. , Statistics Subcommittee of the NCI-EORTC Working Group on Cancer Diagnostics2005. Reporting recommendations for tumor marker prognostic studies (REMARK). J. Natl. Cancer Inst.. 97, 1180–1184. [DOI] [PubMed] [Google Scholar]
- Mengel, M. , von Wasielewski, R. , Wiese, B. , Rüdiger, T. , Müller-Hermelink, H.K. , Kreipe, H. , 2002. Inter-laboratory and inter-observer reproducibility of immunohistochemical assessment of the Ki-67 labelling index in a large multi-centre trial. J. Pathol.. 198, 292–299. [DOI] [PubMed] [Google Scholar]
- Miller, L.D. , Smeds, J. , George, J. , Vega, V.B. , Vergara, L. , Ploner, A. , Pawitan, Y. , Hall, P. , Klaar, S. , Liu, E.T. , Bergh, J. , 2005. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. PNAS. 102, 13550–13555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey, J.S. , Ryan, J.C. , Van Dolah, F.M. , 2006. Microarray validation: factors influencing correlation between oligonucleotide microarrays and real-time PCR. Biol. Proced. Online. 8, 175–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoi, Y. , Kishi, K. , Tanei, T. , Tsunashima, R. , Tominaga, N. , Baba, Y. , Kim, S.J. , Taguchi, T. , Tamaki, Y. , Noguchi, S. , 2011. High genomic grade index associated with poor prognosis for lymph node-negative and estrogen receptor-positive breast cancers and with good response to chemotherapy. Cancer. 117, 472–479. [DOI] [PubMed] [Google Scholar]
- Naoi, Y. , Kishi, K. , Tanei, T. , Tsunashima, R. , Tominaga, N. , Baba, Y. , Kim, S.J. , Taguchi, T. , Tamaki, Y. , Noguchi, S. , 2011. Development of 95-gene classifier as a powerful predictor of recurrences in node-negative and ER-positive breast cancer patients. Breast Cancer Res. Treat.. 128, 633–641. [DOI] [PubMed] [Google Scholar]
- Nielsen, T. , Polley, M.-Y. , Leung, S. , Mastropasqua, M. , Zabaglo, L. , Bartlett, J. , Viale, G. , McShane, L. , Hayes, D. , Dowsett, M. , the International Ki67 in Breast Cancer Working Group of the BIG-NABCG collaboration2012. An international Ki67 reproducibility study. Cancer Res.. 72, (Suppl. 3) [Google Scholar]
- Nielsen, T.O. , Parker, J.S. , Leung, S. , Voduc, D. , Ebbert, M. , Vickery, T. , Davies, S.R. , Snider, J. , Stijleman, I.J. , Reed, J. , Cheang, M.C.U. , Mardis, E.R. , Perou, C.M. , Bernard, P.S. , Ellis, M.J. , 2010. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor–positive breast cancer. Clin. Cancer Res.. 16, 5222–5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niikura, N. , Iwamoto, T. , Masuda, S. , Kumaki, N. , Xiaoyan, T. , Shirane, M. , Mori, K. , Tsuda, B. , Okamura, T. , Saito, Y. , Suzuki, Y. , Tokuda, Y. , 2012. Immunohistochemical Ki67 labeling index has similar proliferation predictive power to various gene signatures in breast cancer. Cancer Sci.. 103, 1508–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik, S. , Shak, S. , Tang, G. , Kim, C. , Baker, J. , Cronin, M. , Baehner, F.L. , Walker, M.G. , Watson, D. , Park, T. , Hiller, W. , Fisher, E.R. , Wickerham, D.L. , Bryant, J. , Wolmark, N. , 2004. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast Cancer. New Engl. J. Med.. 351, 2817–2826. [DOI] [PubMed] [Google Scholar]
