Abstract
Background
Patients with Estrogen Receptor α‐positive (ER+) Inflammatory Breast Cancer (IBC) are less responsive to endocrine therapy compared with ER+ non‐IBC (nIBC) patients. The study of ER+ IBC samples might reveal biomarkers for endocrine resistant breast cancer.
Materials & methods
Gene expression profiles of ER+ samples from 201 patients were explored for genes that discriminated between IBC and nIBC. Classifier genes were applied onto clinically annotated expression data from 947 patients with ER+ breast cancer and validated with RT‐qPCR for 231 patients treated with first‐line tamoxifen. Relationships with metastasis‐free survival (MFS) and progression‐free survival (PFS) following adjuvant and first‐line endocrine treatment, respectively, were investigated using Cox regression analysis.
Results
A metagene of six genes including the genes encoding for 4‐aminobutyrate aminotransferase (ABAT) and Stanniocalcin‐2 (STC2) were identified to distinguish 22 ER+ IBC from 43 ER+ nIBC patients and remained discriminatory in an independent series of 136 patients. The metagene and two genes were not prognostic in 517 (neo)adjuvant untreated lymph node‐negative ER+ nIBC breast cancer patients. Only ABAT was related to outcome in 250 patients treated with adjuvant tamoxifen. Three independent series of in total 411 patients with advanced disease showed increased metagene scores and decreased expression of ABAT and STC2 to be correlated with poor first‐line endocrine therapy outcome. The biomarkers remained predictive for first‐line tamoxifen treatment outcome in multivariate analysis including traditional factors or published signatures. In an exploratory analysis, ABAT and STC2 protein expression levels had no relation with PFS after first‐line tamoxifen.
Conclusions
This study utilized ER+ IBC to identify a metagene including ABAT and STC2 as predictive biomarkers for endocrine therapy resistance.
Keywords: Inflammatory breast cancer, Endocrine therapy resistance, Metastatic disease, <i>ABAT</i> , <i>STC2</i>
Highlights
ER+ inflammatory breast cancer (IBC) can be used as endocrine therapy resistance model.
Molecular characterization revealed ABAT and STC2 as ER+ IBC discriminatory genes.
ABAT and STC2 are novel biomarkers for tamoxifen resistance in advanced breast cancer.
1. Introduction
Breast cancer is the most prevalent form of cancer in women in the United States and Europe (Torres‐Arzayus et al., 2010). The majority of patients with breast cancer bear tumors expressing detectable levels of the Estrogen Receptor (ER). For these patients, targeted therapies are available including strategies directed at the receptor itself, such as tamoxifen and fulvestrant. In addition, estrogen deprivation offers another therapeutic strategy that can be achieved by ovarian ablation, or LHRH analogs, in the premenopausal patient, or with aromatase inhibitors (AIs) in the postmenopausal setting. These therapies are highly effective; adjuvant endocrine therapy has been shown to reduce mortality from ER+ breast cancer to the same degree as adjuvant chemotherapy (Early Breast Cancer Trialists' Collaborative, G, 2005). Unfortunately, part of the patients with ER+ breast cancer show de novo resistance to endocrine therapy, whereas others initially benefit but ultimately relapse due to acquired endocrine resistance (Leary et al., 2010). Predicting, modulating and/or restoring endocrine responsiveness remain important clinical priorities for which molecular targets are urgently needed.
Inflammatory breast cancer (IBC) is a rare (∼5%) but aggressive form of locally advanced breast cancer. At time of diagnosis, virtually all patients with IBC have lymph node metastases and 1/3 of the patients have metastases in distant organs. As a consequence, the prognosis for patients with IBC is dismal (Dawood et al., 2011; Dirix et al., 2006). Analysis of the Surveillance, Epidemiology and End Results (SEER)‐database revealed that IBC is characterized by atypical clinicopathological features (Dawood et al., 2011), including frequent absence of ER protein expression (Hance et al., 2005). Our research group and others have shown that this IBC‐specific clinicopathological profile is corroborated at the molecular level by a distinct gene expression profile (Bertucci et al., 2004; Van Laere et al., 2007a; Van Laere et al., 2005). Exploration of this gene expression profile led to the discovery of pronounced activation of the transcription factor NFkB in IBC (Lerebours et al., 2008; Van Laere et al., 2006) and more recently to the observation that TGFβ‐signaling is repressed (Van Laere et al., 2008). Furthermore, we demonstrated that the IBC‐specific expression profile harbors the molecular traits of aggressive tumor cell behavior in general (Van Laere et al., 2008), including stem cell biology (Van Laere et al., 2010). As such, we consider IBC, although occurring rarely, as a suitable example to elucidate mechanisms responsible for tumor cell dissemination, metastasis and drug resistance in breast cancer in general.
The majority (depending on the reference up to 66%) of patients with IBC lack ER protein expression, but ER+ tumor samples from patients with IBC exist. Clinically, patients with ER+ IBC are less responsive to endocrine treatment as compared to patients with other forms of ER+ breast cancer. In light of molecular heterogeneity and our previous results, we reasoned that studying ER+ IBC focusing on endocrine treatment response might provide new insights into molecular resistance mechanisms of endocrine therapy. In the current study, we evaluated expression profiles from patients with ER+ IBC and nIBC. The purpose of this study was 1) to identify differentially expressed genes between IBC and nIBC, 2) assess their accuracy to predict ER+ IBC, and 3) to define their relationship with endocrine therapy response in clinical samples. Discriminatory genes were identified by gene expression arrays, of which two genes remained deregulated in an independent series of ER+ samples between patients with and without IBC. When applied onto clinically annotated expression series from patients with ER+ breast cancer treated with endocrine therapy either in the adjuvant or advanced setting, decreased expression of these two genes were linked with poor responsiveness to endocrine therapy. These two genes when validated with quantitative real‐time PCR for mRNA expression and with immunohistochemistry for protein expression, demonstrated predictive value only at the mRNA level.
2. Materials and methods
2.1. Study design and patient samples
The present study describes a retrospective analysis performed in accordance with the Code of Conduct of the Federation of Medical Scientific Societies in the Netherlands, Belgium and France, and is reported following the REMARK recommendations (McShane et al., 2006). The local medical ethics committees have approved the study. Follow‐up, tumor staging, and response to therapy was defined by standard International Union Against Cancer (Geneva, Switzerland) classification criteria (Hayward et al., 1978). Samples were recruited from the Translational Cancer Research Unit (TCRU, Antwerp, Belgium), the Institut Poali‐Calmettes (IPC, Marseille France), the Erasmus Medical Center (EMC, Rotterdam, the Netherlands) and the Netherlands Cancer Institute (NKI, Amsterdam, the Netherlands). The ER‐status of the tumors was established by immunohistochemistry (≥10% positive tumor cells) or EIA (≥10 fmol/mg protein) and together with additional clinicopathological characteristics have been described before for each of the series (Bekhouche et al., 2011; Desmedt et al., 2007; Jansen et al., 2013; Kok et al., 2009; Loi et al., 2008; Reijm et al., 2014; Schmidt et al., 2008; Van der Auwera et al., 2010; Van Laere et al., 2007b; Wang et al., 2005).
The mRNA‐datasets used in this study are presented in Table 1 and includes ER+ IBC‐subsets (I–II), endocrine treated subsets (III–VI), and untreated lymph node‐negative (LNN) patients (VII–IX).
Table 1.
Datasets. Detailed information on the datasets used in this study. The datasets are numbered as series I to IX and the subset provides information about its study subject (IBC, treatment outcome, clinical outcome). For each dataset the author and array reference are presented next to the array‐platform.
| Information of datasets used in this study | ||||||
|---|---|---|---|---|---|---|
| Series | Analysis | Subset | Reference | GEO/ArrayExpress | Platform | Total number of cases |
| I | IBC | Discovery | Van Laere et al. | E‐MTAB‐1006 | Affymetrix | 65 |
| II | Validation | Bertucci et al. | NA | Affymetrix | 136 | |
| III | Treatment Outcome | Tamoxifen Adjuvant | Loi et al. | GSE6532 | Affymetrix | 250 |
| IV | Tamoxifen Advanced | Kok et al. | NA | Agilent | 96 | |
| V | Aromatase Inhibitors | Jansen et al. (2013) | GSE41994 | Agilent | 84 | |
| VI | Tamoxifen Advanced | Jansen et al. (2007) | NA | qRT‐PCR | 231 | |
| VII | Clinical Outcome | LNN ER+, untreated | Wang et al. | GSE2034 | Affymetrix | 221 |
| VIII | LNN ER+, untreated | Desmedt et al. | GSE7390 | Affymetrix | 134 | |
| IX | LNN ER+, untreated | Schmidt et al. | GSE11121 | Affymetrix | 162 | |
The discovery and test phase incorporated 2 patient series with ER+ IBC: a) a discovery series (I) of 65 samples from patients with and without ER+ IBC retrieved from the TCRU (E‐MTAB‐1006) and b) an independent test series (II) of samples from 136 patients with and without ER+ IBC that received adjuvant treatment, retrieved from IPC. Samples from patients with ER+ IBC in series I (N = 22) and series II (N = 39) were selected by strictly adhering to the consensus diagnostic criteria (Dawood et al., 2010).
