Abstract
Adeno-associated viruses (AAVs) are commonly used for in vivo gene transfer. Nevertheless, AAVs that provide efficient transduction across specific organs or cell populations are needed. Here, we describe AAV-PHP.eB and AAV-PHP.S, capsids that efficiently transduce the central and peripheral nervous systems, respectively. In the adult mouse, intravenous administration of 1×1011 vector genomes (vg) of AAV-PHP.eB transduced 69% of cortical and 55% of striatal neurons, while 1×1012 vg AAV-PHP.S transduced 82% of dorsal root ganglion neurons, as well as cardiac and enteric neurons. The efficiency of these vectors facilitates robust co-transduction and stochastic, multicolor labeling for individual cell morphology studies. To support such efforts, we provide methods for labeling a tunable fraction of cells without compromising color diversity. Furthermore, when used with cell type-specific promoters, these AAVs provide targeted gene expression across the nervous system and enable efficient and versatile gene manipulation throughout the nervous system of transgenic and non-transgenic animals.
INTRODUCTION
Adeno-associated viruses (AAVs)1 have been extensively used as vehicles for gene transfer to the nervous system enabling gene expression and knockdown, gene editing2,3, circuit modulation4,5, in vivo imaging6,7, disease model development8, and the evaluation of therapeutic candidates for the treatment of neurological diseases9. AAVs are well suited for these applications because they provide safe, long-term expression in the nervous system10,11. Most of these applications rely on local AAV injections into the adult brain to bypass the blood-brain barrier (BBB) and to temporally and spatially restrict transgene expression.
Targeted AAV injections have also been used for gene delivery to peripheral neurons to test strategies for treating chronic pain12,13 and for tracing, monitoring, and modulating specific subpopulations of vagal neurons14,15. Many peripheral neuron populations, however, are difficult to access surgically (e.g., dorsal root ganglia (DRG), nodose ganglia, sympathetic chain ganglia, and cardiac ganglia) or are widely distributed (e.g., the enteric nervous system), thereby limiting methods for genetic manipulation of these targets. Likewise, in the CNS, single localized injections may be insufficient to study circuits in larger species16 or to test gene therapies for diseases that involve the entire nervous system or widely distributed cell populations (e.g., Parkinson’s, Huntington’s, amyotrophic lateral sclerosis, Alzheimer’s, spinal muscular atrophy, Friedreich’s ataxia, and numerous lysosomal storage diseases)9.
Systemic AAV delivery provides a non-invasive alternative for broad gene delivery to the nervous system17; however, the high viral load required and relatively low transduction efficiency have limited wide adoption of this method. Several groups have developed AAVs that enhance gene transfer to the CNS after intravenous delivery. The recently reported AAV-AS capsid18, which utilizes a polyalanine N-terminal extension to the AAV9.4719 VP2 capsid protein, provides higher neuronal transduction, particularly in the striatum, which may have applications for Huntington’s disease. Similarly, the AAV-BR1 capsid20, based on AAV2, may be useful for applications that require more efficient and selective transduction of brain endothelial cells. Using a cell type-specific capsid selection method we developed called CREATE (Cre REcombinase-based AAV Targeted Evolution), we recently identified AAV-PHP.B, a capsid that transduces the majority of neurons and astrocytes across many regions of the adult mouse brain and spinal cord after intravenous injection21. While the efficiency of AAV-PHP.B opens up new possibilities for CNS-wide genetic modification, it requires a substantial dose of vector (e.g., 1 × 1012 vg per adult mouse or higher).
Here, we used CREATE to further evolve AAV-PHP.B for more efficient transduction of neurons throughout the adult mouse brain and spinal cord. We describe a novel enhanced variant of AAV-PHP.B, AAV-PHP.eB, which lowers the viral load required to transduce the majority of CNS neurons. Notably, we also report the characterization of a second capsid variant, AAV-PHP.S, that displays improved tropism towards peripheral neurons, including those in the DRG, cardiac ganglia, and enteric nervous system.
AAVs are also used for the bulk study of neuronal anatomical connectivity and morphology22 and are components of multi-viral strategies for tracing the relationships between bulk inputs and outputs23. At the single-cell level, AAV-based multicolor labeling systems24 have been developed with the goal of improving tracing efforts. However, the lack of control over the labeling density and uniformity of color diversity has been a persistent challenge24. To overcome these challenges, we have developed a two-component viral vector system to stochastically label cells with a wide range of hues while independently controlling the fraction of cells labeled. In addition, using the novel capsids reported here, we expressed a variety of fluorescent reporters under different cell type-specific promoters, supporting the potential use of these vectors for population-wide genetic manipulations of the nervous system in Cre transgenic or wild-type mice.
