Abstract
β‐catenin is a key signal transducer in the canonical WNT pathway and is negatively regulated by ubiquitin‐dependent proteolysis. Through screening of various deubiquitinating enzymes (DUBs), we identified ubiquitin specific protease 4 (USP4) as a candidate for β‐catenin‐specific DUB. The effects of USP4 overexpression or knockdown suggested that USP4 positively controls the stability of β‐catenin and enhances β‐catenin‐regulated transcription. Domain mapping results revealed that the C‐terminal catalytic domain is responsible for β‐catenin binding and nuclear transport. Examination of colon cancer tissues from patients revealed a correlation between elevated expression levels of USP4 and β‐catenin. Consistent with this correlation, USP4 knockdown in HCT116, a colon cancer cell line, reduced invasion and migration activity. These observations indicate that USP4 acts as a positive regulator of the WNT/β‐catenin pathway by deubiquitination and facilitates nuclear localization of β‐catenin. Therefore, we propose that USP4 is a potential target for anti‐cancer therapeutics.
Keywords: Ubiquitin specific protease 4 (USP4), β-catenin, Cancer
Abbreviations
- WNT
wingless-type
- Ub
ubiquitin
- DUB
deubiquitinating enzyme
- USP
ubiquitin-specific protease
- CK1α
casein kinase 1α
- APC
adenomatous polyposis coli
- GSK3β
glycogen synthase kinase 3β
1. Introduction
WNT signaling pathway is involved in many important cellular events including cell movement, cell maintenance, and control of cell polarity during embryonic development (Clevers, 2006; Komiya and Habas, 2008). A recent report also showed that WNT pathway plays an important role in self‐renewal of embryonic stem cells (Holland et al., 2013). The canonical WNT signaling pathway is composed of several key components: WNT, an extracellular signaling protein; Frizzled (Fz) and Low‐density lipoprotein receptor‐related protein 5 or 6 (LRP5/6), cell surface receptors; and β‐catenin, a signal transducing transcriptional activator. In the absence of WNT stimulation, cytosolic β‐catenin is maintained at low levels due to ubiquitin (Ub)‐dependent proteasomal degradation, which is mediated by casein kinase 1α (CK1α) and a destruction complex composed of adenomatous polyposis coli (APC), Axin, and glycogen synthase kinase 3β (GSK3β). In this complex, β‐catenin is phosphorylated at S45 by CKIα, and subsequently by GSK3β at residues S33, S37, and T41. Phosphorylated β‐catenin is recognized by the SCFβ‐TRCP (Skp1‐Cullin1‐F box) complex, an Ub ligase (E3) which links ubiquitin to K19 and K49 of β‐catenin. The destruction complex promotes the proteasomal degradation of polyubiquitinated β‐catenin. Consequently, WNT signaling is negatively regulated by the ubiquitin‐proteasome system (UPS).
The binding of WNT ligands (WNT1 and WNT3a) to their receptors disrupts the function of the destruction complex, which enables β‐catenin to translocate and accumulate in the nucleus. β‐Catenin works in concert with TCF/LEF‐1 (T cell specific transcription factor/Lymphoid enhancer‐binding factor 1) to turn on the expression of target genes such as c‐Myc, Cyclin D1, and Axin. WNT signaling is elaborately regulated at multiple levels by various intra‐ and extracellular signals. Misregulation of the WNT/β‐catenin signaling pathway is correlated with various diseases. In particular, disruption of WNT/β‐catenin signaling causes unregulated cell proliferation and cancer. Therefore, β‐catenin is considered as a target for novel cancer therapeutics (Watanabe and Dai, 2011; Anastas and Moon, 2013). The molecular mechanisms leading to misregulation of WNT signaling and their relevance to cancer pathogenesis have been well characterized in colon cancers (Burgess et al., 2011). Mutations in APC, the most common cause of colon cancers, abolishes Axin binding and ubiquitination of β‐catenin. Mutations in Axin and β‐catenin were also found in colon cancers; though they are not as frequent as APC mutations (Liu et al., 2000; Morin et al., 1997). It is well established that β‐catenin is associated with cancer progression, but it is required for the development of anti‐cancer therapies that target oncogenic β‐catenin to unravel the delicate mechanism by which its activity and stability are regulated.
Cellular stability of proteins is negatively controlled by ubiquitination. K48‐linked ubiquitination which is mediated by Ub‐activating enzyme (E1), Ub‐conjugating enzyme (E2), and Ub‐ligases (E3) directs target proteins to the proteasome for degradation. The removal of Ub‐chains by deubiquitinating enzymes (DUBs), on the other hand, increases the stability of the protein. The WNT/β‐catenin signaling pathway is known to be modulated at the protein level by various DUBs. UBPY/USP8 positively regulates the pathway by modifying Frizzled (Mukai et al., 2010), whereas USP34 deubiquitinates Axin and consequently works as a negative regulator of the WNT pathway (Lui et al., 2011). CYLD, a familial cylindromatosis tumor suppressor, has also been suggested to work as a negative regulator of WNT/β‐catenin signaling through the deubiquitination of Dishevelled proteins (Dvl) (Tauriello et al., 2010). Ub‐dependent proteasomal degradation of β‐catenin is thought to be the most critical control of canonical WNT pathway. Therefore, one would expect that there might be a specific deubiquitination mechanism for β‐catenin. In other prospect, DUB of β‐catenin could stabilize β‐catenin and help to positively regulate WNT/β‐catenin signaling pathway. At present, FAM/USP9X and ubiquitin carboxyl‐terminal hydrolase isozyme L1 (UCH‐L1) are considered as candidate DUBs for β‐catenin (Taya et al., 1999; Bheda et al., 2009). However, their roles in WNT/β‐catenin signaling pathway and physiological relevance to cancer have not been clearly elucidated. Often the fine control of essential signaling pathways involves multiple regulators as in the case of p53 which is regulated by USP7 and USP10 (Lee and Gu, 2010; Li et al., 2002; Yuan et al., 2010). Therefore, we have hypothesized that an unknown DUB(s) may positively regulate WNT/β‐catenin, and they are physiologically relevant to cell proliferation and cancer progression.
To identify a β‐catenin‐specific DUB, we examined deubiquitination of β‐catenin in the presence of various DUBs. Among them, USP4 was shown to deubiquitinate phosphorylated β‐catenin, and further investigated as a candidate DUB for β‐catenin. In previous reports, USP4 was found to deubiquitinate Ro52 (Wada and Kamitani, 2006), ARF‐binding protein 1 (ARF‐BP1) (Zhang et al., 2011), TNF‐receptor‐associated factor (TRAF) (Xiao et al., 2012), TGF‐beta activated kinase 1 (TAK1) (Fan et al., 2011) and pyruvate dehydrogenase kinase isozyme 1 (PDK1) (Uras et al., 2012), and also to form a complex with the S9/Rpn6 subunit, a molecular clamp of the proteasome (Zhao et al., 2012). It is interesting that USP4 has been proposed to be involved in WNT signaling through the deubiquitination of nemo‐like kinase (NLK) and TCF (Zhao et al., 2009). However, the cellular function of USP4 and its relationship to WNT signaling remains largely unclear.
In this study, we investigated the role of USP4 in modulating WNT signaling as a β‐catenin‐specific DUB, and suggested a regulatory mechanism. Our results show that USP4 works as a positive regulator of the WNT/β‐catenin signaling pathway primarily by enhancing the stability of β‐catenin through deubiquitination and by moderate effect on translocation of β‐catenin to the nucleus. We also found that USP4 levels are elevated in correlation with β‐catenin in colorectal cancers. These data collectively propose that USP4 is a potential oncogene.
2. Materials and methods
2.1. Cell culture and transfection
293T and HCT116 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% heat‐inactivated FBS (Gibco) containing penicillin‐streptomycin (Welgene). Cells were incubated in a humid incubator at 37 °C (95% O2 and 5% CO2) and maintained in a subconfluent state, unless otherwise indicated. Plasmids were transfected into cells using ExGen500 and TurboFect reagents (Fermentas), according to the manufacturer's protocol. After incubation for 48–72 h, the cells were harvested and analyzed by immunoblotting or reporter assay.
2.2. Wnt3a‐CM
Wnt3a‐conditioned media (Wnt3a‐CM) and control‐conditioned media (Control‐CM) were prepared according to ATCC instructions. Briefly, Wnt3a‐producing L cells and control L cells were cultured in 100 mm dishes until confluent, and then all cells were used to seed a T75 flask. Medium was exchanged 24 h later and the medium was collected after an additional 24 h. Both Wnt3a‐CM and Control‐CM were filtered using a 0.45 micron filter and stored −80 °C until required for experiments.
2.3. Plasmids
Full‐length and deletion mutants of USP4 were cloned into pDEST‐CMV6 with the SRT tag (TFIGAIATDT) at the N‐terminus (Lee et al., 2001). The catalytic residue mutant (USP4 C311A) and the deletion mutants, USP4 ΔMT‐1 (lacking 767QPQKKKK773) and USP4 ΔMT‐2 (lacking 413KKKP416 and 767QPQKKKK773), were generated using PCR mutagenesis (Figure 5). The Myc‐β‐catenin expression plasmid, TOP flash and FOP flash reporter plasmids were kindly provided by Prof. Choi CY (Sungkyunkwan University, Korea).
Figure 4.