To evaluate discovered genes for their relationship with endocrine treatment outcome, 4 additional data sets (III–VI) were incorporated of patients with ER+ breast cancer treated with endocrine therapy for primary and advanced disease: 3 data sets (IV–VI) of 411 metastatic breast cancer patients in total from EMC, NKI and TRCU; and 1 data set with primary breast cancers of 250 patients (III). The data set of 411 patients with advanced ER+ breast cancer treated with first‐line therapy contained three subsets, one of 96 patients treated with tamoxifen (IV) and one of 84 patients treated with aromatase inhibitors (AIs) (V), and one of 231 patients treated with tamoxifen and profiled using RT‐qPCR for dedicated genes (VI). The RT‐qPCR data set was used as an independent validation series for the genome‐wide expression series. All samples in these cohorts were classified according to the Recurrence Score (RS) (24), the Genomic Grade Index (GGI) (25). For each of these patient series tumor size and histological grade were recorded, in addition to age and menopausal status at start of therapy, dominant site of relapse and disease‐free interval for the RT‐qPCR data set. To assess the prognostic value of the discovered genes, we incorporated also 3 series (VII‐IX) of ER+ tumors from 517 LNN breast cancer patients, who did not receive any type of adjuvant systemic therapy. Details regarding the application of the above classifiers are provided in the Supplementary data file, Tables A.1 and A.2.
The discovered predictive genes were also evaluated for their protein expression pattern in a tissue microarray including cores of ER‐positive primary tumor specimens from a cohort of advanced breast cancer patients who have been treated with first‐line tamoxifen previously described (Reijm et al., 2014). A subset of 110 ER‐positive tumors were explored for their protein staining, i.e. the number of positive cells and the staining intensity, and a staining IHC‐score was calculated to evaluate the relationship between IHC‐score and progression‐free survival.
2.2. Methods
2.2.1. RNA isolation and (genome‐wide) expression profiling
RNA isolation for the samples retrieved from each of the participating centers (TCRU, EMC, NKI and IPC) and quality control was done as described before (Bekhouche et al., 2011, 2005, 2013, 2009, 2007). Genome‐wide expression profiles were available from Affymetrix HGU133A or HGU133plus2 platforms (I–III, VII–IX) and 44k mRNA oligoarrays of Agilent Technologies (IV–V).
Expression analyses were verified by RT‐qPCR (series VI) and were performed for the “IBC‐like” genes (i.e. ABAT, ADAMDEC1, CLEC7A, ETS1, ITK and STC2) to discriminate between IBC and nIBC, for the Recurrence Score genes (i.e. AURKA, BAG1, BCL2, BIRC5, CCNB1, CD68, CTSL2, ERBB2, ESR1, GRB7, GSTM1, MKI67, MMP11, MYBL2, PGR and SCUBE2) and for a panel of reference genes (i.e. HMBS, HPRT1, TBP and B2M). Assay details are provided in Supplementary Table A.1. The cDNA synthesis, quantification and the methodology to ensure PCR specificity have been described previously (Sieuwerts et al., 2005; van Agthoven et al., 2009). RT‐qPCR was performed in a Mx3000P™ Real‐Time PCR System (Agilent, Amsterdam, The Netherlands) using the TaqMan‐based gene expression assays from Applied Biosystems/Life Technologies and SYBR‐based intron‐spanning forward and reverse primer combinations for the other genes. Levels of the target genes, expressed relative to the reference genes were quantified as follows: mRNA target = 2(mean Ct reference genes−mean Ct target genes).
Expression levels of each series (Supplementary Table A.2) were normalized and subsequently harmonized for cross‐platform evaluation and robust regression analyses. To accomplish harmonization of series, Hampel'S M‐Estimators were calculated in SPSS (version 20) for all series and applied to establish the harmonization factor for the genes in each series when using series I as reference (Supplementary Table A.3).
2.2.2. Comparative analysis or ER+ IBC and nIBC expression profiles
Global differences in gene expression between samples from patients with and without ER+ IBC in the discovery series were analyzed using the global test (Goeman et al., 2004) and Principal Component Analysis (PCA). Using the PAM50‐algorithm, each sample in the discovery series was classified according to the molecular subtypes, ER activity, and Risk‐Of‐Relapse (ROR) models based on the molecular subtypes alone (ROR‐S) or in combination with cell proliferation (ROR‐P), as described before (Ellis et al., 2011). In addition, the Recurrence Score (RS) (Paik et al., 2004) and the HOXB13/IL17RB gene expression ratio (Jansen et al., 2007; Ma et al., 2006) were calculated.
2.2.3. Biomarker discovery analysis
We performed Prediction Analysis of Microarrays (PAM) (Tibshirani et al., 2002) to identify a series of biomarkers able to discriminate ER+ IBC from ER+ nIBC samples. The discovery series were randomly divided into training sets of 40 samples and test sets of 25 samples to obtain 10 gene signatures, which were compared to identify common classifiers. In total six classifier genes were shared between these 10 gene signatures, i.e. ABAT, ADAMDEC1, CLEC7A, ETS1, ITK and STC2. PCA was performed onto the ER+ IBC discovery and validation series to evaluate the discriminatory performance of all genes and of the six common classifier genes together. Mutual relationships between these common classifier genes and ER were investigated using the Ingenuity Pathway Analysis (IPA) software.
2.2.4. Construction of an ER+ IBC‐like metagene
The thus identified six genes ABAT, ADAMDEC1, CLEC7A, ETS1, ITK and STC2 were combined into an ER+ IBC‐like metagene. The regression coefficients of each of the genes obtained within the discovery series were used to calculate a score for the metagene in all other series. This metagene score was evaluated as biomarker representing the signature of above six genes.
2.2.5. Diagnostic evaluation of biomarkers and classifiers
The IBC discovery and test sets (series I & II) were used to assess the predictive potential of the biomarkers to identify IBC and nIBC. The biomarkers were evaluated as continuous variables with Receiver Operator Characteristic (ROC) analyses using the STATA statistical package. The ROC analyses were performed to define Area Under Curves (AUC) and assess the discriminatory potential of the biomarkers. Next, ROC analyses were used to select cutoffs with optimal sensitivity and specificity. These cutoffs generated dichotomized biomarkers which were subsequently explored as classifiers using distribution dot‐plots created in STATA and evaluated for their diagnostic effectiveness by SISA (http://www.quantitativeskills.com/sisa/). The distribution dot‐plots illustrated the performance and the number of false positives/negatives when applying the cutoffs. The SISA tool established for the classifiers their accuracy, sensitivity, specificity and Youden's Index of the predictions.
2.2.6. Survival analysis of biomarkers and classifiers
The biomarkers were evaluated for their relationship with survival using Cox regression analyses in two ways: as a continuous variable or dichotomized to a threshold as classifier to distinguish “IBC‐like” and “nIBC‐like”. For the assessment of the relationship with first‐line therapy outcome in advanced disease, Progression‐Free Survival (PFS), defined as the time elapsed between initiation of endocrine therapy and the first detection of disease progression, was considered as endpoint. PFS was censored at 36 months. For the assessment of the relationship with prognosis and adjuvant therapy in early disease, Metastasis‐Free Survival (MFS) was used as endpoint and defined as the time elapsed between the date of diagnosis and the date of distant metastatic relapse. Multivariate Cox regression analyses were performed on each of the endocrine treated advanced disease subsets for PFS (series IV–VI). The models included the biomarkers as continuous variable on the one hand and the published signatures for the Recurrence Score or the GGI on the other hand. Additional multivariate analyses were performed with the base model of clinic‐pathological factors including age and menopausal status at start of therapy, dominant site of relapse, disease free interval (DFI), and the mRNA expression levels of ER (ESR1), PR (PGR), and HER2 (ERBB2). All data computations were done with the STATA statistical package version 12.0. All P‐values are two‐sided and P < 0.05 was considered statistically significant.
2.2.7. Tissue microarrays and immunohistological and evaluation
Tissue microarrays (TMAs) of formalin‐fixed, paraffin‐embedded primary breast tumor specimens were prepared and immunohistochemically stained according to the procedures described previously (Reijm et al., 2014). The staining was performed with the primary monoclonal antibody against ABAT (HPA041690) and STC2 (HPA045372; Atlas Antibodies AB, Stockholm, Sweden). The antibodies were incubated for 1 h (1:100 dilution) after 20 min antigen retrieval at pH6.0. Subsequently, the TMA‐slides were incubated with a secondary antibody and staining was visualized using diaminobenzidine (DAB). ABAT and STC2 protein staining was scored for quantity and intensity. Staining was grouped into standardized categories for the percentage of staining positive cells (0%, 1–20%, 21–50%, 51–75%, or 76–100% of positive cells) and for staining intensity (negative, weak, moderate, strong). Subsequently, the scores for quantity and intensity were multiplied to generate a staining IHC‐score. Based on the IHC‐score, a specimen was classified as negative or positive for ABAT and STC2 protein expression.
3. Results
3.1. Comparative analysis of ER+ IBC and nIBC expression profiles
Using global test analysis and PCA, we observed that ER+ samples from patients with and without IBC exhibited significant differences in their expression profiles that led to segregation of IBC and nIBC samples in the PCA plot of the discovery series (Figure 1A). Evaluation of the Recurrence Score, the HOXB13/IL17RB expression ratio and PAM50‐derived scores for molecular subtypes, ER activity, risk of relapse models (ROR‐S and ROR‐P), indicated decreased ER signaling and sensitivity to endocrine treatment for ER+ samples from IBCs compared to those of nIBCs (Figure 1B).
Figure 1.