RESULTS
Engineered AAV capsids for efficient transduction across the central and peripheral nervous systems
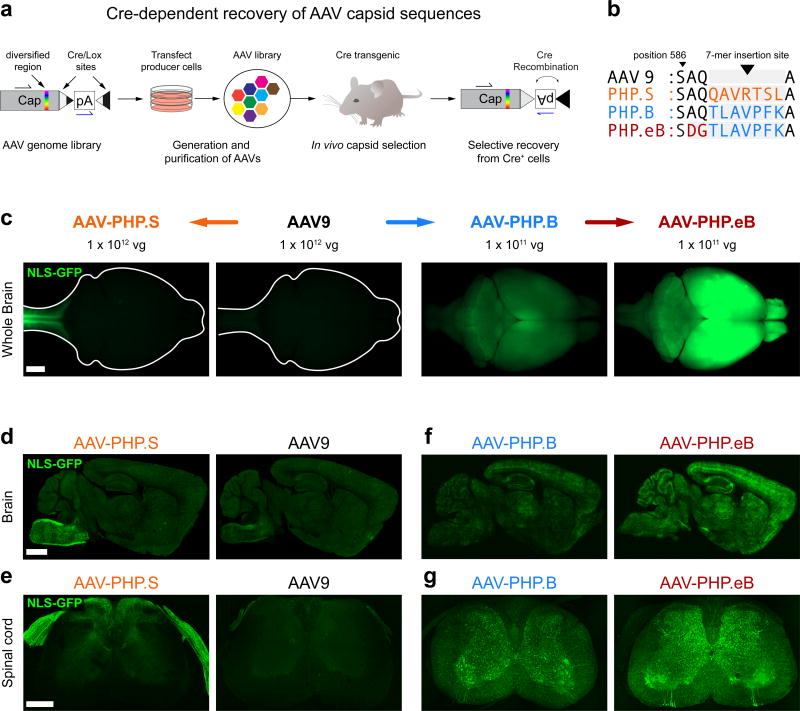
Here we report two novel AAV capsids that provide increased gene transfer to the CNS and the PNS via the vasculature. We developed these vectors by applying the previously published in vivo capsid selection method, CREATE21 (Fig. 1a) to AAV-PHP.B and AAV9 as parental capsids. First, we sought to evolve the previously described AAV-PHP.B vector for more efficient tropism towards the CNS. We generated an AAV capsid library in which the AAV-PHP.B 7-mer targeting sequence was modified by randomizing three consecutive amino acids (AAs) in an overlapping fashion across the 7-mer and flanking AAs (Methods). To provide selective pressure for capsids that transduce both neurons and glia, we subjected the library to parallel in vivo selections in adult Vglut2-IRES-Cre, Vgat-IRES-Cre, and GFAP-Cre mice, which express Cre in glutamatergic neurons, GABAergic neurons and astrocytes, respectively. After two rounds of selection, we found that a variant comprising the sequence DGT in place of the AAV-PHP.B sequence AQTLAVPFK at AA positions 587–589 (Fig. 1b) was enriched in all three transgenic lines (11.1%, 12.1%, and 15.7% of the total recovered sequences, respectively). We used this DGT substitution variant or AAV-PHP.B to package a single-stranded (ss) AAV reporter genome that expresses a nuclear-localized GFP (NLS-GFP) from the constitutive CAG promoter, (ssAAV-CAG-NLS-GFP), and delivered 1 × 1011 vg of the viruses by intravenous injection to adult mice. We found that the DGT substitution variant appeared to transduce the CNS more efficiently than AAV-PHP.B as judged by wide-field fluorescence microscopy of the intact brain (Fig. 1c) and confocal microscopy on thin sections from brain (Fig. 1f) and spinal cord (Fig. 1g). We refer to this enhanced AAV-PHP.B variant as AAV-PHP.eB and present further quantitative characterization below. Second, we performed a separate selection of an AAV9-based 7-mer library in GFAP-Cre mice21. After two rounds of selection, we found a variant (7-mer sequence – QAVRTSL, representing 33.3% of the total sequences recovered; Fig. 1b). As above, we evaluated the capsid against AAV9, its parent capsid, by packaging the ssAAV-CAG-NLS-GFP reporter and delivering the vectors at 1 × 1012 vg/mouse. While the QAVRTSL variant and AAV9 both showed similar sparse transduction of the brain (Fig. 1c, d), the QAVRTSL variant notably appeared to strongly transduce peripheral sensory afferents entering the spinal cord and brain stem (Fig. 1c–e). We refer to this variant as AAV-PHP.S. While unexpected given the selection scheme, the improved tropism for sensory neurons provided by AAV-PHP.S could be useful for widespread gene transfer to cells of the PNS. We explore this possibility below.
Figure 1. Engineered AAV capsids for efficient transduction across the peripheral and central nervous systems.