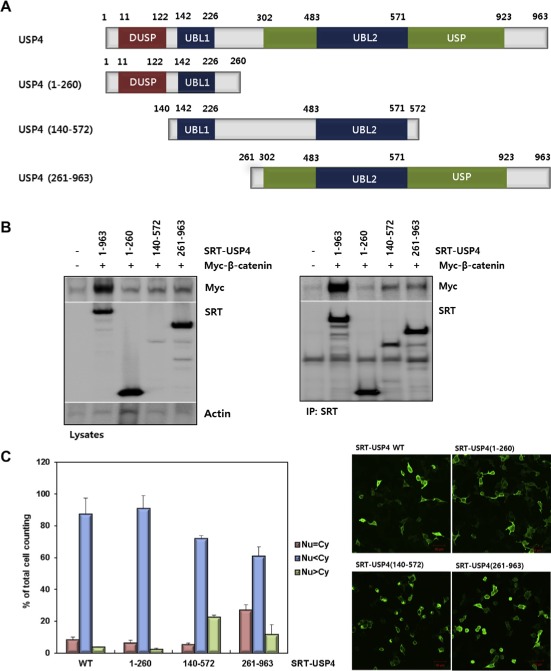
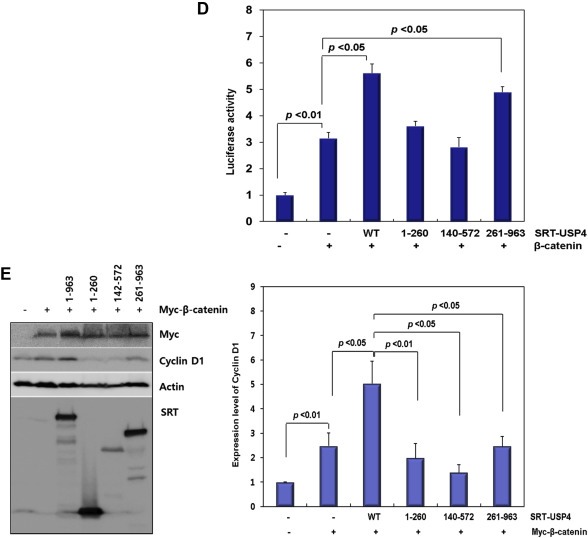
Domain mapping of USP4. (A) The domain structure of USP4 and design of the truncation mutants. Three constructs, USP4(1–260), USP4(140–572), and USP4(261–963), were drawn with residue numbers. The representative domains are also shown (DUSP, domain present in ubiquitin‐specific proteases; UBL, ubiquitin‐like domain; USP, ubiquitin specific proteases domain). (B) Interaction between the truncation mutants of USP4 and β‐catenin in 293T cells. Myc‐β‐catenin was co‐transfected with SRT‐tagged wild‐type (WT) or truncated USP4. Cells lysates were immunoprecipitated with anti‐SRT antibody. USP4 and β‐catenin levels in whole cell lysates and IP eluents were immunoblotted using the indicated antibodies. Actin was used as a loading control. (C) Subcellular localization of USP4 mutants. Each construct was transfected into 293T cells and their localization was quantified by counting the number of cells in three categories as described in Figure 3C (left). Localization of USP4 mutants were also visualized by immunostaining (right). Scale bars: 50 μm. (D) The effect of USP4 mutants on the TOP flash reporter activity. Myc‐β‐catenin was co‐expressed with the wild‐type or each truncation mutant in 293T cells for 48 h. TOP flash luciferase activity was estimated after normalization with pRL‐SV40 activity. (E) The effect of USP4 mutants on the expression of Cyclin D1. Myc‐β‐catenin and Cyclin D1 were immunoblotted with indicated antibodies (left). The band intensity of immunoblots was quantified and displayed as the relative amount to the control (right).
2.4. USP4 knockdown
Small interfering RNAs (siRNAs) were transfected into cells using G‐fectin (Genolution Pharmaceuticals Inc.) or Lipofectamine®RNAiMAX reagent (Invitrogen), according to the manufacturer's protocol. Two USP4 siRNAs were used: siRNA‐1 (5′‐CCAUUUCUGCUUGGCUGUCUCCUUU‐3′, Milojevic et al., 2006) and siRNA‐2 (5′‐CUGCAUAUGCGAAGAACAA‐3′, Xiao et al., 2012).
2.5. Reverse transcription PCR (RT‐PCR) and real time PCR (qPCR)
Total RNA was isolated from cultured cells using Trizol® Reagent (Invitrogen), according to the manufacturer's protocol. Reverse transcription of RNA into cDNA was carried out using 2 μg of RNA incubated for one hour at 42 °C and primed with an oligo (dT) primer (Clontech Laboratories, Inc). Quantitative real‐time PCR (qPCR) was performed using the Thermal Cycler Dice™ Real Time System TAKARA TP800 with SYBR Green PCR master mix (Takara). PCR amplification was performed for 40 cycles of 94 °C for 5 s and 60 °C for 30 s. Relative quantification of mRNA was determined using the ΔΔCt method using TP800 software (Takara) and levels were normalized to GAPDH. The following primers were used in PCR: USP4, 5′‐TACTAAACTGGTACGGCTGTGT‐3′and 5′‐GGGATGTTGAATAGCTTCCGC‐3′(Primer bank, NM_199443); Cyclin D1, 5′‐ATGGAACACCAGCTCCTGTGCTGC‐3′and 5′‐TTCCTCGCAGACCTCCAGCATCC‐3′(NM_053056.2); Axin2, 5′‐CCTGCCACCAAGACCTACAT‐3′, and 5′‐GTTTCCGTGGACCTCACACT‐3′(NM_004655); β‐catenin, 5′‐AAAGCGGCTGTTAGTCACTGG‐3′, and 5′‐CGAGTCATTGCATACTGTCCAT‐3′(Primer bank, NM_001098209); GAPDH, 5′‐CTGGTAAAGTGGATATTGTTGCCAT‐3′, and 5′‐TGGAATCATATTGGAACATGTAAACC‐3′ (NM_002046.4). Each experiment repeated three times independently, unless otherwise mentioned.
2.6. Luciferase reporter assay
293T cells were seeded at 1 × 105 cells per well in 12‐well plates and cultured overnight. The cells were transfected for 24–48 h using transfection reagent (Fermentas) with the indicated plasmids along with the T cell factor (TCF) reporter plasmid (TOP flash) which contains the TCF binding site driving the expression of the firefly luciferase gene. FOP flash containing mutated TCF binding sites and Renilla luciferase reporter vectors were used as controls. The control plasmids were added to maintain equal amounts of total DNA in each transfection. After 48 h post‐transfection, cells were collected for dual specific luciferase reporter assays. Luciferase activity was measured according to the manufacturer's protocol (Promega). To calculate the relative luciferase activity, firefly luciferase activity was divided by Renilla luciferase activity.
2.7. Immunoblot analysis and immunoprecipitation
Cells were cultured, harvested in ice‐cold PBS, and solubilized in RIPA lysis buffer (50 mM Tris, 50 mM NaCl, 0.5% Triton X‐100, and 1 mM EGTA) supplemented with protease inhibitors (1 mM PMSF, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin, and 1 mM DTT). Cell debris was removed by centrifugation at 13,000 rpm for 10 min and the clarified samples were collected. Samples were mixed with 5× sample buffer (187.5 mM Tris–HCl, pH 6.5, 6% SDS, 30% glycerol, 150 mM DTT, and 0.03% bromophenol blue) and boiled for 10 min. The protein concentration was estimated using the Bradford method (Bio‐Rad). The samples were separated on 8 or 10% SDS‐PAGE gels and transferred onto PVDF membranes (GE Healthcare Life Science). The membranes were washed with 0.05% Tween 20 in TBS (140 mM NaCl, and 20 mM Tris, pH 7.4) and blocked with 5% skim milk. Immunoblotting was performed with specific antibodies according to the manufacturer's instructions. Immunoblotting antibodies used for β‐catenin, USP4, ubiquitin, and β‐actin detection were purchased from Santa Cruz, Cyclin D1 from Abcam, HA from R&D system, and GAPDH from Cell Signaling. The antibody against the SRT‐tag was made in the animal facility at Sungkyunkwan University School of Medicine (Korea). To observe the linkage type of Ub, immunoblotting was performed using the chain‐specific ubiquitin antibodies which detect K48‐ (Abcam) and K63‐ (Enzo Life Sciences) linked ubiquitins, respectively. Immunoblots were quantified by densitometric analyses with normalization against a loading control. For immunoprecipitation, USP4 antibody was purchased from Bethyl Laboratories. The immunoblotted membranes probed with the HRP‐conjugated antibody were developed using ECL solution (ELPIS‐Biotech) and exposed to X‐ray film or analyzed with the luminescent image analyzer, LAS‐3000 (Fujifilm). For immunoblotting of the ubiquitinated proteins, harvested cells were boiled for 10 min with denaturation buffer (1% SDS, 5 mM EDTA, 10 mM DTT, 15 U/ml DNase I) and mixed with the cell lysis buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X‐100, 2.5 mM sodium pyrophosphate, 1 mM β‐glycerophosphate, 1 mM Na3VO4, protease inhibitor cocktail, Sigma P8340). The cell lysates were incubated overnight with Agarose A beads (GE Healthcare Life Science) with Ub‐antibodies. Beads were washed with cell lysis buffer and boiled with 3× sample buffer for SDS analysis.
2.8. Preparation of subcellular fractions
For the separation of the nuclear and cytoplasmic fractions, cells were scraped, washed with cold PBS and resuspended in a hypotonic buffer (50 mM HEPES, pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM Na3VO4, 50 mM NaF, 1 mM PMSF, and protease inhibitors cocktail) on ice for 20 min. The cytosolic fraction was obtained by centrifugation at 800 × g for 5 min. For acquiring the nuclear fractions, the pellets were washed with the hypotonic buffer and resuspended in the hypotonic buffer containing 1% Triton X‐100 and 150 mM NaCl. After ice incubation for 20 min, the mixture was sheared by passing 10 times through a 26‐gauge needle and centrifuged at 100,000 × g for 20 min. The supernatants were used for nuclear fraction. All fractionated samples were boiled with 5× sample buffer for 10 min, and then loaded on 8% SDS‐PAGE for immunoblotting. Lamin B (Santa Cruz, M‐20) and α‐tubulin (Santa Cruz, 10D8) were used as nuclear and cytosolic markers, respectively.
2.9. Invasion assay
HCT116 cells were seeded into 6‐well plates in DMEM with 10% FBS. On the following day, cells were transfected with siRNA and cultured for 24 h before being used in the invasion assay. The invasion assay was performed in a 24‐well invasion chamber with an 8 μm pore size polycarbonate membrane, according to the manufacturer's instruction (Millipore). Chambers were rehydrated for 2 h at room temperature. Then, 5 × 105 cells in 300 μl serum‐free media were placed into the upper chamber while the lower chamber was filled with 500 μl media containing 10% FBS. After incubation at 37 °C for 48 h, non‐invading cells present on the membrane surface were removed and cells on the lower surface of the membrane were stained. The stained cells were counted by microscopic observation.
2.10. Migration assay
HCT116 cells were transfected with USP4 siRNA for 24 h and starved in serum‐free media for 24 h before being used in the migration assay. The migration assay was performed for 20 h according to the manufacturer's instruction (Millipore). After non‐migrated cells were removed, cells that had migrated were stained. The stained cells were counted by microscopic observation.