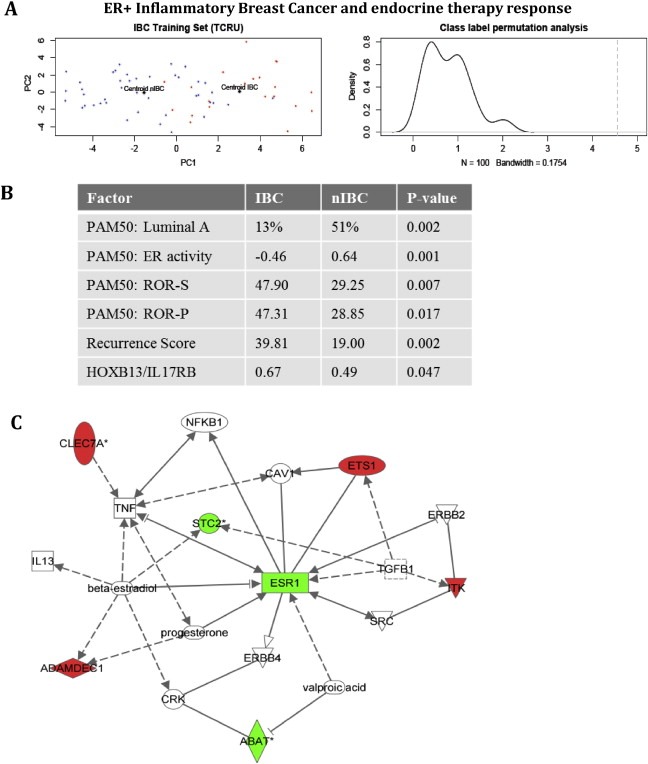
IBC discriminatory genes. Evaluation of IBC and nIBC in the discovery series I (Figure 1A‐B) and of ER+ IBC discriminatory genes (ABAT, ADAMDEC1, CLEC7A, ETS1, ITK, STC2) (Figure 1C). Figure 1A illustrates the Principle Component Analyses of the tumor samples by their gene expression profiles. Red dots denote ER+ IBC samples, blue dots denote ER+ nIBC samples. The centroids for both tumor phenotypes are indicated in black and labeled respectively “Centroid IBC” and “Centroid nIBC”. PCA for the common 6 classifier genes showed an expected segregation of ER+ samples from patients with and without IBC on the 2D scatter plot representation of the 1st (X‐axis) and the 2nd (Y‐axis) principal component. Class label permutation analyses (applying 100 class label permutations) demonstrated that the centroids of the ER+ samples from patients with and without IBC are significantly segregated (Observed Euclidean distance = 4.555, average expected Euclidean distance = 0.890; P > 0.010). Figure 1B presents the results on PAM50 analyses, Recurrence score, and HOXB13/IL17RB. For PAM50, the percentage Luminal A‐type tumors in IBC and nIBC is provided in addition to the ROR‐S, ROR‐P and ER activity scores. The ER activity score ranges from negative to positive, with negative values indicating repressed ER activity. In addition, the RS and the HOXB13/IL17RB gene expression ratio are provided for both tumor types. The reported P‐values result from the comparison of the IBC and nIBC groups with respect to these variables. Figure 1C depicts the network obtained for the 6 IBC discriminatory genes together with the estrogen receptor‐α (ESR1) when evaluated with Ingenuity Pathway Analyses. This exploratory analysis revealed interactions with hormone receptor signaling, inflammation, cell survival, epidermal growth factor signaling, stem cell signaling and TGFβ signaling, indicating a potential involvement for each of these biological features in endocrine resistance. The molecules are color‐coded red if the corresponding gene is overexpressed in ER+ IBC samples and green if the corresponding gene is repressed in ER+ IBC samples. Uncolored nodes are added by the software. Solid lines signify direct gene‐gene interactions, whereas broken lines represent indirect relationships that may require secondary effectors not depicted in the network. All connections are supported by at least one published report or from canonical information stored in the Ingenuity Pathway Knowledge Base.
3.2. Biomarker discovery analysis
As shown above, ER+ IBC samples exhibited molecular characteristics of resistance to endocrine therapy, making their gene expression profiles a potential source for biomarker discovery. Using PAM on 10 alternatively composed training sets of 40 randomly selected ER+ samples from the discovery set (series I), we generated 10 distinct gene signatures distinguishing IBC from nIBC samples. Application of these gene signatures onto corresponding series of the 25 left‐out samples revealed an average sensitivity of 89% (range 71%–100%), specificity of 80% (range 67%–100%) and test error rate of 18% (range 0%–28%). These 10 gene signatures had 6 overlapping genes: ABAT, ADAMDEC1, CLEC7A, ETS1, ITK1 and STC2. Relative to nIBC, ABAT and STC2 expression levels were decreased in IBC whereas the levels of the remaining 4 genes were increased (Table 2 and Figure 2). Exploratory IPA analysis was performed to investigate mutual relationships between the 6 classifier genes and ER (Figure 1C) and suggested their potential involvement in endocrine therapy resistance.
Table 2.
ER+ IBC discriminatory genes. The expression levels in IBC and nIBC, significance and regression coefficients for the genes identified in the biomarker discovery analysis.
| ER+ IBC‐Metagene | ||||
|---|---|---|---|---|
| Median expression level | ||||
| Genes | nIBC (N = 43) | IBC (N = 22) | P‐value | Regression coefficient |
| ABAT | 10.63 | 8.64 | 9.19E‐08 | −0.295 |
| ADAMDEC1 | 6.26 | 9.07 | 1.31E‐03 | 0.566 |
| CLEC7A | 7.24 | 8.12 | 9.55E‐03 | 0.270 |
| ETS1 | 4.09 | 4.99 | 7.09E‐05 | 0.112 |
| ITK | 5.27 | 7.09 | 2.49E‐04 | 0.332 |
| STC2 | 10.8 | 7.64 | 1.70E‐04 | −0.630 |
Figure 2.
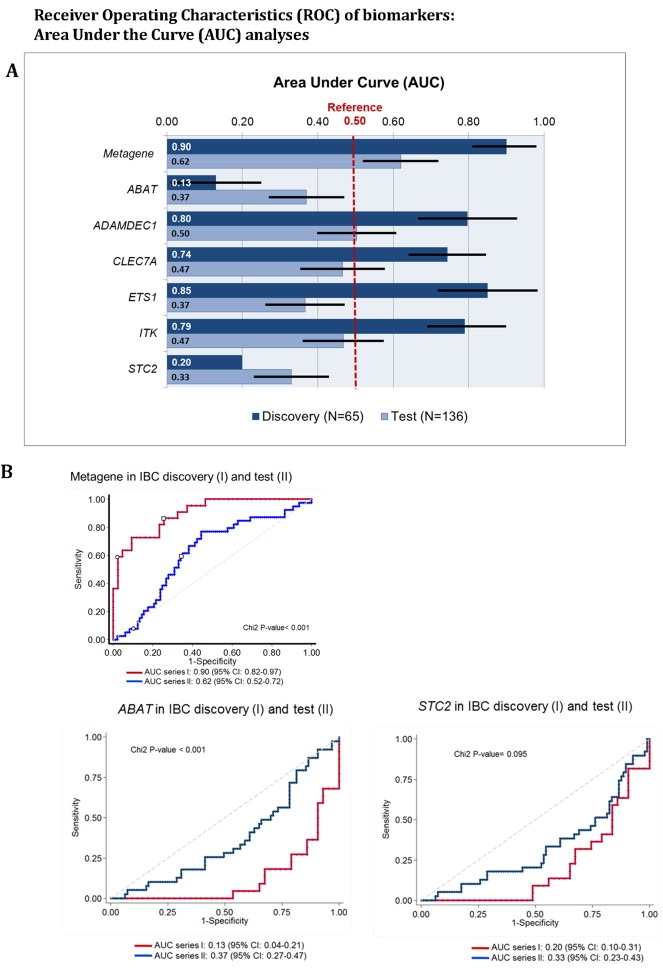
Receiver Operator Characteristics (ROC) Analyses. The ROC‐analyses generate Area Under Curve (AUC) values presented in Figure 2A as measure for the discriminatory potential of the individual genes to predict IBCs and nIBCs correctly within the test (series II) compared to the discovery (series I). Factors with AUC (or their intervals) value 0.5 are not informative. The results show as illustrated with ROC plots in Figure 2B that AUCs for only the metagene, ABAT and STC2 are discriminatory and comparable for the discovery and test.
3.3. Diagnostic effectiveness of biomarkers and classifiers
Next, these six genes were evaluated for their potential to distinguish IBC from nIBC in an independent subset of patients with and without IBC (series II). Only the AUCs for the metagene score, ABAT, and STC2 were discriminatory and comparable in both the discovery and test cohort (Figure 2). The ROC AUCs of these three biomarkers were further explored to establish optimal classifier cutoffs. Distribution dot‐plots in the discovery cohort were used to verify the ER+ IBC classification thresholds, which were set for the metagene score at ≥0.0 and for both ABAT and STC2 at ≤ 10.0 (Figure 3A, C and E). The classifiers, however, exhibited moderate diagnostic effectiveness with regard to ER+ IBC prediction, since only a maximum of 64% accuracy was achieved within the test (Figure 3B, D and F).
Figure 3.
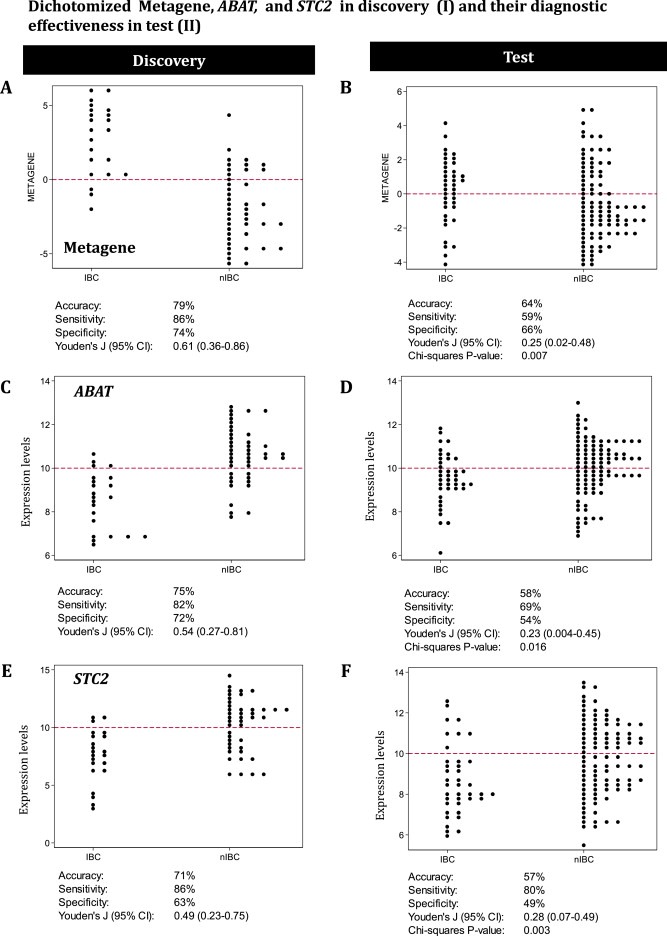
Dot‐plots and diagnostic effectiveness. This figure represents dot‐plots and the diagnostic performance of the biomarkers in the discovery and test series for IBC and nIBC (series I and II). The metagene scores and expression levels of ABAT and STC2 measured in the discovery (series I) were evaluated in dot‐plots to explore the defined thresholds that classify samples as IBC‐like or nIBC‐like. The diagnostic effectiveness of the biomarker IBC classification were evaluated in the independent test (series II).