(a) Schematic of the CREATE selection method. (b) The amino acid (AA) sequences for the 7-mer insertions and flanking sequences for AAV-PHP.S, .B and .eB; the 7-mer and adjacent substitutions are highlighted in colored text. (c–g) ssAAV-CAG-NLS-GFP was packaged into the indicated capsid and intravenously injected into adult mice at 1 × 1012 vg/mouse (AAV9 and AAV-PHP.S; c–e) or 1 × 1011 vg/mouse (AAV-PHP.B and AAV-PHP.eB; c, f, and g). (c) Representative whole-brain fluorescence images after three weeks of expression. (d–g) Representative confocal images of native GFP fluorescence from sagittal brain sections (d and f) and transverse spinal cord sections (e and g) are shown for the indicated capsids. For (d–g), all imaging and display conditions are matched across panel pairs to allow for comparisons. Panels (d) and (f) are 40 µm maximum intensity projections (MIPs) and panels (e) and (g) are 300 µm MIPs. Scale bars for (c–e) are 1 mm.
Together these data indicate that the tropism of AAV9 can be routed towards either more efficient CNS or PNS transduction (Fig. 1c), depending on the AA insertion/modification. A comparison of the AA insertion/modification sequences of AAV-PHP.eB and AAV-PHP.S along with their parent capsids, AAV-PHP.B and AAV9, respectively, is shown in Fig. 1b, and their nucleotide sequences and relative viral production efficiencies are provided in (Supplementary Table 1).
Quantification of transduction by AAV-PHP.eB and AAV-PHP.S
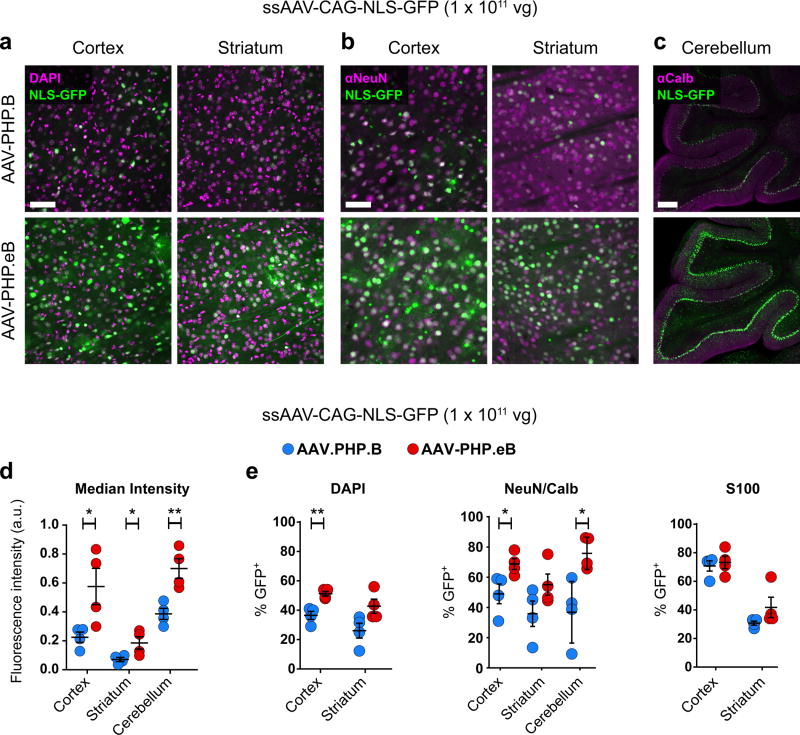
Next, we quantified the transduction efficiency of AAV-PHP.eB and AAV-PHP.B three weeks after intravenous administration of 1 × 1011 vg of ssAAV-CAG-NLS-GFP. First, when compared with AAV-PHP.B, AAV-PHP.eB provided significantly increased (> 2.5-fold) GFP fluorescence per cell nucleus throughout the cortex and striatum and (> 1.8-fold) in cerebellar Purkinje cells (Fig. 2d and Supplementary Fig. 1). To quantify expression in Purkinje cells, an automated cell segmentation and analysis pipeline was developed (Supplementary Fig. 2 and Methods). AAV-PHP.eB also showed an increase over AAV-PHP.B in both the percentage of total cortical cells transduced (DAPI+ cells that express GFP; 51 ± 2% vs. 37 ± 3%; Fig. 2a,e) and the percentage of NeuN+ neurons transduced (69 ± 4% vs. 49 ± 7%; Fig. 2b,e). In the striatum, a similar trend towards increased transduction by AAV-PHP.eB compared to AAV-PHP.B was observed from quantifying both the percentage of DAPI+ and NeuN+ cells that express GFP (DAPI+: 42 ± 5% vs. 26 ± 5%; Fig. 2a,e; NeuN+: 55 ± 7% vs 36 ± 8%, Fig. 2b,e). In the cerebellum, we observed an increase in the percentage of Calbindin+ Purkinje cells that expressed GFP in mice injected with AAV-PHP.eB as compared with AAV-PHP.B (76 ± 5% vs. 37 ± 10%, Fig. 2c,e). Both vectors transduced a similar percentage of S100+ glia in the cortex as well as the striatum (Fig. 2e and Supplementary Fig. 3).