2.11. Immunostaining
293T cells were seeded on glass coverslips in 12‐well plates and transfected with the corresponding plasmids the next day. After 48 h incubation, the cells were fixed with 3.7% paraformaldehyde for 30 min, permeabilized by 0.1% Triton X‐100 in PBS (T‐PBS) for 15 min, and blocked with 1% BSA in T‐PBS for 30 min. The cells on coverslips were incubated with primary antibodies overnight at 4 °C. After washing with T‐PBS, the cells were incubated with secondary antibodies at RT for 2 h. FITC‐conjugated anti‐mouse antibody and Alexa Fluor® 564‐conjugated anti‐rabbit antibody (Invitrogen) were used for immunodetection. The cells were washed again and mounted with Vectashield medium with DAPI (Vector Lab). Immunostained cells were analyzed by a fluorescence microscope (ZEISS).
2.12. Preparation of tissue samples and RT‐PCR
Human samples were obtained from the Chonnam National University Hwasun Hospital (Korea) and the use of these specimens was authorized by the Institutional Review Board of the Catholic University of Korea, College of Medicine (MC13SISI0140). Consent was obtained from the patients to use the tissue specimens for research purposes. Freshly frozen colorectal adenocarcinomas derived from primary resection specimens were prepared to analyze the expression level of USP4. Corresponding normal colonic tissues derived from the same patients were also prepared as paired samples. Total RNA was isolated from the 30 paired tissues (normal and tumor) using Trizol (Invitrogen). cDNA was synthesized using SuperScript III reverse transcriptase (Invitrogen) and RT‐qPCR was performed using power SYBR®Green PCR Master Mix (Applied Biosystems, USA). The primers used in this study were as follows: USP4, 5′‐CCTGGGCTCTGTGGACTTG‐3′ and 5′‐TGTTGATTTCGGCTTCATACTC‐3′; β‐catenin, 5′‐CTTGGACTGAGACTGCTGATCTTG‐3′ and 5′‐CACCAGAGTGAAAAGAACGATAGCTA‐3′; Cyclin D1, 5′‐CCGTCCATGCGGAAGATC‐3′ and 5′‐ATGGCCAGCGGGAAGAC‐3′; GAPDH; 5′‐CTGGTAAAGTGGATATTGTTGCCAT‐3′, and 5′‐TGGAATCATATTGGAACATGTAAACC‐3′. The quantity of each mRNA was normalized using a GAPDH mRNA standard and the data are represented as fold change between normal and tumor samples. Chi‐square test was used for calculating the p‐value.
2.13. Immunohistochemistry
Expression of the indicated proteins in colon cancers was evaluated by immunohistochemistry using tissue microarray slides from Super Bio Chips (Korea) that contained evaluable specimens derived from 40 primary invasive colon cancers, 10 distant metastases, and 10 normal adjacent‐to‐cancer samples. For detection of the indicated proteins, the Ventana BenchMark XT's UltraView DAB kit was used to detect the target antigens in tissue using a chromogenic substrate (DAB). Primary antibodies against anti‐β‐catenin (1:800) and USP4 (1:50) were purchased from Santa Cruz. The scoring criteria for immunohistochemistry were as follow; negative, no staining or <5% tumor cells with staining; weak positive, <30% tumor cells with faint staining; strong positive, >30% tumor cells with strong staining.
2.14. Statistical analysis
Results were normalized by control value and represented as mean ± standard error of mean. All experiments were repeated at least three times, unless otherwise mentioned. Statistical comparisons between groups were determined by Student's t‐test. If p‐values are less than 0.05, they are considered to be statistically significant and indicated in the graph.
3. Results
3.1. USP4 positively controls the stability of β‐catenin by deubiquitination
The initial screening was performed for DUBs that have strong deubiquitination activity in 293T cells. The amount of total ubiquitinated proteins was monitored by immunoblotting after transfection of DUB‐encoding plasmids (Supplementary Figure 1A). Among the tested DUBs, USP3, 4, 7, 11, and 18 showed high DUB activity, as seen from the decreased HA‐Ub bands (Supplementary Figure 1A). Subsequently they were subjected to DUB assay using phospho‐β‐catenin as a substrate. We first confirmed the proteasomal degradation of phospho‐β‐catenin by examining changes in the levels of ubiquitin‐conjugated phospho‐β‐catenin and total β‐catenin by the treatment of MG‐132, a proteasome inhibitor (Supplementary Figure 1B). Then, the level of ubiquitinated phospho‐β‐catenin was examined in the whole cell lysates of DUB‐expressing cells after MG132 treatment (Supplementary Figure 1C). In USP4‐transfected cells, the high molecular weight bands corresponding to polyubiquitinated phospho‐β‐catenin were significantly diminished, suggesting that USP4 has DUB activity for β‐catenin. We further investigated the physical interaction between β‐catenin and USP4 through co‐immunoprecipitation. The interaction of endogenous β‐catenin and USP4 was confirmed in 293T and HCT116 cells, a colon cancer cell line harboring stable β‐catenin that has a mutation at S45 phosphorylation site (Figure 1A). Also the transfected SRT‐tagged USP4 was shown to interact with both endogenous and transfected exogenous β‐catenin (Supplementary Figure 1D).
Figure 1.
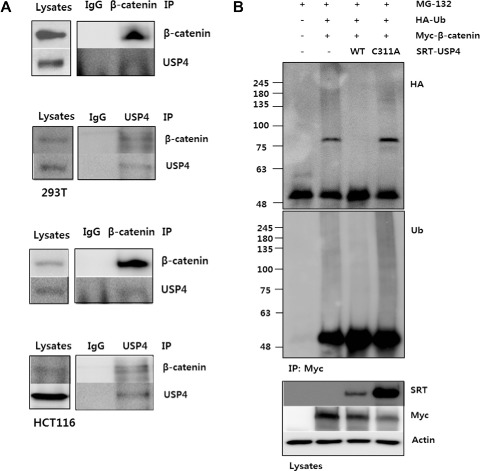
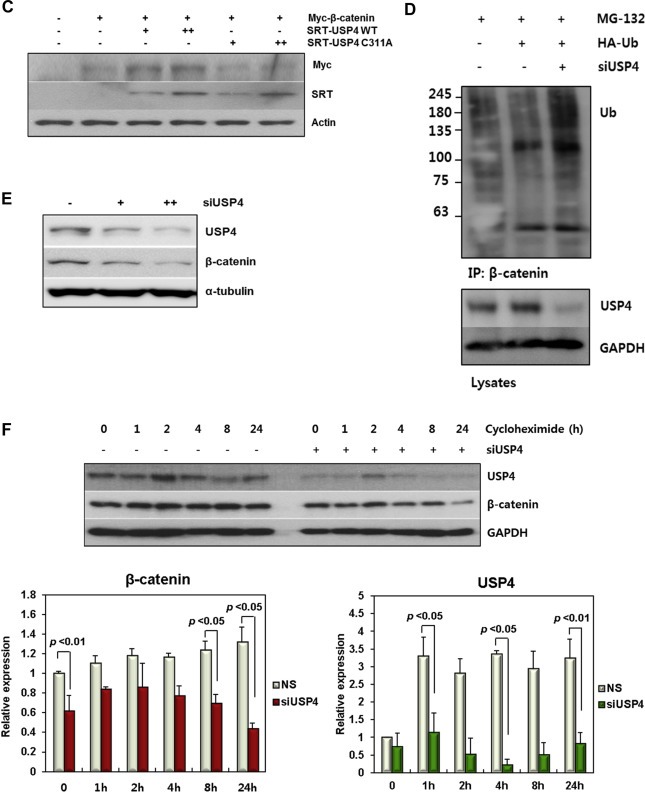
USP4 binds β‐catenin controls its stability. (A) Interaction between USP4 and β‐catenin in 293T and HCT116 cells. Four hours prior to harvest, 10 μM MG132 was added to inhibit proteasomal degradation. The cells lysates were immunoprecipitated using anti‐β‐catenin and anti‐USP4 antibodies, respectively. The cell lysates and IP eluents were immunoblotted using anti‐β‐catenin and anti‐SRT antibodies. IgG was used for control. (B) DUB activity of USP4 and its active‐site mutant C311A on Ub‐β‐catenin. 293T cells transfected with indicated DNAs were culture for 24 h. MG132 (10 μM) was added 4 h before harvest. The cell lysates was immunoprecipitated with anti‐Myc antibody under denaturing condition. The ubiquitination level of immunoprecipitants was measured by immunoblotting using anti‐HA and anti‐Ub antibodies, respectively. (C) The effect of wild type and C311A USP4 on β‐catenin stability. Wild‐type or C311A SRT‐USP4 (+: 0.2 μg, ++: 0.5 μg) was co‐transfected with Myc‐β‐catenin (0.5 μg) into 293T cells. After 48 h the levels of Myc‐β‐catenin and SRT‐USP4 were evaluated by immunoblotting. (D) The effect of USP4 siRNA (siUSP4) on ubiquitination of β‐catenin. 293T cells, transfected with siUSP4 and HA‐Ub were treated with 10 μM MG132 for 4 h prior to harvest. The harvested cells were denatured by SDS and immunoprecipitated using anti‐β‐catenin antibody. The ubiquitinated β‐catenin was detected by SDS‐PAGE and immunoblotting. USP4 level in whole cell lysates was also examined by immunoblotting. (E) The effect of siUSP4 on β‐catenin stability. USP4 was knocked down using siUSP4 (+: 20 pM, ++: 50 pM) in 293T cells. After 48 h, USP4 and β‐catenin levels were analyzed by immunoblotting. α‐tubulin was used as a loading control. (F) The effect of cycloheximide and USP4 on β‐catenin stability. 293T cells were transfected with 60 pM siUSP4 and after 48 h treated with 100 μg/ml cycloheximide for the indicated times. Scrambled siRNA‐transfected cells were used as a control (NS). Immunoblots of endogenous USP4 and β‐catenin are shown. GAPDH was used as a loading control. The bar graph shows the band intensities of USP4 and β‐catenin. Each sample were normalized relative to the loading control.