3.4. Biomarkers in early disease: prognosis and adjuvant tamoxifen
The metagene score, ABAT, and STC2 were subsequently evaluated as continuous variable for their relationship with MFS with regard to prognosis and outcome after adjuvant tamoxifen (Table 3). The prognostic value was determined on three series of in total 517 (neo)adjuvant systemic treatment naïve patients with ER+ LNN breast cancer (series VII–IX). None of the biomarkers, assessed by microarrays, were prognostic when evaluated for all 517 patients. The biomarkers were also evaluated in 250 ER+ patients treated with adjuvant tamoxifen (series III). Decreased expression of ABAT (HR = 0.73; 95% CI = 0.61–0.87; P = 0.001) showed a significant correlation with poor MFS in these tamoxifen treated patients, whereas STC2 and the metagene score had no association with MFS after tamoxifen.
Table 3.
Biomarkers and outcome. This table provides the results of the univariate Cox regression analyses performed for MFS and PFS to determine the prognostic and predictive value of the metagene, ABAT and STC2 in the different patient series.
| Clinical setting | Series | N | outcomea | Biomarkers as continuous variable: | |||||
|---|---|---|---|---|---|---|---|---|---|
| Metagene score | ABAT | STC2 | |||||||
| HR (95%CI)b | P | HR (95%CI) | P | HR (95%CI) | P | ||||
| Early Disease | |||||||||
| Prognosis | VII–IX | 517 | MFS | 0.99 (0.92–1.08) | 0.967 | 0.93 (0.82–1.06) | 0.298 | 1.01 (0.92–1.11) | 0.867 |
| Adjuvant tamoxifen | III | 250 | MFS | 1.09 (0.99–1.20) | 0.084 | 0.73 (0.61–0.87) | 0.001 | 0.93 (0.83–1.04) | 0.216 |
| Advanced Disease | |||||||||
| First‐line tamoxifen | IV | 96 | PFS | 1.13 (1.03–1.25) | 0.012 | 0.80 (0.69–0.93) | 0.004 | 0.90 (0.79–1.03) | 0.117 |
| First‐line aromatase inhibitors | V | 84 | PFS | 1.28 (1.13–1.45) | <0.001 | 0.74 (0.60–0.91) | 0.004 | 0.76 (0.66–0.88) | <0.001 |
| First‐line tamoxifen | VI | 231 | PFS | 1.09 (1.04–1.14) | <0.001 | 0.85 (0.80–0.92) | <0.001 | 0.93 (0.88–0.98) | 0.004 |
Outcome defined by metastasis free survival (MFS) or by progression free survival (PFS).
Hazard Ratio (HR) and its 95% confidence interval (CI) for MFS in early disease and PFS in advanced disease.
3.5. Biomarkers and first‐line endocrine therapy for advanced disease
The metagene score, ABAT, and STC2 were also evaluated for their relation with PFS on microarray based series of patients with advanced disease treated with first‐line tamoxifen (IV, N = 96) or aromatase inhibitors (V, N = 84), and validated with RT‐qPCR on an independent series of patients treated with first‐line tamoxifen (VI, N = 231). As continuous variables, increased metagene scores and decreased expression of ABAT and STC2 were correlated with poor treatment outcome in all three series of patients, except for STC2 in series IV (Table 3). As classifiers, apart from STC2 in series IV, all three biomarkers showed significant associations with PFS for all series in the Kaplan–Meier survival analyses (Figure 4).
Figure 4.
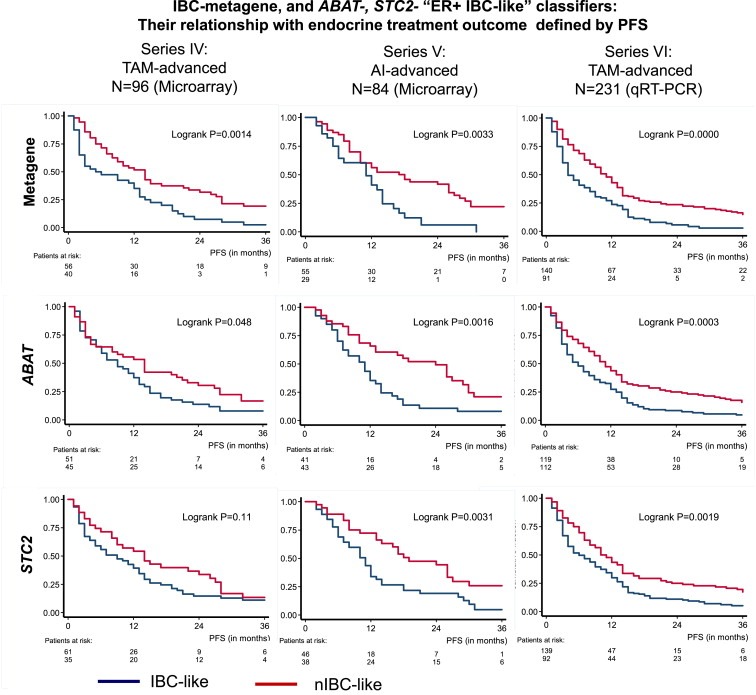
Kaplan–Meier Analyses for outcome after endocrine treatment The metagene, ABAT and STC2 as IBC/nIBC classifiers and their relation with PFS as measure for treatment outcome in advanced disease after first‐line tamoxifen (series IV and VI) and aromatase inhibitors (series V).
3.6. Biomarkers and published signatures
The biomarkers were compared for their relation with PFS in advanced disease (series IV, V, VI) with published signatures for Recurrence Score and GGI (Table 4). When compared to Recurrence Score and GGI only ABAT remained significantly associated with PFS in all series (except for GGI in series V). In contrast, the metagene score and STC2 were only independent from Recurrence Score and GGI in AI‐treated patients (series V). In summary, especially ABAT expression levels were independently associated with PFS when compared to published signatures separately and validated with RT‐qPCR.
Table 4.
Biomarkers and published signatures in advanced disease. The metagene, ABAT and STC2 were compared with the published signatures for Recurrence Score and Genomic Grade Index (GGI) for their relationship with PFS in advanced disease after treatment with tamoxifen (series IV and VI) or aromatase inhibitors (series V).
| Univariate | Bivariate (biomarker together with published signature evaluated) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Metagene score | ABAT | STC2 | |||||||
| Signature | N | HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P |
| Series IV. Tam (N = 96) | |||||||||
| Recurrence Score | |||||||||
| low | 35 | 1.00 | 1.11 (0.99–1.24) | 0.085 | 0.84 (0.71–0.99) | 0.036 | 0.94 (0.82–1.09) | 0.430 | |
| moderate | 12 | 1.01 (0.49–2.08) | 0.977 | ||||||
| high | 49 | 1.61 (1.00–2.60) | 0.050 | ||||||
| GGI | 96 | 0.99 (0.90–1.10) | 0.899 | 1.14 (1.03–1.26) | 0.010 | 0.80 (0.69–0.93) | 0.005 | 0.90 (0.78–1.03) | 0.113 |
| TAM78 | 62 | 2.34 (1.34–4.11) | 0.003 | 1.11 (0.98–1.27) | 0.102 | 0.86 (0.71–1.04) | 0.130 | 0.94 (0.79–1.11) | 0.455 |
| ROR‐S | |||||||||
| low | 33 | 1.00 | 1.10 (0.97–1.24) | 0.141 | 0.85 (0.72–1.01) | 0.065 | 0.96 (0.83–1.10) | 0.534 | |
| moderate | 30 | 1.42 (0.82–2.46) | 0.216 | ||||||
| high | 33 | 1.93 (1.14–3.25) | 0.014 | ||||||
| ROR‐P | |||||||||
| low | 23 | 1.00 | 1.13 (1.01–1.26) | 0.039 | 0.80 (0.67–0.96) | 0.018 | 0.91 (0.78–1.06) | 0.230 | |
| moderate | 37 | 0.93 (0.53–1.65) | 0.816 | ||||||
| high | 36 | 1.34 (0.77–2.33) | 0.307 | ||||||
| Series V. AI (N = 84) | |||||||||
| Recurrence Score | |||||||||
| low | 18 | 1.00 | 1.23 (1.08–1.42) | 0.003 | 0.77 (0.62–0.95) | 0.013 | 0.79 (0.67–0.93) | 0.004 | |
| moderate | 7 | 1.85 (0.64–5.36) | 0.259 | ||||||
| high | 59 | 2.86 (1.34–6.07) | 0.006 | ||||||
| GGI | 84 | 3.64 (1.87–7.08) | <0.001 | 1.19 (1.03–1.38) | 0.019 | 0.84 (0.67–1.05) | 0.124 | 0.84 (0.71–0.99) | 0.043 |
| TAM78 | 84 | 1.80 (1.08–3.00) | 0.023 | 1.26 (1.11–1.44) | 0.001 | 0.74 (0.60–0.93) | 0.008 | 0.76 (0.65–0.88) | <0.001 |
| ROR‐S | |||||||||
| Low | 24 | 1.00 | 1.19 (1.03–1.37) | 0.016 | 0.82 (0.66–1.03) | 0.087 | 0.83 (0.71–0.97) | 0.018 | |
| moderate | 31 | 1.20 (0.61–2.36) | 0.603 | ||||||
| high | 29 | 3.20 (1.65–6.21) | 0.001 | ||||||
| ROR‐P | |||||||||
| low | 18 | 1.00 | 1.19 (1.03–1.38) | 0.017 | 0.80 (0.64–1.00) | 0.050 | 0.82 (0.71–0.96) | 0.015 | |
| moderate | 38 | 1.79 (0.84–3.82) | 0.132 | ||||||
| high | 28 | 3.94 (1.81–8.59) | 0.001 | ||||||
| Series VI. Tam (N = 231), qRT‐PCR | |||||||||
| Recurrence Score | |||||||||
| low | 65 | 1.00 | 1.05 (0.99–1.11) | 0.134 | 0.88 (0.80–0.97) | 0.011 | 0.96 (0.90–1.02) | 0.196 | |
| moderate | 13 | 1.90 (1.00–3.60) | 0.048 | ||||||
| high | 131 | 2.39 (1.70–3.37) | <0.001 | ||||||
| GGI | 226 | 1.53 (1.29–1.81) | <0.001 | 1.06 (1.01–1.12) | 0.032 | 0.89 (0.81–0.97) | 0.009 | 0.94 (0.89–1.01) | 0.057 |
3.7. Biomarkers and clinicopathological predictors
Multivariate analyses were performed by adding the biomarkers separately to a base model of traditional clinicopathological factors for endocrine therapy in advanced disease (Table 5). The model included age, menopausal status, dominant site of relapse, disease‐free interval and mRNA expression levels for ESR1, PGR, and ERBB2. These multivariate analyses showed that low ABAT levels were significantly related with poor PFS in the series of 96 patients treated with tamoxifen (series IV (HR = 0.78, P = 0.027)), whereas high metagene scores (HR = 1.24, P = 0.005) and low STC2 levels (HR = 0.79, P = 0.011) were associated with poor PFS in the series of 84 patients treated with AI (series V). All three biomarkers were independently related with PFS in the RT‐qPCR validation series of 231 patients treated with tamoxifen (series VI).