Figure 2. AAV-PHP.eB transduces several CNS regions more efficiently than AAV-PHP.B.
ssAAV-PHP.B:CAG-NLS-GFP or ssAAV-PHP.eB:CAG-NLS-GFP was intravenously injected into adult mice at 1 × 1011 vg/mouse. Native GFP fluorescence was assessed after three weeks of expression. (a and b) Representative images of native GFP fluorescence (green) and DAPI staining (a, magenta) or NeuN staining (b, magenta) in the cortex and striatum. (c) Representative images of Calbindin immunohistochemistry (IHC) (magenta) in the cerebellum. (d) Mean GFP intensity in all DAPI+ nuclei in the cortex (t6 = 2.688; P = 0.0361) and striatum (t6 = 2.536; P = 0.0443) or in Calbindin+ cells in the cerebellum (t6 = 4.007; P = 0.0071). (e) Quantification of cell transduction. From left to right: The percentage of DAPI+ cells that express GFP in the cortex (t6 = 4.669; P = 0.0034) and striatum (t6 = 2.390; P = 0.0541). The percentage of NeuN+ cells that express GFP in the cortex (t6 = 2.662; P = 0.037) and in the striatum (t6 = 1.764; P = 0.128). The percentage of Calbindin+ cerebellar Purkinje cells that express GFP (t6 = 3.328; P = 0.039). The percentage of S100+ cells that express GFP in the cortex (t6 = 0.4422; P = 0.6738) and striatum (t6 = 1.512; P = 0.1814). (d and e) For quantification: n = 4 mice per group, mean ± SEM, unpaired, two-tailed t-test (***P ≤ 0.001; **P ≤ 0.01; *P ≤ 0.05; n.s., P ≥ 0.05). All images were single-plane confocal images of native GFP fluorescence. Scale bars are 50 µm (a and b) and 100 µm (c).
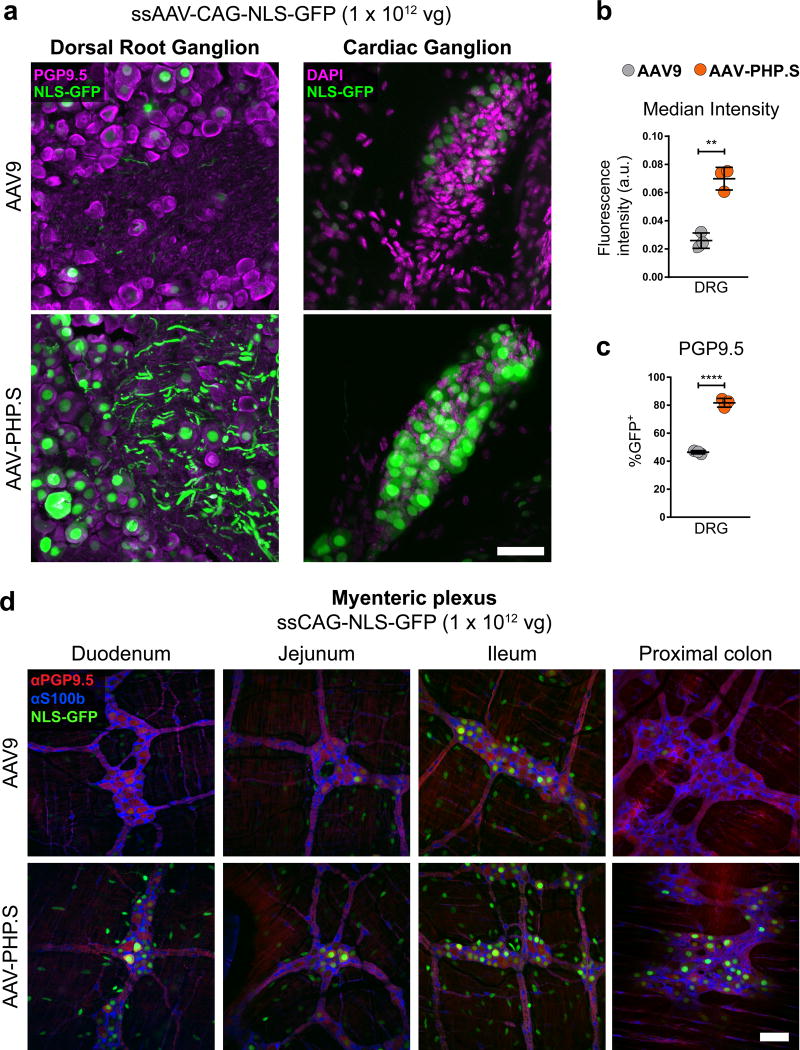
Next, we examined the transduction efficiency of AAV-PHP.S compared with AAV9, a vector known to provide broad transduction of the PNS and many peripheral organs after intravenous delivery17,25,26. We packaged ssAAV-CAG-NLS-GFP into AAV9 or AAV-PHP.S, administered the viruses to adult mice by intravenous injection at 1 × 1012 vg/mouse, and then assessed GFP expression after three weeks. In the DRG, AAV-PHP.S provided a significant increase in transduction over AAV9 (Fig. 3a) as quantified both by native GFP fluorescence per cell nucleus (> 2.6-fold) (Fig. 3b) and by the percentage of cells positive for PGP9.5, a marker for neurons in the PNS, that co-expressed GFP (82 ± 2% vs. 46 ± 0.7%) (Fig. 3c). AAV-PHP.S also transduced the cardiac ganglia (Fig. 3a) and cells within the enteric nervous system of the small intestine and colon (Fig. 3d and Supplementary Video 1) with high efficiency. To determine the identity of enteric