USP4 is a cysteine protease, and C311 has been identified as the catalytic cysteine by bioinformatic analyses. Therefore, a USP4 mutant bearing a Cys‐to‐Ala substitution at 311 (C311A) was generated as a catalytically defective mutant for comparison with wild‐type. To verify the DUB activity of USP4 on β‐catenin and confirm the role of C311, the ubiquitination level of β‐catenin was compared by anti‐ubiquitin antibody in cells transfected with wild‐type and mutant USP4 under denaturing (Figure 1B) and non‐denaturing conditions (Supplementary Figure 1E). The high molecular‐weight β‐catenin bands corresponding to ubiquitinated forms clearly diminished in cells transfected with SRT‐USP4. However, these bands were virtually unaffected by C311A USP4, suggesting that C311 is the key catalytic residue for the deubiquitination of β‐catenin. C311A mutant was shown to retain the β‐catenin binding activity (Supplementary Figure 1F). These results suggest that USP4 mediates the deubiquitination of β‐catenin and C311 is essential as a catalytic residue. To further characterize the chain‐specificity of DUB activity of USP4,the immunoprecipitated proteins were immunoblotted using the antibody specific to K48‐linked Ub chains (Supplementary Figure 1G). USP4‐catalyzed removal of K48‐linked Ub from β‐catenin was confirmed in both denaturation and non‐denaturation conditions.
Since K48‐linked poly‐Ub chains are responsible for Ub‐dependent proteasomal degradation, we expected that USP4 contributes to the control of β‐catenin stability and cellular abundance. To investigate the function of USP4 relevant to the stability of β‐catenin, USP4 and β‐catenin expression plasmids were co‐transfected into 293T cells and the β‐catenin levels were examined (Figure 1C). Co‐transfection with USP4 increased the amount of β‐catenin in a dose‐dependent manner, but the C311A mutant did not change the β‐catenin level. However, the mRNA level of β‐catenin was not substantially altered by USP4 expression (Supplementary Figure 1H). These results indicate that β‐catenin level is not transcriptionally regulated by USP4, but its stability is controlled in protein level by the catalytic activity of USP4.
To corroborate its stabilizing effect on β‐catenin, USP4 expression was knocked down in HCT116 cells using two USP4‐targeting siRNAs (siUSP4‐1 and siUSP4‐2). Both siRNAs were able to reduce USP4 protein and mRNA levels (Supplementary Figure 1I). siUSP4‐1 was more effective, so used in subsequent experiments to suppress USP4 expression, unless otherwise specified. When USP4 was reduced by siUSP4, ubiquitinated forms of β‐catenin were increased under denaturing (Figure 1D) and non‐denaturing conditions (Supplementary Figure 1J). Similar results were observed when the gels were immonoblotted using antibody specific to K48‐linked ubiquitin (Supplementary Figure 1K). Conclusively, siUSP4 reduced the stability of β‐catenin in a dose‐dependent manner by siUSP4 (Figure 1E). These results further confirmed that USP4 controls the stability of β‐catenin by deubiquitination.
The effect of USP4 on the stability of β‐catenin was further tested by treating transfected cells with cycloheximide to block protein synthesis (Figure 1F). β‐catenin levels decreased over time in cycloheximide‐treated cells, and the cellular degradation of β‐catenin was noticeably faster when USP4 was knocked down (Figure 1F). These results confirmed that USP4 is important for regulating the cellular stability of β‐catenin.
3.2. USP4 enhances the transcriptional activity of β‐catenin
In canonical WNT signaling, the transcriptional activity of TCF and expression of its target genes are promoted by the binding of β‐catenin to TCF where it functions as a transcriptional co‐activator. To evaluate the role of USP4 in β‐catenin activity, we examined the transcriptional activity of TCF/β‐catenin using the TOP flash reporter assay in USP4‐transfected 293T cells. Co‐expression of SRT‐USP4 with Myc‐β‐catenin increased reporter activity about 2‐fold compared to expression of Myc‐β‐catenin alone, but SRT‐C311A‐USP4 expression resulted in much less of an effect (Figure 2A and Supplementary Figure 2A). On the other hand, TOP flash activity diminished when siUSP4 was transfected (Figure 2B and Supplementary Figure 2B). A similar effect of siUSP4 was also observed when Myc‐β‐catenin was overexpressed (Figure 2B and Supplementary Figure 2B).
Figure 1.
(continued).
Accordingly, USP4 significantly enhanced the expression of the WNT/β‐catenin downstream target genes, Cyclin D1 and Axin2 (Figure 2C). USP4 knockdown also caused substantial and dose‐dependent decreases in the mRNA levels of the downstream target genes of WNT/β‐catenin (Figure 2D and Supplementary Figure 2C). These results confirm that USP4 knockdown suppresses the transcriptional activity of β‐catenin, thereby supporting the suggestion that USP4 functions as a positive regulator of the WNT/β‐catenin pathway.
To determine whether regulation of β‐catenin by USP4 is responsive to a WNT signal, luciferase activity was assayed in the presence of Wnt3a conditioned media (Wnt3a‐CM) (Figure 2E and Supplementary Figure 2D). As is well known, Wnt3a‐CM treated cells showed higher luciferase activity than non‐treated cells since the WNT signal enhances the activity of β‐catenin. The stimulating effect of the WNT signal was also severely affected by USP4 knockdown (Figure 2E). Specifically, reporter activity decreased about 3‐fold in USP4 knockdown cells when the assay was performed in the presence of Wnt3a‐CM.
3.3. USP4 also affects β‐catenin activity by localizing β‐catenin to the nucleus
To investigate the action mechanism of USP4, further analysis was carried out by transfecting 293T cells with Myc‐S33A‐β‐catenin. This mutant, lacking the GSK3β phosphorylation site, is not susceptible to ubiquitination and subsequent proteasomal degradation, and thus it exists in a stable form in cells. Accordingly, Myc‐S33A‐β‐catenin stimulated reporter activity to a greater extent than did wild‐type β‐catenin (Figure 2F vs. Figure 2A). Interestingly, the effect of Myc‐S33A‐β‐catenin was reduced by USP4 knockdown (Figure 2F), while the stability of the mutant β‐catenin was maintained (Supplementary Figure 2E). The similar results were also observed in HCT116 cells; siUSP4 transfection in HCT116 cells reduced the reporter activity in a dose dependent manner (Supplementary Figure 2F). This observation raised the possibility that USP4‐dependent activation of β‐catenin is not only caused by the catalytic activity of USP4 but also mediated by another mechanism such as a nuclear transport of β‐catenin.
It is known that β‐catenin is transported into the nucleus as a complex with other transporters such as Rac1 (Wu et al., 2008) or directly binds to the nuclear pore complex using its armadillo repeats (Sharma et al., 2012) since it has no NLS sequence (Fagotto et al., 1998). Therefore, we hypothesized that USP4 shuttles to the nucleus and has a role in transporting β‐catenin to the nucleus. A model involving shuttling of USP4 to the nucleus was also supported by mass spectrometric analysis which revealed associations of USP4 with importin as well as with exportin (Supplementary Table 1).
Initially, we investigated the localization of USP4 using immunofluorescence and immunoblotting before and after transfecting SRT‐USP4 in 293T cells (Figure 3 and Supplementary Figure 3). As reported previously (Soboleva et al., 2005), USP4 was present in both the nucleus and cytoplasm, however we observed that USP4 was more prominent in the cytoplasm (Figure 3A–C). Subsequently, we studied the effect of USP4 on the localization of β‐catenin by transfection of USP4. The results of both immunofluorescence and immunoblotting revealed that SRT‐USP4 affects the localization of β‐catenin by increasing the amount of β‐catenin in the nucleus upon USP4 expression (Figure 3A and B). In contrast, when USP4 level was decreased by siUSP4, its effect on the nuclear localization of β‐catenin was less significant (Figure 3D and E). These results consistently revealed that USP4 affects the localization of β‐catenin to the nucleus (Figure 3A and Supplementary Figure 3A). Their nuclear transport and localization was more evident when cells were treated with Leptomycin B (LMB), an inhibitor of CRM1‐dependent nuclear export (Supplementary Figure 3B). In this condition, the nuclear localization of both USP4 and β‐catenin were increased by LMB.
Figure 2.
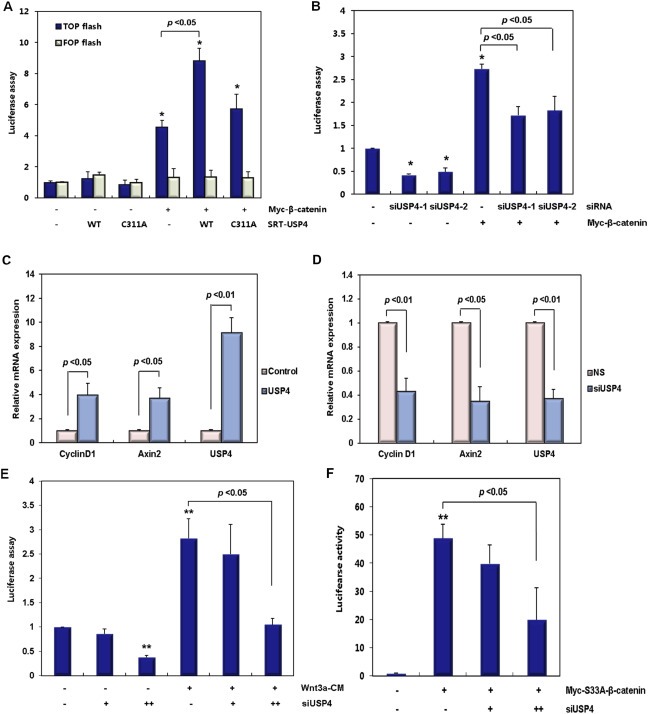
USP4 controls the activity of β‐catenin. (A) The effect of USP4 and its mutant on the TOP flash reporter activity. 293T cells were co‐transfected with SRT‐USP4 (wild type and C311A) and Myc‐β‐catenin expression plasmids, along with TOP flash reporter plasmid which has a β‐catenin‐induced TCF reporter. FOP flash activity was also examined to determine the specific effects on TOP flash activity in 293T cells. (B) The effect of siUSP4 on the TOP flash reporter activity. USP4 was knocked down with 20 pM of USP4 siRNAs (siUSP4‐1 and siUSP4‐2) and TOP flash activity was measured. The cells were then transfected with a Myc‐β‐catenin expression plasmid or the parental vector. (C) The effect of USP4 and its mutant on the expression of β‐catenin target gene. The mRNA level of β‐catenin target genes was examined in 293T cells. Twenty four hours after the transfection with SRT‐USP4, the mRNA levels of Cyclin D1, Axin2, and USP4 were quantified by real time‐PCR and normalized to GAPDH. (D) The effect of siUSP4 on the expression of β‐catenin target genes. 293T cells were transfected with 70 pM siUSP4 for 48 h and the levels of indicted mRNAs was quantified by real time‐PCR with normalization to GAPDH. (E) The effect of siUSP4 on the transactivation activity of β‐catenin in the presence of WNT signal. siUSP4 (+: 10 pM, ++: 30 pM) was transfected into 293T cells along with the TOP flash reporter. After 24 h, the culture media was exchanged with control‐CM or Wnt3a‐CM. The cells were incubated for another 24 h prior to measurement of reporter activity. (F) The effect of siUSP4 on the transactivation activity of S33A β‐catenin. 293T cells were transfected with siUSP4 (+: 40 pM, ++: 90 pM). After 24 h, the Myc‐S33A‐β‐catenin expression plasmid was transfected along with the TOP flash reporter plasmid. After 48 h of incubation, the reporter assay was performed. In each graph, the results were normalized to either empty vectors or siRNA negative controls. The statistical significance of the normalized values was indicated by “*” (p < 0.05) and “**” (p < 0.01). When the data is statistically significant, their p‐values are indicated in the graphs.