Table 5.
Biomarkers and clinico‐pathological factors in advanced disease. This table provides the results of the uni‐ and multivariate Cox regression analyses for PFS performed on the advanced disease patient series (series IV–VI). The biomarkers were separately added to the base model of clinico‐pathological factors in multivariate analysis.
| Clinicopathological factors | Series IV. Tam (N = 96) | Series V. AI (N = 84) | Series VI. Tam (N = 231), qRT‐PCR | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Univariate (N = 96) | Multivariate (N = 91) | N | Univariate (N = 84) | Multivariate (N = 82) | N | Univariate (N = 231) | Multivariate (N = 231) | |||||||
| HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | ||||
| Age | |||||||||||||||
| <55 | 34 | 1.00 | 1.00 | 18 | 1.00 | 1.00 | 91 | 1.00 | 1.00 | ||||||
| 56–70 | 43 | 0.75 (0.46–1.22) | 0.250 | 1.09 (0.43–2.79) | 0.855 | 32 | 1.07 (0.56–2.05) | 0.839 | 0.86 (0.36–2.02) | 0.727 | 81 | 0.78 (0.57–1.07) | 0.123 | 0.83 (0.53–1.31) | 0.427 |
| >70 | 19 | 0.90 (0.49–1.65) | 0.742 | 1.21 (0.44–3.38) | 0.712 | 34 | 0.75 (0.38–1.46) | 0.390 | 0.59 (0.23–1.48) | 0.260 | 59 | 0.69 (0.48–0.98) | 0.038 | 0.69 (0.43–1.12) | 0.134 |
| Menopausal status | |||||||||||||||
| Premenopausal | 29 | 1.00 | 1.00 | 7 | 1.00 | 1.00 | 61 | 1.00 | 1.00 | ||||||
| Postmenopausal | 67 | 0.81 (0.51–1.30) | 0.388 | 0.99 (0.39–2.54) | 0.988 | 76 | 0.99 (0.43–2.30) | 0.978 | 1.29 (0.41–4.04) | 0.662 | 170 | 0.80 (0.59–1.09) | 0.155 | 1.14 (0.72–1.80) | 0.586 |
| Dominant site of relapse | |||||||||||||||
| Bone | 41 | 1.00 | 1.00 | 46 | 1.00 | 1.00 | 120 | 1.00 | 1.00 | ||||||
| LRR or viscera | 53 | 0.81 (0.52–1.26) | 0.345 | 0.78 (0.46–1.32) | 0.357 | 38 | 0.79 (0.48–1.31) | 0.367 | 0.86 (0.49–1.49) | 0.586 | 111 | 0.91 (0.69–1.20) | 0.518 | 0.93 (0.70–1.23) | 0.600 |
| Disease free interval | |||||||||||||||
| <1 year | 17 | 1.00 | 1.00 | 14 | 1.00 | 1.00 | 65 | 1.00 | 1.00 | ||||||
| 2–3 years | 24 | 0.43 (0.22–0.84) | 0.013 | 0.43 (0.21–0.88) | 0.021 | 28 | 0.64 (0.32–1.27) | 0.204 | 0.84 (0.39–1.79) | 0.650 | 100 | 0.58 (0.42–0.80) | 0.001 | 0.55 (0.40–0.77) | <0.001 |
| >3 years | 52 | 0.36 (0.20–0.64) | 0.001 | 0.33 (0.18–0.63) | 0.001 | 41 | 0.38 (0.19–0.75) | 0.005 | 0.44 (0.21–0.91) | 0.027 | 66 | 0.47 (0.32–0.68) | <0.001 | 0.46 (0.32–0.67) | <0.001 |
| mRNA levels as continuous variable: | |||||||||||||||
| ESR1 | 96 | 0.84 (0.73–0.95) | 0.007 | 0.81 (0.68–0.96) | 0.016 | 84 | 0.67 (0.32–1.40) | 0.285 | 1.03 (0.46–2.35) | 0.935 | 231 | 0.91 (0.85–0.97) | 0.003 | 0.93 (0.86–1.00) | 0.037 |
| PGR | 95 | 0.94 (0.86–1.03) | 0.217 | 0.99 (0.88–1.10) | 0.807 | 84 | 0.55 (0.37–0.80) | 0.002 | 0.61 (0.40–0.94) | 0.026 | 231 | 0.90 (0.84–0.97) | 0.004 | 0.92 (0.85–1.00) | 0.043 |
| HER2 | 96 | 1.05 (0.89–1.25) | 0.550 | 1.13 (0.94–1.37) | 0.198 | 84 | 1.17 (0.69–1.98) | 0.563 | 1.40 (0.79–2.50) | 0.253 | 231 | 1.13 (1.00–1.27) | 0.044 | 1.07 (0.95–1.20) | 0.279 |
| Added to the model | Added to the model | Added to the model | |||||||||||||
| Metagene score | 96 | 1.29 (1.13–1.46) | <0.001 | 1.09 (0.95–1.24) | 0.210 | 84 | 1.28 (1.13–1.45) | <0.001 | 1.24 (1.07–1.44) | 0.005 | 231 | 1.09 (1.04–1.14) | <0.001 | 1.08 (1.02–1.14) | 0.009 |
| ABAT | 96 | 0.80 (0.69–0.94) | 0.005 | 0.78 (0.63–0.97) | 0.027 | 84 | 0.74 (0.60–0.91) | 0.004 | 0.83 (0.65–1.05) | 0.127 | 231 | 0.85 (0.80–0.92) | <0.001 | 0.87 (0.79–0.96) | 0.004 |
| STC2 | 96 | 0.90 (0.79–1.03) | 0.117 | 0.99 (0.83–1.18) | 0.903 | 84 | 0.76 (0.66–0.88) | <0.001 | 0.79 (0.67–0.95) | 0.011 | 231 | 0.93 (0.88–0.98) | 0.004 | 0.93 (0.87–0.99) | 0.017 |
3.8. Biomarkers and protein expression
In an exploratory study, ABAT and STC2 protein expression were examined in 110 ER‐positive primary breast cancer specimens (Figure 5A). Evaluation of quantity and intensity separately showed for both proteins no significant relationships with PFS (Supplementary Figure 1). The quantity and intensity scores were multiplied to generate an IHC‐score for the staining and classified 77 specimens (69%) as ABAT‐positive and 78 specimens (70%) as STC2‐positive (Figure 5B). These dichotomized IHC‐scores were not related with PFS, i.e. not for ABAT (HR = 0.79; 95% CI = 0.51–1.23; P = 0.30) and not for STC2 (HR = 0.93; 95% CI = 0.60–1.44; P = 0.74).
Figure 5.
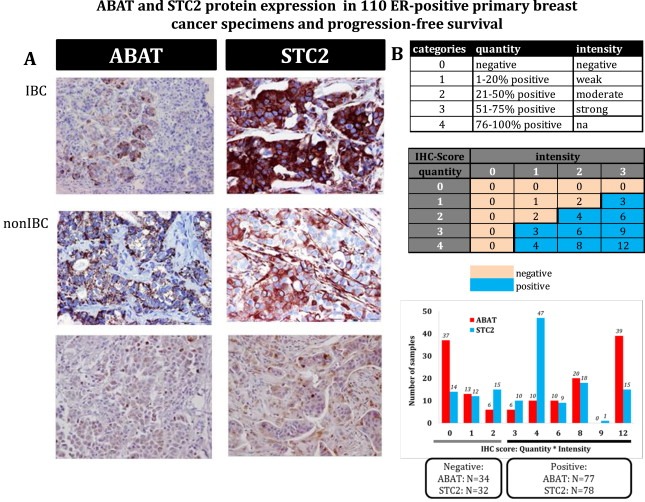
ABAT and STC2 protein expression The expression of ABAT and STC2 protein was evaluated with immunohistochemistry in 110 ER‐positive primary tumor specimens of advanced breast cancer patients treated with first‐line tamoxifen. In Figure 5A representative samples are shown for ABAT and STC2 staining in IBC and nIBC patients. Figure 5B demonstrates the staining categories for quantity, intensity, and IHC‐scores, and the distribution of IHC‐scores for ABAT and STC2. The IHC‐scores were dichotomized into positive and negative scores, identifying 77 ABAT‐positive and 78 STC2‐positive specimens. Both ABAT and STC2 protein expression had no relationship with progression free survival.