nervous system cells transduced by AAV-PHP.S, we performed immunohistochemistry (IHC) for the neuronal marker PGP9.5 and the astrocyte marker S100b. The majority of GFP+ cells within the myenteric plexus (Fig. 3d) and submucosal plexus (Supplementary Fig. 4) of the small intestine and colon co-localized to PGP9.5+ cells with sparse and faint transduction seen in S100b+ cells (Supplementary Video 1). Notably, we also observed robust transduction by AAV-PHP.S of cells in many organs, including the liver, lungs, heart, and stomach cells within the muscle and myenteric plexus layers (Supplementary Fig. 4).
Figure 3. AAV-PHP.S efficiently transduces peripheral neurons.
ssAAV-PHP.S:CAG-NLS-GFP or ssAAV9:CAG-NLS-GFP was intravenously injected into adult mice at 1 × 1012 vg/mouse. Native GFP fluorescence was assessed after three weeks of expression. (a) Representative images of GFP expression and neuronal PGP9.5 (left, magenta) or DAPI (right, magenta) staining in a dorsal root ganglion (DRG, left) and cardiac ganglion (right). (b) Quantification of the mean GFP fluorescence intensity per cell with AAV-PHP.S or AAV9 (t4 = 7.814; P = 0.0014) (c) Quantification of the percentage of PGP9.5+ cells transduced (t4 = 18.29; P < 0.0001) (d) Representative images of GFP expression, with neuronal PGP9.5 (red) and astrocyte S100b (blue) staining in the myenteric plexus of the duodenum, jejunum, lleum and proximal colon. In (b and c) n = 3 independent animals per group, mean ± SEM, unpaired, two-tailed t-test. For representative images (a; n = 3 animals) and (d; n = 2 animals). All imaging and display conditions are matched in the GFP channel across panel pairs. Scale bars in (a and d) are 50 µm.
AAV-PHP.eB and AAV-PHP.S enable efficient co-transduction of neurons
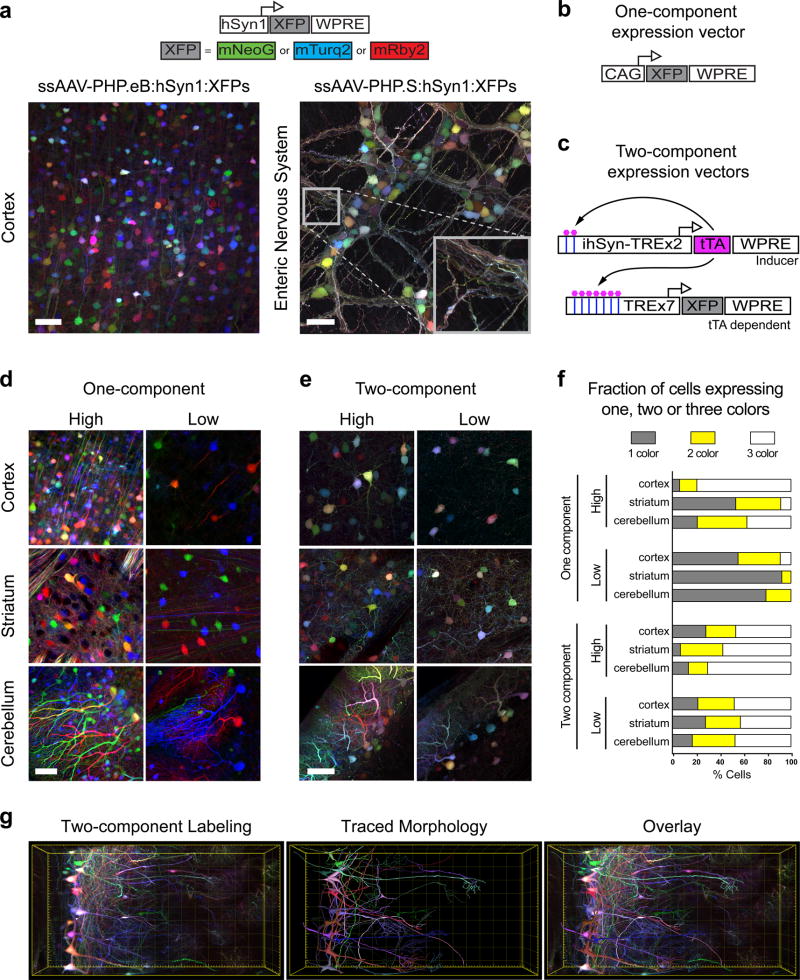
The increased transduction efficiency and enhanced expression per cell observed with both AAV-PHP.eB and AAV-PHP.S, relative to their parental capsids, suggested that a large fraction of cells are transduced by multiple AAV genomes when administered at the above doses. The transduction efficiency of these vectors may therefore enable applications that require expression from more than one AAV genome. To test this, we co-administered a cocktail of three AAV genomes with the single fluorescent proteins (XFPs) mRuby2, mNeonGreen, and mTurquoise2 expressed from the human Synapsin (hSyn1) promoter, which limits expression to neurons. The three-vector mix (AAV-hSyn1-3XFP) was