3.4. Domain mapping revealed that USP4(261–963) is responsible for USP4 activity
USP4 contains four distinctive domains: a domain present in ubiquitin‐specific proteases (DUSP), two ubiquitin‐like domains (UBL1 and UBL2, respectively) and an ubiquitin specific proteases domain (USP). In order to identify the domains responsible for the interaction and localization of β‐catenin, three deletion mutants were constructed (Figure 4A): USP4(1–260) covers DUSP and UBL1, USP4(140–572) contains UBL1 and UBL2, and USP4(261–963) harbors USP which overlaps with UBL2 (Figure 4A). The interaction of each mutant with β‐catenin was examined by immunoprecipitation (Figure 4B). The wild type USP4 and USP4(261–963) were associated with Myc‐β‐catenin, while USP4(1–260) showed only weak interaction (Figure 4B). Besides, USP4(261–963) primarily contributed to the transcriptional activity of β‐catenin, since highest luciferase reporter activity and the expression level of target gene, Cyclin D1, were observed in USP4(261–963)‐transfected cells (Figure 4D and E). Although USP4(140–572) still had β‐catenin binding activity, it showed weak effect on transcriptional activation, possibly due to the condensation in nucleus (Figure 4C). Taken these results together, it was concluded that β‐catenin activity is primarily associated with the C‐terminal catalytic domain of USP4. However, the N‐terminus of USP4(1–260) seems to be necessary for full USP4 activity because reporter activity and the target gene expression were not 100% recapitulated by the C‐terminal domain alone (Figure 4D and E).
Figure 3.
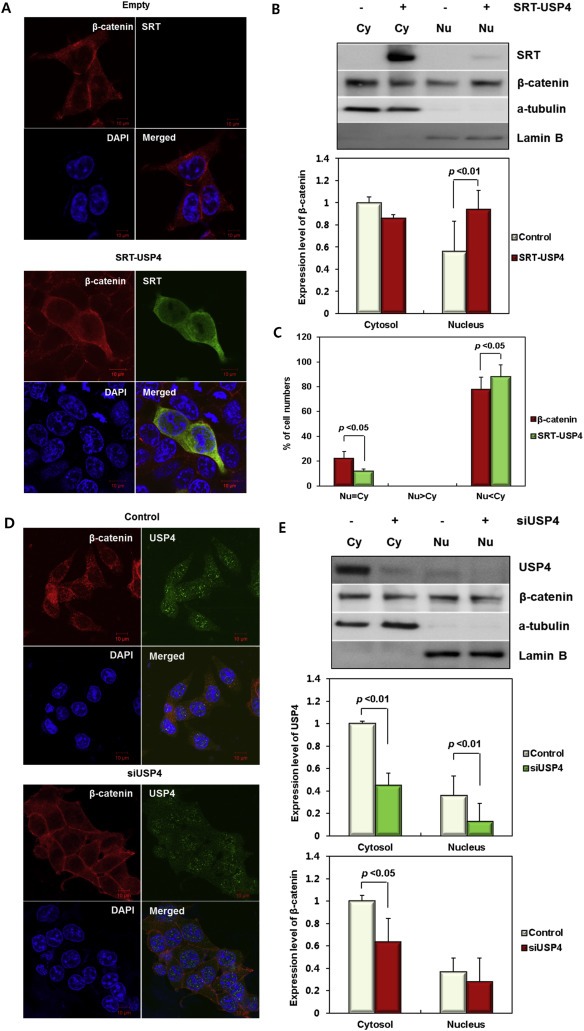
Cellular localization of USP4 and β‐catenin. (A) Subcellular localization of SRT‐USP4 and endogenous β‐catenin in 293T cells. The location of each protein was examined by immunofluorescence using anti‐SRT (green) and anti‐β‐catenin (red) antibodies, respectively. Nuclei (blue) were stained with DAPI. (B) β‐catenin in subcellular fractions. 293T cells were transfected with SRT‐USP4 for 24 h, and then whole cells lysates were fractionated into cytoplasmic (Cy) and nuclear (Nu) fractions. Top: The amount of SRT‐USP4 and endogenous β‐catenin in each fraction was analyzed by immunoblotting. Bottom: The band intensity of the β‐catenin immunoblot was quantified and presented in bar graphs. (C) Subcellular localization of USP4 and β‐catenin. 293T cells were immunostained with anti‐SRT and anti‐β‐catenin antibodies and the number of co‐immunostained cells were counted in the following three categories: Nu < Cy, proteins dominant in the cytoplasmic fraction; Nu > Cy, proteins dominant in the nuclear fraction; Nu = Cy, almost equal distribution in both fractions. The results are displayed in a bar graph with % values. (D) Subcellular localization of endogenous USP4 and β‐catenin in HCT116 cells. The cultured HCT116 cells were transfected with siUSP4, and immunostained using anti‐USP4 (green) and anti‐β‐catenin (red) antibodies. (E) USP4 and β‐catenin in subcellular fractions. The lysates of siUSP4‐transfected HCT116 cells were fractionated. USP4 and β‐catenin in cytoplasmic (Cy) and nuclear (Nu) fractions were immunoblotted (Top) and the band intensities were quantified (Bottom). Scale bar = 10 µm (A, D). The quantified band intensity was normalized to α‐tubulin for cytosolic fraction and Lamin B for nuclear fraction, respectively, and displayed as the relative amount to the proteins in the cytosolic fractions of control (B, E).
We further investigated which domains of USP4 are involved in the cellular localization of USP4 and β‐catenin (Supplementary Figure 3A). USP4(1–260) and β‐catenin were localized mostly in the cytoplasm, while USP4(261–963) had the localization pattern similar to the wild‐type. USP4 mutants and β‐catenin showed the similar localization pattern when the nuclear export was inhibited by LMB (Supplementary Figure 3B). These results suggest the association of the C‐terminus of USP4(261–963) with a subset of β‐catenin in the nucleus as evidenced by the observed binding (Figure 4B).
3.5. Nuclear localization sequences of USP4
From this study, we postulate that the nuclear localization sequences (NLS) present in the C‐terminus of USP4(261–963) control the location of USP4 (Figure 5A), as previously proposed (Soboleva et al., 2005). To further investigate the role of the NLS, we constructed SRT‐USP4 mutants lacking proposed NLS sequences (ΔMT‐1 and ΔMT‐2), as shown in Figure 5A. However, ΔMT‐1 was utilized for the majority of subsequent analyses due to rapid degradation of ΔMT‐2 (Supplementary Figure 4A). Changes in the cellular distribution of β‐catenin in response to the expression of USP4 ΔMT‐1 were examined by immunofluorescence (Figure 5B, top) and quantified by counting the number of cells in which a given subcellular localization of the proteins was seen (Figure 5B, bottom). While β‐catenin and wild‐type SRT‐USP4 were distributed both in the nucleus and in the cytoplasm, mutations in the signal sequences of USP4 clearly hindered nuclear transport, as the cytoplasmic localization of ΔMT‐1 was increased (Figure 5B). Furthermore, a similar cellular redistribution of β‐catenin was seen with ΔMT‐1. To investigate whether USP4 works as a general activator of nuclear localization, USP4 was double‐stained with eIF3a, a cytoplasmic protein which can be accumulated in nucleus by nuclear transport similar to β‐catenin (Saletta et al., 2010). Although USP4 slightly enhanced the expression of eIF3a, it did not increase the nuclear localization of eIF3a (Supplementary Figure 3C), which partially supports that the nuclear localization effect of USP4 is specific to β‐catenin.
Figure 5.