4. Discussion
Inflammatory breast cancer is a rare (∼5%) but highly aggressive form of locally advanced breast cancer with an elevated invasive and metastatic potential. It is characterized by clinical and pathological characteristics atypical for breast cancer in general, amongst others a low frequency of ER positivity. In the past, we showed that the molecular portrait of IBC indeed contained fingerprints of aggressive tumor cell behavior in breast cancer in general (Van Laere et al., 2008). Patients with IBC bearing ER expressing tumor cells constitute approximately 30% of all IBC cases and endocrine treatment in these patients is observed to be poorly effective. The molecular profile of samples from patients with ER+ IBC could provide additional hints towards unraveling the molecular biology associated with resistance to endocrine treatment. In this study, we demonstrate that, at least at the molecular level, ER+ IBC is characterized by features associated with endocrine resistance. For instance, the recurrence score and the HOXB13/IL17RB gene expression ratio are both significantly elevated in ER+ IBC compared with ER+ nIBC. Several studies have shown that elevated levels for both parameters are highly predictive of endocrine therapy resistance (Dowsett et al., 2010; Jansen et al., 2007; Ma et al., 2004; Paik et al., 2004). In addition, application of the PAM50‐algorithm (Tibshirani et al., 2002) revealed a remarkably low frequency of Luminal A‐type samples, which are shown to be more frequently responsive to endocrine treatment compared with their Luminal B‐type counterparts. This hypothesis is supported by the observation that samples from tumors with a Luminal B‐phenotype frequently exhibit high Recurrence Scores (Fan et al., 2006).
Using repetitive prediction analysis to obtain robust predictors, we identified a metagene of six genes consisting of ABAT, ADAMDEC1, CLEC7A, ETS1, ITK and STC2 to discriminate ER+ IBC from ER+ nIBC samples within the discovery series. These biomarkers were each verified and demonstrated that only the metagene and the genes ABAT and STC2 remained predictive in the test series. The metagene is a slightly better predictor than the single genes, however, its performance in the other series was largely dictated by ABAT and STC2. It is intriguing that ABAT and STC2, as 2 genes not yet linked to inflammation, discriminate between ER+ IBC and nIBC. A role in the inflammatory response, however, is less likely to be established since both ABAT and STC2 are down‐regulated in IBC compared to nIBC.
Recently, both ABAT and STC2 were described in the 100 rules used in the Absolute Intrinsic Molecular Subtyping (AIMS) (Paquet and Hallett, 2015). AIMS enables subtyping from gene expression profile at mRNA expression levels of an individual sample without the need of large, diverse, and normalized datasets. These findings indicate that both genes are highly relevant in molecular subtyping. Moreover, the molecular subtyping of breast cancer becomes more and more important in the clinical management of patients. The results of our study may therefore contribute with regard to endocrine treatment decision making.
Although our study demonstrated only that both ABAT and STC2 are just biomarkers, literature suggests for both a role in ER signaling. ABAT (MIM: 137150) has been identified as a luminal‐like gene with an ER‐binding site within 20 kb distance from the transcription start site (Krijgsman et al., 2011). This gene is incorporated in Agendia's BluePrint assay, an 80‐gene molecular subtyping profile developed in 200 breast cancer specimens and validated in four independent cohorts. ABAT encodes for 4‐aminobutyrate aminotransferase, an enzyme responsible for the catabolism of gamma‐aminobutyric acid (GABA), which might be involved in the hormonal regulation and pathogenesis of breast cancer (Opolski et al., 2000). Moreover, comparative metabolomics demonstrated alterations in glutamine and beta‐alanine metabolism along with low ABAT expression with shortened survival in ER+ and ER− breast cancer (Budczies et al., 2013).
Bouras and colleagues have shown that STC2 (MIM: 603665) as an estrogen responsive gene is co‐expressed with ER (Bouras et al., 2002). In fact, independent studies have indeed shown that STC2 is a dynamic marker of estrogen‐driven pathway activation and that constitutive expression after serum withdrawal negatively affects breast cancer cell growth, cell viability and cell migration (Raulic et al., 2008; Urruticoechea et al., 2008)). Therefore, reduced expression of STC2 in breast cancer cells enables survival and cell growth in the absence of estrogen, thereby contributing to endocrine treatment resistance. Of note, in a recent effort to redefine the molecular portraits of IBC on an extended series of 137 samples, repressed STC2 expression levels were observed in IBC in a molecular subtype‐independent manner (Van Laere et al., 2010). Future studies are needed to provide functional evidence that ABAT and STC2 are mechanistically involved in endocrine therapy response.
Based upon above considerations and findings we evaluated ABAT and STC2 further in different datasets to determine their relationship with prognosis and treatment outcome to adjuvant and first‐line endocrine therapy (i.e. tamoxifen and aromatase inhibitors). Both biomarkers were not prognostic, whereas only decreased levels of ABAT were associated with shorter MFS after adjuvant tamoxifen. In the advanced disease setting, decreased expression of ABAT and STC2 characterized patients with reduced PFS under either tamoxifen‐ or AI‐based endocrine therapy. Particularly in patients treated with first‐line tamoxifen these “ER+ IBC‐like” predictors were associated with sensitivity to endocrine treatment in an independent data set profiled with an alternative technology. The latter is important as it proves that determination of ABAT and STC2 expression levels is also applicable with standard PCR technologies, making the “bench‐to‐bedside” transition more feasible. We also explored whether ABAT and STC2 protein expression might be applicable as predictive biomarkers in immunohistochemical assays, however, our findings showed no relationship with tamoxifen outcome.
Combining these biomarkers with clinico‐pathological factors in multivariate analyses demonstrated that ABAT remained significantly associated with response to tamoxifen in two independent patient series, whereas the metagene and STC2 was only independent within the qRT‐PCR validation. Moreover, only ABAT remained independent predictive for tamoxifen in both patient series when combined with the published signatures Recurrence Score and GGI. All these multivariate analyses on different patient series summarized indicate ABAT as a robust predictor for the response to tamoxifen.
In conclusion, this study has identified an increased metagene score and decreased expression of ABAT and STC2 in IBC, and correlated the metagene and low expression of the genes with poor tamoxifen treatment outcome in the advanced setting as shown with qRT‐PCR in an independent validation. ABAT and STC2 protein expression were not informative with regard to treatment outcome. Further studies on the classifier genes are needed to elucidate the mechanism of therapy resistance.
Funding
This work was supported in part by the Translational Research Grant 220‐2008, Stichting Tegen Kanker (to LD) and Top Institute Pharma, Leiden, The Netherlands, Projects T3‐108 and T3‐502 (to EB and MJ).
Conflict of interest
None of the authors has a conflict of interest.
Authors' contributions
LS and MJ contributed equally. MJ, LS, AS and SvL designed the study. FB, IS, PR, SL, PvD, MK, CvD, JM, PV, JF, LD, and EB participated in the study design, provided tissues and clinical follow‐up data and helped to draft the manuscript. AS, CvL, DRA, ML, CvD, MT and KR carried out experiments and performed (statistical) data analyses. MJ, LS, AS, EB and SvL prepared the manuscript.
Supporting information
The following are the supplementary data related to this article:
Supplementary Figure 1 Kaplan–Meier Analyses of ABAT and STC2 protein expression for outcome after tamoxifen treatment. Quantity (Figure 1A) and intensity (Figure 1B) staining, and IHC‐scores (Figure 1C) for ABAT and STC2 protein expression in 110 ER‐positive primary tumor specimens and their relation with PFS as measure for treatment outcome in advanced disease after first‐line tamoxifen. The frequency of positive cells indicated by quantity was categorized into 0% (0), 1–20% (1), 21–50% (2), 51–75% (3) and 76–100% (4). Staining intensity was grouped into negative (0), weak (1), moderate (2), and strong (3). The IHC‐score was dichotomized into negative (0) and positive (1).
Supplementary data
Supplementary data
Acknowledgments
The authors are especially grateful to Marion Meijer‐van Gelder, Marcel Smid, and Vanja de Weerd for their contribution and technical support. The authors thank also the surgeons, pathologists, and internists of the Dutch Cancer Institute, the St. Clara Hospital, Ikazia Hospital, St. Franciscus Gasthuis at Rotterdam, and Ruwaard van Putten Hospital at Spijkenisse, and GZA Hospitals St‐Augustinus at Antwerp for the supply of tumor tissues and/or assistance in the collection of clinical follow‐up data.
Supplementary data 1.
1.1.
Supplementary data related to this article can be found online at http://dx.doi.org/10.1016/j.molonc.2015.02.006.
Jansen Maurice P.H.M., Sas Leen, Sieuwerts Anieta M., Van Cauwenberghe Caroline, Ramirez-Ardila Diana, Look Maxime, Ruigrok-Ritstier Kirsten, Finetti Pascal, Bertucci François, Timmermans Mieke M., van Deurzen Carolien H.M., Martens John W.M., Simon Iris, Roepman Paul, Linn Sabine C., van Dam Peter, Kok Marleen, Lardon Filip, Vermeulen Peter B., Foekens John A., Dirix Luc, Berns Els M.J.J., Van Laere Steven, (2015), Decreased expression of ABAT and STC2 hallmarks ER-positive inflammatory breast cancer and endocrine therapy resistance in advanced disease, Molecular Oncology, 9, doi: 10.1016/j.molonc.2015.02.006.