packaged into AAV-PHP.eB or AAV-PHP.S and injected into adult wild-type mice. Co-administration of these vectors resulted in dense labeling of cells with multiple colors in the brain when delivered with AAV-PHP.eB and in the enteric nervous system with AAV-PHP.S (Fig. 4a). Because cells transduced by a single vector would express only one of the three XFPs, and thereby be red, green or blue, the wide color diversity seen in both the brain and the enteric nervous system demonstrates that these novel capsids enable the independent and stochastic co-transduction of neurons by multiple viral vectors.
Figure 4. AAV-PHP.eB and AAV-PHP.S transfer multiple genomes per cell and enable tunable multicolor labeling.
(a) Co-administration of a cocktail of three ssAAV vectors that each express one of three XFPs (top, schematic) from the hSyn1 promoter labels individual cells with a range of hues. Representative images of the cortex (left, AAV-PHP.eB; total dose: 4 × 1011 vg) and the enteric nervous system (right, AAV-PHP.S; total dose: 1 × 1012 vg). The inset highlights the dense network of neurites labeled with a range of hues. Schematics show one-component (b) and tTA-TRE-based two-component systems (c). Expression from the TRE-XFP vectors is dependent on co-transduction with a tTA (tet-off) inducer vector that contains a positive feedback loop to increase tTA expression (ssAAV-PHP.eB:ihSyn-tTA). (d) Representative images using the one-component expression system with a 3XFP cocktail expressed from the CAG promoter at an equal dose per XFP (high: total dose = 2 × 1011 vg; low: total dose = 1 × 1010 vg). (e) Representative images using the two-component system: 1 × 1012 total of the TRE-3XFP cocktail and a high (1 × 1011 vg) or low (1 × 1010 vg) dose of the tTA inducer. (f) Percentages of individual cells that express one, two, or all three XFPs. (g) MIP (left) of a coronal section from the olfactory bulb of a Tbx21-Cre mouse transduced with a cocktail of ssAAV-PHP.eB:TRE-DIO-3XFP (1 × 1012 vg total dose) and sAAV-PHP.eB:ihSyn-tTA (1 × 1010 vg). The morphologies of local neuronal dendritic arbors were traced with NeuTube and imported into Imaris v8.3 for 3D rendering (middle) and overlay (right). Scale bars (d and e) are 50 µm and are MIPs of 40 µm z-stacks. The total volume in (g) is 0.042mm3. All images were acquired on tissue extracted three weeks after intravenous AAV delivery in adult mice, except for (a) where the expression was assessed after 11 days.