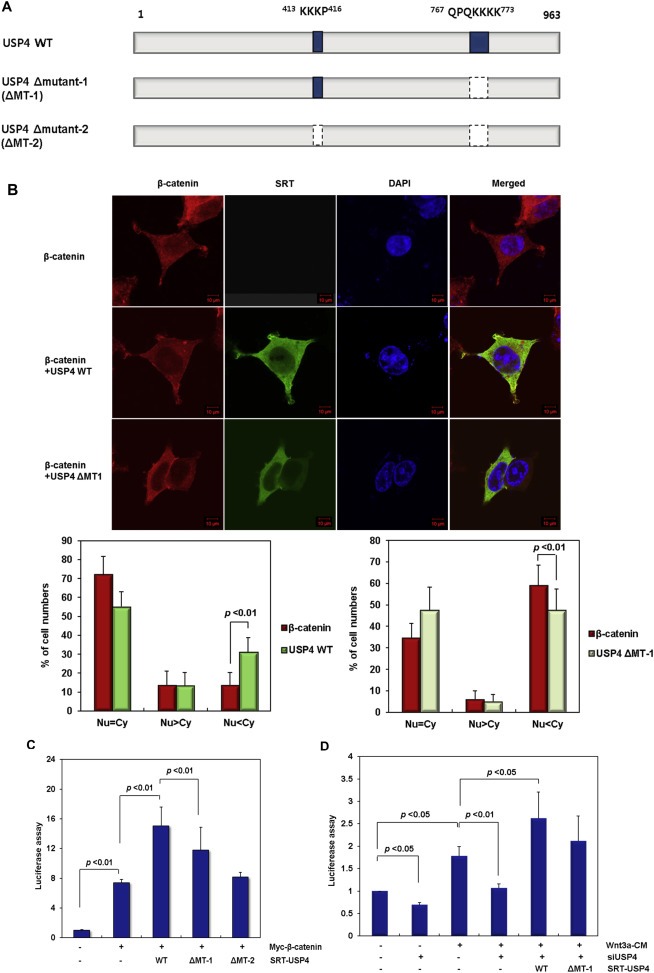
The role of nuclear localization sequence (NLS) in USP4. (A) Construction of the USP4 mutants lacking the NLS sequence (ΔMT‐1 and ΔMT‐2). (B) Subcellular localization of ΔMT‐1. After transfection of wild‐type USP4 or ΔMT‐1 along with Myc‐β‐catenin, the location of each protein was examined by immunofluorescence using anti‐SRT (green) and anti‐β‐catenin (red) antibodies, respectively (Top). Nuclei were stained with DAPI (blue). The cellular localization of the expressed proteins was examined in 100 immunostained cells and their relative abundance is represented by percentage bar graphs (bottom) using the same criteria in Figure 3C. (C) The effect of the NLS deletion mutants on the TOP flash reporter activity in the presence of WNT signal. The plasmids encoding wild‐type USP4, ΔMT‐1 and ΔMT‐2 were transfected into 293T cells along with Myc‐β‐catenin. After 48 h, TOP flash activity was measured. (D) The effect of NLS deletion mutant of USP4 on the TOP flash reporter activity in siUSP4‐treated HCT116 cells. HCT116 cells were transfected with siUSP4 for 6 h, followed by complementation with the wild‐type (WT) or ΔMT‐1 USP4. After 24 h, the cells were cultured in control‐CM or Wnt3a‐CM for an additional 24 h before the reporter assay.
We further investigated whether the cellular distribution of USP4 affects the transcriptional activity of β‐catenin. Reporter activity was lower in ΔMT‐1 expressing cells than in wild‐type USP4 expressing cells. Transcriptional activity of β‐catenin decreased to a greater degree with the expression of ΔMT‐2, possibly due to the degradation of USP4 (Figure 5C and Supplementary Figure 4A). The assay results revealed that transcriptional activity increases in proportion to the amount of USP4 in the nucleus, which suggests that USP4 enhances β‐catenin activity by facilitating its transport from the cytoplasm to the nucleus. To further confirm the role of USP4 in activation of β‐catenin, we measured the transcriptional activity of β‐catenin in HCT116 cells by expressing the wild‐type and ΔMT‐1 USP4 in siUSP4 expressing cells in the presence of a Wnt3a‐CM (Figure 5D and Supplementary Figure 4B). The reporter activity that was suppressed by siUSP4 transfection was significantly restored upon induction of wild‐type USP4. ΔMT‐1 also was able to increase the transcriptional activity of β‐catenin, though these effects were less than those caused by wild‐type USP4. However, the DUB activity of ΔMT‐1 was comparable to that of wild type, implying that the DUB and localization activities are independent (Supplementary Figure 4C).
3.6. USP4 is associated with colon cancer
Our results revealed that USP4 expression correlates with WNT/β‐catenin signaling by enhancing the transcriptional activity of β‐catenin. Therefore, we hypothesized that an elevated expression level of USP4 may contribute to the development of colon cancer. To verify its relevance to cancer, invasion and migration activities of the siUSP4‐treated HCT116 cells were examined (Figure 6A and B). USP4 knockdown significantly decreased the number of invasive cells in a concentration dependent manner (Figure 6A), and the number of migratory cells was also reduced (Figure 6B). In these assays, the decrease in USP4 levels was confirmed by immunoblotting (Figure 6A and B bottom). Consistent with this finding, the expression levels of both MMP‐1 and MMP‐9, representative markers of migration and invasion in human colon cancers (Zucker and Vacirca, 2004), were clearly decreased by siUSP4 (Figure 6C). These results suggest that USP4 is closely associated with invasion and migration of colon cancer cells.
Figure 4.
(continued).
Figure 6.
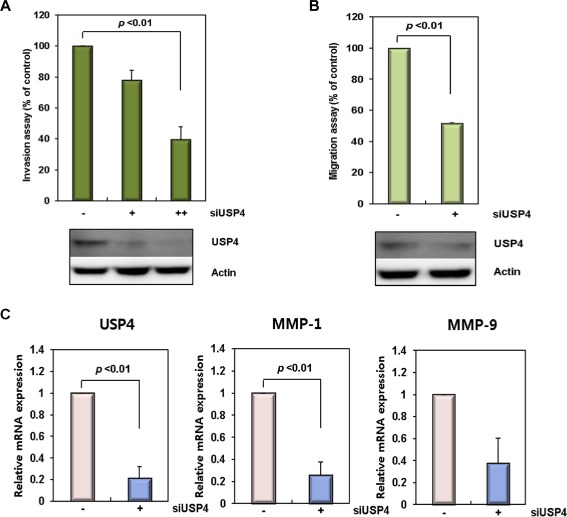
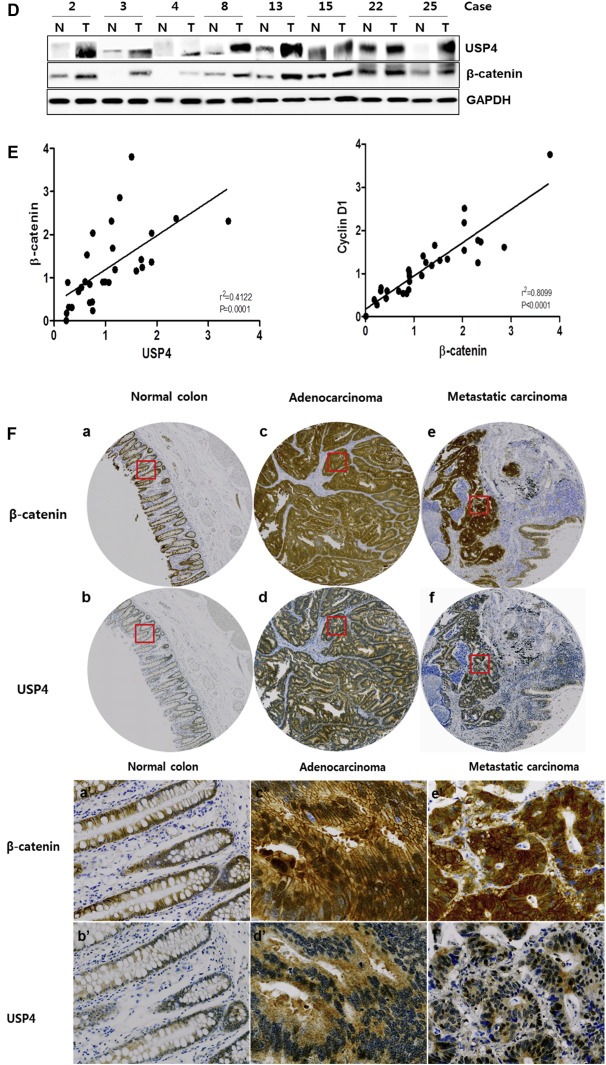
USP4 is correlated with β‐catenin in colon cancers. (A) Invasion activity of siUSP4‐treated colon cancer cells. HCT116 cells were treated with USP4 siRNA (siUSP4; +: 40 pM, ++: 90 pM) for 24 h, and in vitro cell invasion assays were performed for 48 h. (B) Cell migration ability of siUSP4‐treated colon cancer cells. HCT116 cells were treated with 90 pM siUSP4 for 48 h and assayed for their migration ability for 20 h. The invasion and migration activities of the siUSP4 transfected cells were determined relative to those of scrambled siRNA‐treated cells. The expression level of USP4 was analyzed by immunoblotting. Data shown represent the average values from three independent experiments. (C) Expression of migration and invasion markers in the siUSP4‐treated colon cancer cells. The expression level of USP4, MMP‐1, and MMP‐9 was examined 24 h after siUSP4 treatment in HCT116 cells. The mRNA levels were quantified by real time‐PCR with normalization to GAPDH. (D) The correlation between the expression levels of USP4 and β‐catenin, and Cyclin D1 and β‐catenin in colon cancer tissues. The mRNA levels of USP4, β‐catenin, and Cyclin D1 were measured by quantitative real time‐PCR according to an internal calibrator using the 2−△△CT method with normalization to GAPDH. This experiment was repeated twice. (E) Expression of USP4 and β‐catenin in colon cancer tissues. The levels of USP4 and β‐catenin proteins in colon cancer and corresponding normal tissues were measured by immunoblotting using GAPDH as a loading control. T, colon cancer samples; N, normal samples. (F) Expression of β‐catenin (a, c, e) and USP4 proteins (b, d, f) in normal tissues (a, b), primary (c, d), and metastatic colon cancers (e, f) (top). The representative regions of each sample (red boxes, top a–f) are magnified (bottom, a′–f′). In normal colon tissues, β‐catenin (a and a′) and USP4 (b and b′) showed expression in mucosa. Staining of colon adenocarcinoma revealed high levels of β‐catenin (c and c′) and USP4 (d and d′). β‐catenin (e and e′) and USP4 (f and f′) were also actively expressed in metastatic carcinoma that had metastasized from the colon to the ovary. Both adenocarcinoma and metastatic carcinoma tissues were taken from the same patient.
Figure 6.
(continued).
To further investigate the association of USP4 and β‐catenin in colon cancers, we examined 30 human colon specimens (cancer and normal). The correlation between USP4 and β‐catenin was confirmed at the protein level: eight out of eleven samples with increased β‐catenin also showed elevated USP4 (Figure 6D). Furthermore, the expression levels of USP4, β‐catenin and Cyclin D1 were also investigated. As expected, the mRNA levels of β‐catenin and Cyclin D1 were significantly associated, supporting the idea that increased β‐catenin correlates with the expression of Cyclin D1, as well as with cancer progression in the examined cancer specimens (Figure 6E). Interestingly, mRNA levels of USP4 were elevated in the majority of specimens with increased β‐catenin mRNA (Figure 6E).