- Parker, J.S. , Mullins, M. , Cheang, M.C.U. , Leung, S. , Voduc, D. , Vickery, T. , Davies, S. , Fauron, C. , He, X. , Hu, Z. , Quackenbush, J.F. , Stijleman, I.J. , Palazzo, J. , Marron, J.S. , Nobel, A.B. , Mardis, E. , Nielsen, T.O. , Ellis, M.J. , Perou, C.M. , Bernard, P.S. , 2009. Supervised risk predictor of breast Cancer based on intrinsic subtypes. JCO. 27, 1160–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawitan, Y. , Bjöhle, J. , Amler, L. , Borg, A.-L. , Egyhazi, S. , Hall, P. , Han, X. , Holmberg, L. , Huang, F. , Klaar, S. , Liu, E.T. , Miller, L. , Nordgren, H. , Ploner, A. , Sandelin, K. , Shaw, P.M. , Smeds, J. , Skoog, L. , Wedrén, S. , Bergh, J. , 2005. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res.. 7, R953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou, C.M. , Sørlie, T. , Eisen, M.B. , van de Rijn, M. , Jeffrey, S.S. , Rees, C.A. , Pollack, J.R. , Ross, D.T. , Johnsen, H. , Akslen, L.A. , Fluge, O. , Pergamenschikov, A. , Williams, C. , Zhu, S.X. , Lønning, P.E. , Børresen-Dale, A.L. , Brown, P.O. , Botstein, D. , 2000. Molecular portraits of human breast tumours. Nature. 406, 747–752. [DOI] [PubMed] [Google Scholar]
- Polley, M.-Y.C. , Leung, S.C.Y. , McShane, L.M. , Gao, D. , Hugh, J.C. , Mastropasqua, M.G. , Viale, G. , Zabaglo, L.A. , Penault-Llorca, F. , Gown, A.M. , Symmans, W.F. , Piper, T. , Mehl, E. , Enos, R.A. , Hayes, D.F. , Dowsett, M. , Nielsen, T.O. , 2013. An international Ki67 reproducibility study. JNCI J. Natl. Cancer Inst. Djt. 306, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyal, F. , Bollet, M.A. , Caly, M. , Gentien, D. , Carpentier, S. , Peyro-Saint-Paul, H. , Pierga, J.-Y. , Cottu, P. , Dieras, V. , Sigal-Zafrani, B. , Vincent-Salomon, A. , Sastre-Garau, X. , 2012. Respective prognostic value of genomic grade and histological proliferation markers in early stage (pN0) breast carcinoma. PLoS ONE. 7, e35184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørlie, T. , Tibshirani, R. , Parker, J. , Hastie, T. , Marron, J.S. , Nobel, A. , Deng, S. , Johnsen, H. , Pesich, R. , Geisler, S. , Demeter, J. , Perou, C.M. , Lønning, P.E. , Brown, P.O. , Børresen-Dale, A.-L. , Botstein, D. , 2003. Repeated observation of breast tumor subtypes in independent gene expression data sets. PNAS. 100, 8418–8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiriou, C. , Pusztai, L. , 2009. Gene-expression signatures in breast cancer. N. Engl. J. Med.. 360, 790–800. [DOI] [PubMed] [Google Scholar]
- Sotiriou, C. , Wirapati, P. , Loi, S. , Harris, A. , Fox, S. , Smeds, J. , Nordgren, H. , Farmer, P. , Praz, V. , Haibe-Kains, B. , Desmedt, C. , Larsimont, D. , Cardoso, F. , Peterse, H. , Nuyten, D. , Buyse, M. , Van de Vijver, M.J. , Bergh, J. , Piccart, M. , Delorenzi, M. , 2006. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J. Natl. Cancer Inst.. 98, 262–272. [DOI] [PubMed] [Google Scholar]
- Stuart-Harris, R. , Caldas, C. , Pinder, S.E. , Pharoah, P. , 2008. Proliferation markers and survival in early breast cancer: a systematic review and meta-analysis of 85 studies in 32,825 patients. Breast. 17, 323–334. [DOI] [PubMed] [Google Scholar]
- Tang, P. , Skinner, K.A. , Hicks, D.G. , 2009. Molecular classification of breast carcinomas by immunohistochemical analysis: are we ready?. Diagn. Mol. Pathol.. 18, 125–132. [DOI] [PubMed] [Google Scholar]