References
- Bekhouche, I. , Finetti, P. , Adelaide, J. , Ferrari, A. , Tarpin, C. , Charafe-Jauffret, E. , Charpin, C. , Houvenaeghel, G. , Jacquemier, J. , Bidaut, G. , Birnbaum, D. , Viens, P. , Chaffanet, M. , Bertucci, F. , 2011. High-resolution comparative genomic hybridization of inflammatory breast cancer and identification of candidate genes. PLoS One 6, e16950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertucci, F. , Finetti, P. , Rougemont, J. , Charafe-Jauffret, E. , Nasser, V. , Loriod, B. , Camerlo, J. , Tagett, R. , Tarpin, C. , Houvenaeghel, G. , Nguyen, C. , Maraninchi, D. , Jacquemier, J. , Houlgatte, R. , Birnbaum, D. , Viens, P. , 2004. Gene expression profiling for molecular characterization of inflammatory breast cancer and prediction of response to chemotherapy. Cancer Res. 64, 8558–8565. [DOI] [PubMed] [Google Scholar]
- Bouras, T. , Southey, M.C. , Chang, A.C. , Reddel, R.R. , Willhite, D. , Glynne, R. , Henderson, M.A. , Armes, J.E. , Venter, D.J. , 2002. Stanniocalcin 2 is an estrogen-responsive gene coexpressed with the estrogen receptor in human breast cancer. Cancer Res. 62, 1289–1295. [PubMed] [Google Scholar]
- Budczies, J. , Brockmoller, S.F. , Muller, B.M. , Barupal, D.K. , Richter-Ehrenstein, C. , Kleine-Tebbe, A. , Griffin, J.L. , Oresic, M. , Dietel, M. , Denkert, C. , Fiehn, O. , 2013. Comparative metabolomics of estrogen receptor positive and estrogen receptor negative breast cancer: alterations in glutamine and beta-alanine metabolism. J. Proteomics 94, 279–288. [DOI] [PubMed] [Google Scholar]
- Dawood, S. , Merajver, S.D. , Viens, P. , Vermeulen, P.B. , Swain, S.M. , Buchholz, T.A. , Dirix, L.Y. , Levine, P.H. , Lucci, A. , Krishnamurthy, S. , Robertson, F.M. , Woodward, W.A. , Yang, W.T. , Ueno, N.T. , Cristofanilli, M. , 2011. International expert panel on inflammatory breast cancer: consensus statement for standardized diagnosis and treatment. Ann. Oncol. 22, 515–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawood, S. , Ueno, N.T. , Valero, V. , Andreopoulou, E. , Hsu, L. , Lara, J. , Woodward, W. , Buchholz, T.A. , Hortobagyi, G.N. , Cristofanilli, M. , 2010. Incidence of and survival following brain metastases among women with inflammatory breast cancer. Ann. Oncol. 21, 2348–2355. [DOI] [PubMed] [Google Scholar]
- Desmedt, C. , Piette, F. , Loi, S. , Wang, Y. , Lallemand, F. , Haibe-Kains, B. , Viale, G. , Delorenzi, M. , Zhang, Y. , d'Assignies, M.S. , Bergh, J. , Lidereau, R. , Ellis, P. , Harris, A.L. , Klijn, J.G. , Foekens, J.A. , Cardoso, F. , Piccart, M.J. , Buyse, M. , Sotiriou, C. , Consortium, T. , 2007. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin. Cancer Res. 13, 3207–3214. [DOI] [PubMed] [Google Scholar]
- Dirix, L.Y. , Van Dam, P. , Prove, A. , Vermeulen, P.B. , 2006. Inflammatory breast cancer: current understanding. Curr. Opin. Oncol. 18, 563–571. [DOI] [PubMed] [Google Scholar]
- Dowsett, M. , Cuzick, J. , Wale, C. , Forbes, J. , Mallon, E.A. , Salter, J. , Quinn, E. , Dunbier, A. , Baum, M. , Buzdar, A. , Howell, A. , Bugarini, R. , Baehner, F.L. , Shak, S. , 2010. Prediction of risk of distant recurrence using the 21-gene recurrence score in node-negative and node-positive postmenopausal patients with breast cancer treated with anastrozole or tamoxifen: a TransATAC study. J. Clin. Oncol. 28, 1829–1834. [DOI] [PubMed] [Google Scholar]
- Early Breast Cancer Trialists' Collaborative, G 2005. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet 365, 1687–1717. [DOI] [PubMed] [Google Scholar]
- Ellis, M.J. , Suman, V.J. , Hoog, J. , Lin, L. , Snider, J. , Prat, A. , Parker, J.S. , Luo, J. , DeSchryver, K. , Allred, D.C. , Esserman, L.J. , Unzeitig, G.W. , Margenthaler, J. , Babiera, G.V. , Marcom, P.K. , Guenther, J.M. , Watson, M.A. , Leitch, M. , Hunt, K. , Olson, J.A. , 2011. Randomized phase II neoadjuvant comparison between letrozole, anastrozole, and exemestane for postmenopausal women with estrogen receptor-rich stage 2 to 3 breast cancer: clinical and biomarker outcomes and predictive value of the baseline PAM50-based intrinsic subtype–ACOSOG Z1031. J. Clin. Oncol. 29, 2342–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, C. , Oh, D.S. , Wessels, L. , Weigelt, B. , Nuyten, D.S. , Nobel, A.B. , van't Veer, L.J. , Perou, C.M. , 2006. Concordance among gene-expression-based predictors for breast cancer. N. Engl. J. Med. 355, 560–569. [DOI] [PubMed] [Google Scholar]
- Goeman, J.J. , van de Geer, S.A. , de Kort, F. , van Houwelingen, H.C. , 2004. A global test for groups of genes: testing association with a clinical outcome. Bioinformatics 20, 93–99. [DOI] [PubMed] [Google Scholar]
- Hance, K.W. , Anderson, W.F. , Devesa, S.S. , Young, H.A. , Levine, P.H. , 2005. Trends in inflammatory breast carcinoma incidence and survival: the surveillance, epidemiology, and end results program at the National Cancer Institute. J. Natl. Cancer Inst. 97, 966–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward, J.L. , Rubens, R.D. , Carbone, P.P. , Heuson, J.C. , Kumaoka, S. , Segaloff, A. , 1978. Assessment of response to therapy in advanced Breast-Cancer. Br. J. Cancer 38, 201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, M.P. , Foekens, J.A. , van Staveren, I.L. , Dirkzwager-Kiel, M.M. , Ritstier, K. , Look, M.P. , Meijer-van Gelder, M.E. , Sieuwerts, A.M. , Portengen, H. , Dorssers, L.C. , Klijn, J.G. , Berns, E.M. , 2005. Molecular classification of tamoxifen-resistant breast carcinomas by gene expression profiling. J. Clin. Oncol. 23, 732–740. [DOI] [PubMed] [Google Scholar]
- Jansen, M.P. , Knijnenburg, T. , Reijm, E.A. , Simon, I. , Kerkhoven, R. , Droog, M. , Velds, A. , van Laere, S. , Dirix, L. , Alexi, X. , Foekens, J.A. , Wessels, L. , Linn, S.C. , Berns, E.M. , Zwart, W. , 2013. Hallmarks of aromatase inhibitor drug resistance revealed by epigenetic profiling in breast cancer. Cancer Res. 73, 6632–6641. [DOI] [PubMed] [Google Scholar]
- Jansen, M.P. , Sieuwerts, A.M. , Look, M.P. , Ritstier, K. , Meijer-van Gelder, M.E. , van Staveren, I.L. , Klijn, J.G. , Foekens, J.A. , Berns, E.M. , 2007. HOXB13-to-IL17BR expression ratio is related with tumor aggressiveness and response to tamoxifen of recurrent breast cancer: a retrospective study. J. Clin. Oncol. 25, 662–668. [DOI] [PubMed] [Google Scholar]
- Kok, M. , Linn, S.C. , Van Laar, R.K. , Jansen, M.P. , van den Berg, T.M. , Delahaye, L.J. , Glas, A.M. , Peterse, J.L. , Hauptmann, M. , Foekens, J.A. , Klijn, J.G. , Wessels, L.F. , Van't Veer, L.J. , Berns, E.M. , 2009. Comparison of gene expression profiles predicting progression in breast cancer patients treated with tamoxifen. Breast Cancer Res. Treat. 113, 275–283. [DOI] [PubMed] [Google Scholar]
- Krijgsman, O. , Roepman, P. , Zwart, W. , Carroll, J.S. , Tian, S. , de Snoo, F.A. , Bender, R.A. , Bernards, R. , Glas, A.M. , 2012. A diagnostic gene profile for molecular subtyping of breast cancer associated with treatment response. Breast Cancer Res. Treat. 133, 37–47. [DOI] [PubMed] [Google Scholar]
- Leary, A.F. , Drury, S. , Detre, S. , Pancholi, S. , Lykkesfeldt, A.E. , Martin, L.A. , Dowsett, M. , Johnston, S.R. , 2010. Lapatinib restores hormone sensitivity with differential effects on estrogen receptor signaling in cell models of human epidermal growth factor receptor 2-negative breast cancer with acquired endocrine resistance. Clin. Cancer Res. 16, 1486–1497. [DOI] [PubMed] [Google Scholar]
- Lerebours, F. , Vacher, S. , Andrieu, C. , Espie, M. , Marty, M. , Lidereau, R. , Bieche, I. , 2008. NF-kappa B genes have a major role in inflammatory breast cancer. BMC Cancer 8, 41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loi, S. , Haibe-Kains, B. , Desmedt, C. , Wirapati, P. , Lallemand, F. , Tutt, A.M. , Gillet, C. , Ellis, P. , Ryder, K. , Reid, J.F. , Daidone, M.G. , Pierotti, M.A. , Berns, E.M. , Jansen, M.P. , Foekens, J.A. , Delorenzi, M. , Bontempi, G. , Piccart, M.J. , Sotiriou, C. , 2008. Predicting prognosis using molecular profiling in estrogen receptor-positive breast cancer treated with tamoxifen. BMC Genomics 9, 239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, X.J. , Hilsenbeck, S.G. , Wang, W. , Ding, L. , Sgroi, D.C. , Bender, R.A. , Osborne, C.K. , Allred, D.C. , Erlander, M.G. , 2006. The HOXB13:IL17BR expression index is a prognostic factor in early-stage breast cancer. J. Clin. Oncol. 24, 4611–4619. [DOI] [PubMed] [Google Scholar]