We next examined the potential relationship of β‐catenin with USP4 in other sets of primary cancer tissues by immunohistochemical staining (Supplementary Table 2). In normal colorectal tissues, both β‐catenin and USP4 were expressed at low levels (Figure 6F‐ab), whereas their expression level was highly increased in adenocarcinoma specimens (Figure 6F‐cd). Among 40 colorectal carcinoma tissues examined, 36 and 29 cases of strong positive expression were observed for β‐catenin (90%) and USP4 (72.5%), respectively. In metastatic carcinoma, high levels of β‐catenin and USP4 were also observed (Figure 6F‐ef). Immunohistochemical analysis of colon cancer tissues supported the idea that the expression of β‐catenin and USP4 are closely associated with colorectal cancer.
4. Discussion
Because the degradation of β‐catenin is regulated via serial phosphorylation and ubiquitination in the cytosol, post‐translational modification is critical for its stability, and accordingly, the determination of cellular fate. We showed that USP4 deubiquitinates β‐catenin (Figure 1B and D). However, USP4 had no effect on the stability of GSK3β, a negative regulator of β‐catenin (Supplementary Figure 5A), suggesting that USP4 enhances the stability and activity of β‐catenin not by affecting β‐catenin regulators but by working as a deubiquitinating enzyme of β‐catenin. As a consequence, the expression levels of WNT/β‐catenin target genes were also regulated by USP4 (Figure 2). The deubiquitinating activity and stabilizing effects of USP4 were abolished when the catalytic residue Cys311 was mutated to Ala (Figure 1B and Supplementary Figure 1E). These results clearly demonstrate that the stability of β‐catenin is regulated by the catalytic activity of USP4.
USP4 has three representative domains, DUSP in the N‐terminus (11–122), UBL domains (142–226 and 483–571), and the USP catalytic domain in the C‐terminus (302–923). In this study, to investigate the role of each domains in the interaction with β‐catenin, three constructs, USP4(1–260), USP4(140–572), and USP4(261–963), were designed. We observed that both USP4(140–572) and USP4(261–963) bind to β‐catenin, thus the second UBL domain (483–571) seems to have a major role in interaction with β‐catenin (Figure 4). Considering reporter activity and target gene expression (Figure 4C and D), USP4(261–963) which contains the UBL2 and USP domain is likely to perform a major role in the β‐catenin activation possibly by direct interaction and deubiquitination. USP4(1–260) appears to have an auxiliary role, since the transcription activity of β‐catenin was not fully recovered by the USP domain only. Consistently, it was reported that DUSP(1–122) is involved in substrate recognition (DeSalle et al., 2001; Song et al., 2010) and that DUSP–UBL domain (11–226) is necessary for the full catalytic turnover of USP4 (Clerici et al., 2014). These reports suggest that USP4(1–260) possibly plays a certain role in recruiting substrates to the USP domain.
According to our novel finding that USP4 acts as the β‐catenin DUB, the transcriptional activity of β‐catenin that is stabilized by the mutation of phosphorylation site cannot be regulated by USP4‐mediated deubiquitination. However, stabilized β‐catenin was partially affected by the USP4 expression level (Figure 2F), and the catalytically defective C311A USP4 was able to activate β‐catenin to some extent (Figure 2A). These results suggested that USP4 might control the activity of β‐catenin via another modulatory mechanism. We have proven that USP4 facilitates the nuclear localization of β‐catenin to some extent (2, 4), which possibly contributes to the enhancement of β‐catenin activity. However, DUB activity seems to perform a primary regulatory role, considering that the decrease of β‐catenin by USP4 knockdown was more significant in the cytosol than in the nucleus (Figure 3D).
So far, several DUBs have been proposed to play roles in WNT signaling by modulating components of the β‐catenin destruction complex (Clague et al., 2012). For example, USP34 increases the stability of Axin, thereby promoting its nuclear accumulation (Lui et al., 2011). FAM (USP9x) has been reported to stabilize β‐catenin through cytoplasmic colocalization, although its effect on the transcriptional activity of β‐catenin was not investigated (Taya et al., 1999). UCHL‐1 is also known to control β‐catenin stability as a DUB (Bheda et al., 2009). However, UCHL‐1 might not have a distinctive role in colon cancers since it is silenced by promoter methylation in many colon cell lines and even in primary colorectal cancers (Okochi‐Takada et al., 2006). Under our experimental conditions, no significant UCHL‐1 DUB activity was measured for β‐catenin, whereas USP4 showed stronger DUB activity (Supplementary Figure 5B). Among DUB enzymes, USP4 shares 60.5% sequence identity with USP15 (Baker et al., 1999), making it tempting to hypothesize that USP15 may also be a functional homolog of USP4. However, USP15 showed only marginal DUB activity on β‐catenin (Supplementary Figure 5B). Therefore, among the DUBs that have been proposed to directly or indirectly affect the stability of β‐catenin, USP4 is most likely to be the main DUB for β‐catenin under our experimental conditions.
USP4 is known to participate in diverse cellular functions and regulates multiple pathways by deubiquitinating non‐proteolytic ubiquitin chains for a functional modulation (Hou et al., 2013; Wang et al., 2013) involved in DNA repair, endocytosis, and kinase‐mediated signaling responses (Luna‐Vargas et al., 2011; Wang et al., 2012). However, USP4 is likely to be associated with K48‐linked proteolytic degradation of β‐catenin rather than functional modulation of β‐catenin by eliminating K63‐chain from β‐catenin. Although K63‐linked ubiquitin modification of β‐catenin, which has a role in β‐catenin stabilization instead of degradation, was reported in MCF7, human breast cancer cells (Gerard et al., 2012), we could not detect the K63‐Ub chain on β‐catenin in this study (data not shown), possibly due to the difference in cell types.
A recent study in SW480 cells proposed that USP4 is a tumor suppressor (Zhao et al., 2009) by demonstrating that USP4 knockdown results in hyper‐activation of canonical WNT signaling through a mechanism involving a NLK–TCF4 interaction. In the present study, we identified USP4 as a DUB for β‐catenin using an overexpression library of DUBs in 293T cells (Supplementary Figure 1B). The USP4 DUB activity and its role as a positive regulator of β‐catenin were further confirmed by knockdown experiments in HCT116 cells. Moreover, we observed that the invasion activity of HCT116 cells was suppressed by USP4 knockdown (Figure 6A). Therefore, we proved that USP4 has oncogenic properties in HCT116 cells. Interestingly, we were also able to observe that USP4 knockdown in SW480 cells increased invasion activity under the same experimental conditions as used in HCT116 cells (Supplementary Figure 5C), suggesting that the role of USP4 is different depending on cell types. Although β‐catenin is activated in both cell lines, as a result of APC and β‐catenin mutations in SW480 and HCT116, respectively, one wild‐type allele of β‐catenin is retained in HCT116 cells (Sekine et al., 2002). Therefore, the contrasting results from SW480 and HCT116 cells may be partially explained by the differential regulation of β‐catenin during cancer pathogenesis.
β‐Catenin can be activated through increased stability or nuclear transport, and aberrant activation of β‐catenin is frequently found in many colon cancers. This study provides evidence in support of the relationship between USP4 and β‐catenin in colon cancers: USP4 expression is elevated in most of the colon cancer tissues which show higher levels of β‐catenin (Figure 6D–F), and the knockdown of USP4 compromised the invasion and migration activities of colon cancer cells (Figure 6A and B).
5. Conclusion
Our study provides substantial evidences for the regulatory roles of USP4 on the ubiquitination and localization of β‐catenin. In conclusion, we suggest that USP4 is a potential oncogene that amplifies the WNT/β‐catenin signal, and that it can be considered as a promising target for anti‐cancer therapeutics.
Supporting information
The following are the supplementary data related to this article:
Supplementary data
Supplementary data
Acknowledgments
This work was supported by National Research Foundation of Korea Grant funded by the Korean Government (NRF‐2012M3A9C6049939 and NRF‐2013R1A1A2058986). We thank Dr. Young Kyu Park and Dr. Seong Yeob Ryu, Department of Gastroenterologic Surgery, Chonnam National University Hwasun Hospital, 160, Ilsim‐ri, Hwasun‐eup, Hwasun‐gun, Jeollanam‐do, 519‐809, Korea, for providing the colorectal cancer samples with clinical information. We also thank Professors Cheol Yong Choi, Jin‐Hyun Ahn and Eek‐hoon Jho for helpful discussion.
Supplementary data 1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2015.06.006.
Yun Sun-Il, Kim Hyeon Ho, Yoon Jung Hwan, Park Won Sang, Hahn Myong-Joon, Kim Hee Cheol, Chung Chin Ha, Kim Kyeong Kyu, (2015), Ubiquitin specific protease 4 positively regulates the WNT/β‐catenin signaling in colorectal cancer, Molecular Oncology, 9, doi: 10.1016/j.molonc.2015.06.006.