Tunable density multiplexed gene expression
Multicolor labeling by AAVs can benefit single-cell morphology and tracing studies27. However, direct local virus injections22,24 may not provide uniform color diversity because of the non-uniform distribution of vector copies radiating outward from the injection site24. We predicted that systemically delivered AAVs may offer a solution to this challenge because the distribution of labeled cells and color diversity would be more uniform. Indeed, when we delivered three separate XFPs expressed from the CAG promoter (AAV-CAG-3XFP; Fig. 4b) we observed a wide range of hues in the cortex, striatum, and cerebellum (Fig. 4d, left). However, this strategy, where the expression of the transgene is controlled in cis, couples the expression level, labeling density, and the color diversity to the virus dose. As expected, when we lowered the virus dose we observed that the density and color diversity were reduced (i.e., most cells were labeled by primary colors – red, green, or blue – likely reflecting single transduction events) (Fig. 4d, right). Therefore, to achieve stochastic, multicolor labeling while reducing the fraction of labeled cells, we sought a two-component AAV system in which the expression of XFPs in each cell is dependent on a separate inducer vector that is co-transduced at a variable dose. In this way, color diversity could be decoupled from the labeling density. We developed two inducer systems: one that uses the tetracycline (tet)-inducible system – a tet-off transactivator (tTA) and a tet-responsive element (TRE)28 (Fig. 4c) – and a second that uses the FLPo-FRT system29 (Supplementary Fig. 5). Importantly, both systems allow for cell type-specific expression using FLEX/DIO vectors and the extensive collection of Cre transgenic animals available (e.g., JAX, ABI, and GENSAT).
The tTA-TRE based system relies on co-administration of three separate vectors with XFP expression driven by a TRE containing promoter, along with an adjustable dose of an AAV-tTA expression vector. To evaluate this approach, we co-administered ssAAV-TRE-mNeonGreen along with a ssAAV-CAG-tTA vector. We delivered ssAAV-TRE-mNeonGreen at the same dose across all animals while varying the dose of the ssAAV-CAG-tTA inducer. We observed a dose-dependent reduction in the density of cells expressing mNeonGreen two weeks after intravenous delivery of the two vectors (Supplementary Fig. 6). Similar results were achieved with the two-component FLPo-FRT system (Supplementary Fig. 5). Although both systems are viable, we focused on the tTA-TRE system because (1) the short (417 bp) TRE promoter is compatible with longer transgenes, and (2) the positive-feedback loop in the AAV-tTA vector provides high levels of expression (Supplementary Fig. 7). However, care should be taken with long-term use of the tTA-TRE overexpression system to avoid the potential toxicity seen with overexpression of the tTA30.
We next sought to test the tTA-TRE system with multicolor labeling. Using the same 3XFP cocktail described above (Fig. 4a), we generated three ssAAV-TRE-XFP genomes and delivered the vectors at a high dose to all animals, along with either 1 × 1011 or 1 × 1010 vg/animal of a neuron specific inducer vector (ssAAV-PHP.eB:hSyn1-tTA). Three weeks after intravenous administration we observed robust and inducer dose-dependent labeling (Fig. 4e). Even at the lower inducer dose, and in contrast with the single-component expression system (Fig. 4d), many cells expressed more than one of the three XFP transgenes. We quantified the fraction of cells that contained each XFP and found that, with the two-component system at a low inducer dose, at least 42% of the labeled cells within the cortex, striatum, and the Purkinje cell population expressed all three XFPs. In comparison, in the one-component system, less than 10% of the labeled cells in the cortex and 1% of cells in the striatum and the Purkinje cell population expressed all three XFPs (Fig. 4f). Finally, we utilized the two-component system to trace mitral cells in the olfactory bulb of an adult Tbx21-Cre mouse, a transgenic line that labels mitral and tufted cells in the olfactory bulb. Three weeks after intravenous administration of 1 × 1012 vg/animal of a cocktail of TRE-DIO-XFPs and 1 × 1010 vg/animal of AAV-ihSyn1-tTA, a positive feedback inducible tTA driven by the hSyn promoter, we observed multicolor labeling at a density that facilitated segmentation of several local dendritic arbors after tissue clearing (Fig. 4g).