- Tobin, N.P. , Lundgren, K.L. , Conway, C. , Anagnostaki, L. , Costello, S. , Landberg, G. , 2012. Automated image analysis of cyclin D1 protein expression in invasive lobular breast carcinoma provides independent prognostic information. Hum. Pathol.. 43, 2053–2061. [DOI] [PubMed] [Google Scholar]
- Toyoda, A. , Yokota, A. , Saito, T. , Kawana, H. , Higashi, M. , Suzuki, Y. , Tanaka, T. , Kitagawa, M. , Harigaya, K. , 2011. Overexpression of human ortholog of mammalian enabled (hMena) is associated with the expression of mutant p53 protein in human breast cancers. Int. J. Oncol.. 38, 89–96. [PubMed] [Google Scholar]
- Van't Veer, L.J. , Dai, H. , van de Vijver, M.J. , He, Y.D. , Hart, A.A.M. , Mao, M. , Peterse, H.L. , van der Kooy, K. , Marton, M.J. , Witteveen, A.T. , Schreiber, G.J. , Kerkhoven, R.M. , Roberts, C. , Linsley, P.S. , Bernards, R. , Friend, S.H. , 2002. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 415, 530–536. [DOI] [PubMed] [Google Scholar]
- Van de Vijver, M.J. , He, Y.D. , van't Veer, L.J. , Dai, H. , Hart, A.A.M. , Voskuil, D.W. , Schreiber, G.J. , Peterse, J.L. , Roberts, C. , Marton, M.J. , Parrish, M. , Atsma, D. , Witteveen, A. , Glas, A. , Delahaye, L. , van der Velde, T. , Bartelink, H. , Rodenhuis, S. , Rutgers, E.T. , Friend, S.H. , Bernards, R. , 2002. A gene-expression signature as a predictor of survival in breast Cancer. New Engl. J. Med.. 347, 1999–2009. [DOI] [PubMed] [Google Scholar]
- Varga, Z. , Diebold, J. , Dommann-Scherrer, C. , Frick, H. , Kaup, D. , Noske, A. , Obermann, E. , Ohlschlegel, C. , Padberg, B. , Rakozy, C. , Sancho Oliver, S. , Schobinger-Clement, S. , Schreiber-Facklam, H. , Singer, G. , Tapia, C. , Wagner, U. , Mastropasqua, M.G. , Viale, G. , Lehr, H.-A. , 2012. How reliable is Ki-67 immunohistochemistry in grade 2 breast carcinomas? A QA study of the Swiss Working Group of Breast- and Gynecopathologists. PLoS ONE. 7, e37379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga, Z. , Sinn, P. , Fritzsche, F. , von Hochstetter, A. , Noske, A. , Schraml, P. , Tausch, C. , Trojan, A. , Moch, H. , 2013. Comparison of EndoPredict and oncotype DX test results in hormone receptor positive invasive breast Cancer. PLoS ONE. 8, e58483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vittinghoff, D.V.G. , Shiboski Eric, Stephen C. , 2009. By Eric Vittinghoff – Regression Methods in Biostatistics first ed. Springer-Verlag; New York, LLC: [Google Scholar]
- Weigelt, B. , Mackay, A. , A'hern, R. , Natrajan, R. , Tan, D.S.P. , Dowsett, M. , Ashworth, A. , Reis-Filho, J.S. , 2010. Breast cancer molecular profiling with single sample predictors: a retrospective analysis. Lancet Oncol.. 11, 339–349. [DOI] [PubMed] [Google Scholar]
- Wirapati, P. , Sotiriou, C. , Kunkel, S. , Farmer, P. , Pradervand, S. , Haibe-Kains, B. , Desmedt, C. , Ignatiadis, M. , Sengstag, T. , Schutz, F. , 2008. Meta-analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res.. 10, R65 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following are the supplementary data related to this article:
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary Figure S1Exclusion criteria and treatment information for the Uppsala and Stockholm cohorts.
Supplementary Figure S2Kaplan–Meier analysis of Ki67 and gene expression signatures in the Stockholm dataset.