References
- Anastas, J.N. , Moon, R.T. , 2013. WNT signaling pathways as therapeutic targets in cancer. Nat. Rev. Cancer. 13, 11–26. [DOI] [PubMed] [Google Scholar]
- Baker, R.T. , Wang, X.W. , Woollatt, E. , White, J.A. , Sutherland, G.R. , 1999. Identification, functional characterization, and chromosomal localization of USP15, a novel human ubiquitin-specific protease related to the UNP oncoprotein, and a systematic nomenclature for human ubiquitin-specific proteases. Genomics. 59, 264–274. [DOI] [PubMed] [Google Scholar]
- Bheda, A. , Yue, W. , Gullapalli, A. , Whitehurst, C. , Liu, R. , Pagano, J.S. , Shackelford, J. , 2009. Positive reciprocal regulation of ubiquitin C-terminal hydrolase L1 and beta-catenin/TCF signaling. PLoS One. 4, e5955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess, A.W. , Faux, M.C. , Layton, M.J. , Ramsay, R.G. , 2011. Wnt signaling and colon tumorigenesis – a view from the periphery. Exp. Cell. Res. 317, 2748–2758. [DOI] [PubMed] [Google Scholar]
- Clague, M.J. , Coulson, J.M. , Urbé, S. , 2012. Cellular functions of the DUBs. J. Cell. Sci. 125, 277–286. [DOI] [PubMed] [Google Scholar]
- Clerici, M. , Luna-Vargas, M.P. , Faesen, A.C. , Sixma, T.K. , 2014. The DUSP-Ubl domain of USP4 enhances its catalytic efficiency by promoting ubiquitin exchange. Nat. Commun. 5, 5399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers, H. , 2006. Wnt/beta-catenin signaling in development and disease. Cell. 127, 469–480. [DOI] [PubMed] [Google Scholar]
- DeSalle, L.M. , Latres, E. , Lin, D. , Graner, E. , Montagnoli, A. , Baker, R.T. , Pagano, M. , Loda, M. , 2001. The de-ubiquitinating enzyme Unp interacts with the retinoblastoma protein. Oncogene. 20, 5538–5542. [DOI] [PubMed] [Google Scholar]
- Fagotto, F. , Glück, U. , Gumbiner, B.M. , 1998. Nuclear localization signal-independent and importin/karyopherin-independent nuclear import of beta-catenin. Curr. Biol. 8, 181–190. [DOI] [PubMed] [Google Scholar]
- Fan, Y.H. , Yu, Y. , Mao, R.F. , Tan, X.J. , Xu, G.F. , Zhang, H. , Lu, X.B. , Fu, S.B. , Yang, J. , 2011. USP4 targets TAK1 to downregulate TNFα-induced NF-κB activation. Cell Death Differ. 18, 1547–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerard, B. , Sanders, M.A. , Visscher, D.W. , Tait, L. , Shekhar, M.P. , 2012. Lysine 394 is a novel Rad6B-induced ubiquitination site on beta-catenin. Biochim. Biophys. Acta. 1823, 1686–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland, J.D. , Klaus, A. , Garratt, A.N. , Birchmeier, W. , 2013. Wnt signaling in stem and cancer stem cells. Curr. Opin. Cell Biol. 25, 254–264. [DOI] [PubMed] [Google Scholar]
- Hou, X. , Wang, L. , Zhang, L. , Pan, X. , Zhao, W. , 2013. Ubiquitin-specific protease 4 promotes TNF-α-induced apoptosis by deubiquitination of RIP1 in head and neck squamous cell carcinoma. FEBS Lett. 587, 311–316. [DOI] [PubMed] [Google Scholar]
- Komiya, Y. , Habas, R. , 2008. Wnt signal transduction pathways. Organogenesis. 4, 68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.R. , Chang, Y.Y. , Hahn, M.J. , 2001. Development of a new epitope tag recognized by a monoclonal antibody to Rickettsia typhi . Biotechniques. 31, 541–545. [DOI] [PubMed] [Google Scholar]
- Lee, J.T. , Gu, W. , 2010. The multiple levels of regulation by p53 ubiquitination. Cell Death Differ. 17, 86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M. , Chen, D. , Shiloh, A. , Luo, J. , Nikolaev, A.Y. , Qin, J. , Gu, W. , 2002. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 416, 648–653. [DOI] [PubMed] [Google Scholar]
- Liu, W. , Dong, X. , Mai, M. , Seelan, R.S. , Taniguchi, K. , Krishnadath, K.K. , Halling, K.C. , Cunningham, J.M. , Boardman, L.A. , Qian, C. , Christensen, E. , Schmidt, S.S. , Roche, P.C. , Smith, D.I. , Thibodeau, S.N. , 2000. Mutations in AXIN2 cause colorectal cancer with defective mismatch repair by activating beta-catenin/TCF signaling. Nat. Genet. 26, 146–147. [DOI] [PubMed] [Google Scholar]
- Lui, T.T. , Lacroix, C. , Ahmed, S.M. , Goldenberg, S.J. , Leach, C.A. , Daulat, A.M. , Angers, S. , 2011. The ubiquitin-specific protease USP34 regulates axin stability and Wnt/β-catenin signaling. Mol. Cell. Biol. 31, 2053–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna-Vargas, M.P. , Faesen, A.C. , van Dijk, W.J. , Rape, M. , Fish, A. , Sixma, T.K. , 2011. Ubiquitin-specific protease 4 is inhibited by its ubiquitin-like domain. EMBO Rep. 12, 365–372. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Milojevic, T. , Reiterer, V. , Stefan, E. , Korkhov, V.M. , Dorostkar, M.M. , Ducza, E. , Ogris, E. , Boehm, S. , Freissmuth, M. , Nanoff, C. , 2006. The ubiquitin-specific protease Usp4 regulates the cell surface level of the A2A receptor. Mol. Pharmacol. 69, 1083–1094. [DOI] [PubMed] [Google Scholar]
- Morin, P.J. , Sparks, A.B. , Korinek, V. , Barker, N. , Clevers, H. , Vogelstein, B. , Kinzler, K.W. , 1997. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 275, 1787–1790. [DOI] [PubMed] [Google Scholar]
- Mukai, A. , Yamamoto-Hino, M. , Awano, W. , Watanabe, W. , Komada, M. , Goto, S. , 2010. Balanced ubiquitylation and deubiquitylation of Frizzled regulate cellular responsiveness to Wg/Wnt. EMBO J. 29, 2114–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okochi-Takada, E. , Nakazawa, K. , Wakabayashi, M. , Mori, A. , Ichimura, S. , Yasugi, T. , Ushijima, T. , 2006. Silencing of the UCHL1 gene in human colorectal and ovarian cancers. Int. J. Cancer. 119, 1338–1344. [DOI] [PubMed] [Google Scholar]
- Saletta, F. , Suryo Rahmanto, Y. , Richardson, D.R. , 2010. The translational regulator eIF3a: the tricky eIF3 subunit. Biochim. Biophys. Acta. 1806, 275–286. [DOI] [PubMed] [Google Scholar]
- Sekine, S. , Shibata, T. , Sakamoto, M. , Hirohashi, S. , 2002. Target disruption of the mutant beta-catenin gene in colon cancer cell line HCT116: preservation of its malignant phenotype. Oncogene. 21, 5906–5911. [DOI] [PubMed] [Google Scholar]
- Sharma, M. , Jamieson, C. , Johnson, M. , Molloy, M.P. , Henderson, B.R. , 2012. Specific armadillo repeat sequences facilitate β-catenin nuclear transport in live cells via direct binding to nucleoporins Nup62, Nup153, and RanBP2/Nup358. J. Biol. Chem. 287, 819–831. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Soboleva, T.A. , Jans, D.A. , Johnson-Saliba, M. , Baker, R.T. , 2005. Nuclear-cytoplasmic shuttling of the oncogenic mouse UNP/USP4 deubiquitylating enzyme. J. Biol. Chem. 280, 745–752. [DOI] [PubMed] [Google Scholar]
- Song, E.J. , Werner, S.L. , Neubauer, J. , Stegmeier, F. , Aspden, J. , Rio, D. , Harper, J.W. , Elledge, S.J. , Kirschner, M.W. , Rape, M. , 2010. The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes Dev. 24, 1434–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauriello, D.V. , Haegebarth, A. , Kuper, I. , Edelmann, M.J. , Henraat, M. , Canninga-van Dijk, M.R. , Kessler, B.M. , Clevers, H. , Maurice, M.M. , 2010. Loss of the tumor suppressor CYLD enhances Wnt/beta-catenin signaling through K63-linked ubiquitination of Dvl. Mol. Cell. 37, 607–619. [DOI] [PubMed] [Google Scholar]
- Taya, S. , Yamamoto, T. , Kanai-Azuma, M. , Wood, S.A. , Kaibuchi, K. , 1999. The deubiquitinating enzyme Fam interacts with and stabilizes β-catenin. Genes Cells. 4, 757–767. [DOI] [PubMed] [Google Scholar]
- Uras, I.Z. , List, T. , Nijman, S.M. , 2012. Ubiquitin-specific protease 4 inhibits mono-ubiquitination of the master growth factor signaling kinase PDK1. PLoS One. 7, e31003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada, K. , Kamitani, T. , 2006. Oncogenic protein UnpEL/Usp4 deubiquitinates Ro52 by its isopeptidase activity. Biochem. Biophys. Res. Commun. 342, 253–258. [DOI] [PubMed] [Google Scholar]
- Wang, G. , Gao, Y. , Li, L. , Jin, G. , Cai, Z. , Chao, J.I. , Lin, H.K. , 2012. K63-linked ubiquitination in kinase activation and cancer. Front. Oncol. 2, 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Zhao, W. , Zhang, M. , Wang, P. , Zhao, K. , Zhao, X. , Yang, S. , Gao, C. , 2013. USP4 positively regulates RIG-I-mediated antiviral response through deubiquitination and stabilization of RIG-I. J. Virol. 87, 4507–4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, K. , Dai, X. , 2011. Winning WNT: race to Wnt signaling inhibitors. Proc. Natl. Acad. Sci. U. S. A. 108, 5929–5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X. , Tu, X. , Joeng, K.S. , Hilton, M.J. , Williams, D.A. , Long, F. , 2008. Rac1 activation controls nuclear localization of beta-catenin during canonical Wnt signaling. Cell. 133, 340–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, N. , Li, H. , Luo, J. , Wang, R. , Chen, H. , Chen, J. , Wang, P. , 2012. Ubiquitin-specific protease 4 (USP4) targets TRAF2 and TRAF6 for deubiquitination and inhibits TNFα-induced cancer cell migration. Biochem. J. 441, 979–986. [DOI] [PubMed] [Google Scholar]
- Yuan, J. , Luo, K. , Zhang, L. , Cheville, J.C. , Lou, Z. , 2010. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell. 140, 384–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Berger, F.G. , Yang, J. , Lu, X. , 2011. USP4 inhibits p53 through deubiquitinating and stabilizing ARF-BP1. EMBO J. 30, 2177–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, B. , Schlesiger, C. , Masucci, M.G. , Lindsten, K. , 2009. The ubiquitin specific protease 4 (USP4) is a new player in the Wnt signaling pathway. J. Cell. Mol. Med. 13, 1886–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, B. , Velasco, K. , Sompallae, R. , Pfirrmann, T. , Masucci, M.G. , Lindsten, K. , 2012. The ubiquitin specific protease-4 (USP4) interacts with the S9/Rpn6 subunit of the proteasome. Biochem. Biophys. Res. Commun. 427, 490–496. [DOI] [PubMed] [Google Scholar]
- Zucker, S. , Vacirca, J. , 2004. Role of matrix metalloproteinases (MMPs) in colorectal cancer. Cancer Metastasis Rev. 23, 101–117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following are the supplementary data related to this article:
Supplementary data
Supplementary data