- Ma, X.J. , Wang, Z. , Ryan, P.D. , Isakoff, S.J. , Barmettler, A. , Fuller, A. , Muir, B. , Mohapatra, G. , Salunga, R. , Tuggle, J.T. , Tran, Y. , Tran, D. , Tassin, A. , Amon, P. , Wang, W. , Enright, E. , Stecker, K. , Estepa-Sabal, E. , Smith, B. , Younger, J. , Balis, U. , Michaelson, J. , Bhan, A. , Habin, K. , Baer, T.M. , Brugge, J. , Haber, D.A. , Erlander, M.G. , Sgroi, D.C. , 2004. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell 5, 607–616. [DOI] [PubMed] [Google Scholar]
- McShane, L.M. , Altman, D.G. , Sauerbrei, W. , Taube, S.E. , Gion, M. , Clark, G.M. , Statistics Subcommittee of, N.C.I.E.W.G.o.C.D2006. REporting recommendations for tumor MARKer prognostic studies (REMARK). Breast Cancer Res. Treat. 100, 229–235. [DOI] [PubMed] [Google Scholar]
- Opolski, A. , Mazurkiewicz, M. , Wietrzyk, J. , Kleinrok, Z. , Radzikowski, C. , 2000. The role of GABA-ergic system in human mammary gland pathology and in growth of transplantable murine mammary cancer. J. Exp. Clin. Cancer Res. 19, 383–390. [PubMed] [Google Scholar]
- Paik, S. , Shak, S. , Tang, G. , Kim, C. , Baker, J. , Cronin, M. , Baehner, F.L. , Walker, M.G. , Watson, D. , Park, T. , Hiller, W. , Fisher, E.R. , Wickerham, D.L. , Bryant, J. , Wolmark, N. , 2004. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 351, 2817–2826. [DOI] [PubMed] [Google Scholar]
- Paquet, E.R. , Hallett, M.T. , 2015. Absolute assignment of breast cancer intrinsic molecular subtype. J. Natl. Cancer Inst. 107, 357 [DOI] [PubMed] [Google Scholar]
- Raulic, S. , Ramos-Valdes, Y. , DiMattia, G.E. , 2008. Stanniocalcin 2 expression is regulated by hormone signalling and negatively affects breast cancer cell viability in vitro. J. Endocrinol. 197, 517–529. [DOI] [PubMed] [Google Scholar]
- Reijm, E.A. , Timmermans, A.M. , Look, M.P. , Meijer-van Gelder, M.E. , Stobbe, C.K. , van Deurzen, C.H. , Martens, J.W. , Sleijfer, S. , Foekens, J.A. , Berns, P.M. , Jansen, M.P. , 2014. High protein expression of EZH2 is related to unfavorable outcome to tamoxifen in metastatic breast cancer. Ann. Oncol. 25, 2185–2190. [DOI] [PubMed] [Google Scholar]
- Schmidt, M. , Bohm, D. , von Torne, C. , Steiner, E. , Puhl, A. , Pilch, H. , Lehr, H.A. , Hengstler, J.G. , Kolbl, H. , Gehrmann, M. , 2008. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res. 68, 5405–5413. [DOI] [PubMed] [Google Scholar]
- Sieuwerts, A.M. , Meijer-van Gelder, M.E. , Timmermans, M. , Trapman, A.M. , Garcia, R.R. , Arnold, M. , Goedheer, A.J. , Portengen, H. , Klijn, J.G. , Foekens, J.A. , 2005. How ADAM-9 and ADAM-11 differentially from estrogen receptor predict response to tamoxifen treatment in patients with recurrent breast cancer: a retrospective study. Clin. Cancer Res. 11, 7311–7321. [DOI] [PubMed] [Google Scholar]
- Tibshirani, R. , Hastie, T. , Narasimhan, B. , Chu, G. , 2002. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc. Natl. Acad. Sci. U. S. A. 99, 6567–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Arzayus, M.I. , Zhao, J. , Bronson, R. , Brown, M. , 2010. Estrogen-dependent and estrogen-independent mechanisms contribute to AIB1-mediated tumor formation. Cancer Res. 70, 4102–4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urruticoechea, A. , Aguilar, H. , Sole, X. , Capella, G. , Martin, L.A. , Dowsett, M. , Germa-Lluch, J.R. , 2008. Pre-clinical validation of early molecular markers of sensitivity to aromatase inhibitors in a mouse model of post-menopausal hormone-sensitive breast cancer. Breast Cancer Res. Treat. 109, 463–470. [DOI] [PubMed] [Google Scholar]
- van Agthoven, T. , Sieuwerts, A.M. , Meijer-van Gelder, M.E. , Look, M.P. , Smid, M. , Veldscholte, J. , Sleijfer, S. , Foekens, J.A. , Dorssers, L.C. , 2009. Relevance of breast cancer antiestrogen resistance genes in human breast cancer progression and tamoxifen resistance. J. Clin. Oncol. 27, 542–549. [DOI] [PubMed] [Google Scholar]
- Van der Auwera, I. , Yu, W. , Suo, L. , Van Neste, L. , van Dam, P. , Van Marck, E.A. , Pauwels, P. , Vermeulen, P.B. , Dirix, L.Y. , Van Laere, S.J. , 2010. Array-based DNA methylation profiling for breast cancer subtype discrimination. PLoS One 5, e12616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Laere, S. , Beissbarth, T. , Van der Auwera, I. , Van den Eynden, G. , Trinh, X.B. , Elst, H. , Van Hummelen, P. , van Dam, P. , Van Marck, E. , Vermeulen, P. , Dirix, L. , 2008. Relapse-free survival in breast cancer patients is associated with a gene expression signature characteristic for inflammatory breast cancer. Clin. Cancer Res. 14, 7452–7460. [DOI] [PubMed] [Google Scholar]
- Van Laere, S. , Limame, R. , Van Marck, E.A. , Vermeulen, P.B. , Dirix, L.Y. , 2010. Is there a role for mammary stem cells in inflammatory breast carcinoma?: a review of evidence from cell line, animal model, and human tissue sample experiments. Cancer 116, 2794–2805. [DOI] [PubMed] [Google Scholar]
- Van Laere, S. , Van der Auwera, I. , Van den Eynden, G. , Van Hummelen, P. , van Dam, P. , Van Marck, E. , Vermeulen, P.B. , Dirix, L. , 2007. Distinct molecular phenotype of inflammatory breast cancer compared to non-inflammatory breast cancer using Affymetrix-based genome-wide gene-expression analysis. Br. J. Cancer 97, 1165–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Laere, S. , Van der Auwera, I. , Van den Eynden, G.G. , Fox, S.B. , Bianchi, F. , Harris, A.L. , van Dam, P. , Van Marck, E.A. , Vermeulen, P.B. , Dirix, L.Y. , 2005. Distinct molecular signature of inflammatory breast cancer by cDNA microarray analysis. Breast Cancer Res. Treat. 93, 237–246. [DOI] [PubMed] [Google Scholar]
- Van Laere, S.J. , Van der Auwera, I. , Van den Eynden, G.G. , Elst, H.J. , Weyler, J. , Harris, A.L. , van Dam, P. , Van Marck, E.A. , Vermeulen, P.B. , Dirix, L.Y. , 2006. Nuclear factor-kappaB signature of inflammatory breast cancer by cDNA microarray validated by quantitative real-time reverse transcription-PCR, immunohistochemistry, and nuclear factor-kappaB DNA-binding. Clin. Cancer Res. 12, 3249–3256. [DOI] [PubMed] [Google Scholar]
- Van Laere, S.J. , Van der Auwera, I. , Van den Eynden, G.G. , van Dam, P. , Van Marck, E.A. , Vermeulen, P.B. , Dirix, L.Y. , 2007. NF-kappaB activation in inflammatory breast cancer is associated with oestrogen receptor downregulation, secondary to EGFR and/or ErbB2 overexpression and MAPK hyperactivation. Br. J. Cancer 97, 659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Klijn, J.G. , Zhang, Y. , Sieuwerts, A.M. , Look, M.P. , Yang, F. , Talantov, D. , Timmermans, M. , Meijer-van Gelder, M.E. , Yu, J. , Jatkoe, T. , Berns, E.M. , Atkins, D. , Foekens, J.A. , 2005. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 365, 671–679. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following are the supplementary data related to this article:
Supplementary Figure 1 Kaplan–Meier Analyses of ABAT and STC2 protein expression for outcome after tamoxifen treatment. Quantity (Figure 1A) and intensity (Figure 1B) staining, and IHC‐scores (Figure 1C) for ABAT and STC2 protein expression in 110 ER‐positive primary tumor specimens and their relation with PFS as measure for treatment outcome in advanced disease after first‐line tamoxifen. The frequency of positive cells indicated by quantity was categorized into 0% (0), 1–20% (1), 21–50% (2), 51–75% (3) and 76–100% (4). Staining intensity was grouped into negative (0), weak (1), moderate (2), and strong (3). The IHC‐score was dichotomized into negative (0) and positive (1).
Supplementary data
Supplementary data