A versatile AAV toolbox with cell-type specific promoters
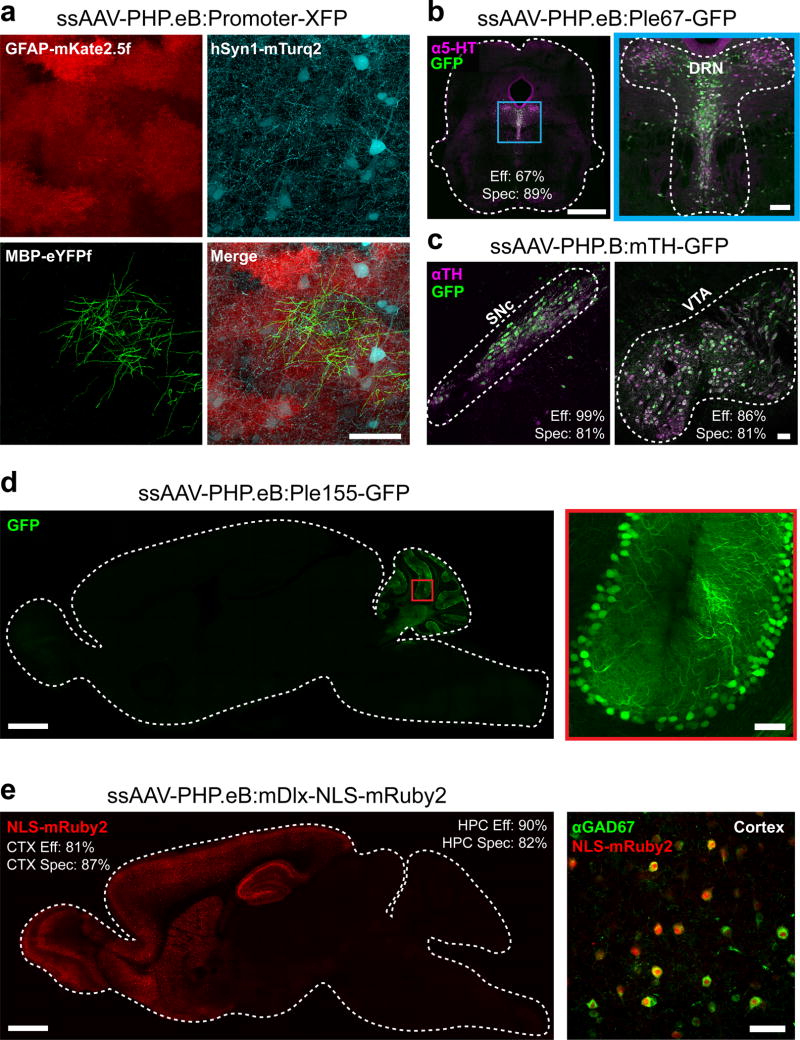
A further advantage of systemic AAV administration is that it provides a means to transduce populations of specific cell types even if the cells are distributed broadly. As a proof-of-concept that AAV-PHP.eB can be used to achieve cell type-restricted expression across the brain, we co-administered three ssAAV genomes that each expressed a spectrally distinct XFP controlled by a different cell type-specific promoter. In this way, we were able to independently express distinct transgenes in neurons (hSyn1), astrocytes (GFAP), and oligodendrocytes (MBP) within the same animal (Fig. 5a). We also packaged AAV vectors containing previously published promoters that restrict reporter expression to catecholaminergic cells (mouse tyrosine hydroxylase; mTH), serotonergic cells (FEV promoter; Ple67), Purkinje cells (PCP2 promoter; Ple155)31, or GABAergic interneurons within the forebrain (mouse distal-less homeobox; mDlx5/6)32 and administered these intravenously (Fig. 5b–e). We provide quantification of the specificity and efficiency of reporter transduction using these gene regulatory elements in (Supplementary Table 2).
Figure 5. AAV-PHP.eB can be used with gene regulatory elements to achieve cell type-restricted gene expression throughout the brain.
(a) Images show co-transduction of the cortex 3 weeks after co-administration of 3 vectors with 3 different promoters driving distinct XFPs (intravenous injection of AAV-PHP.eB at 1 × 1012 vg per viral vector). (b) A vector providing GFP expression driven by a FEV/serotoninergic neuron-specific promoter (ssAAV-PHP.eB:Ple67-GFP) was intravenously delivered at 1 × 1012 vg and co-localized to serotonin (5-hydroxytryptamine, 5-HT, magenta) expressing cells in the dorsal raphe nucleus (DRN) outlined in blue and expanded for detail (right). (c) A vector providing GFP expression from a mouse tyrosine hydroxylase (TH) promoter (ssAAV-PHP.B:mTH-GFP) was intravenously injected at 1 × 1012 vg, and imaging with IHC for TH (magenta) was performed after 2 weeks of expression. Images show the substantia nigra pars compacta (SNc, left) and ventral tegmental area (VTA, right). (d) A vector with a Purkinje cell-selective promoter (Ple155, Pcp2) driving GFP (ssAAV-PHP.eB:Ple155-GFP) was intravenously injected at 1 × 1012 vg and expression was examined at 4 weeks. A whole sagittal section (left) shows native GFP fluorescence (green) in the cerebellum (left) in cells with the morphology of Purkinje cells (see higher resolution of the red boxed region, right). (e) A vector with a forebrain GABAergic interneuron-specific enhancer driving nuclear-localized mRuby2 (AAV-PHP.eB:mDlx-NLS-mRuby2) was intravenously injected at 3 × 1011 vg and expression was examined at 8 weeks (CTX: cortex, HPC: hippocampus). Native mRuby2 expression within the forebrain (red, left). Co-localization was assessed by IHC for GABAergic cells (GAD67+, green, right). The scale bars in (a; b, right; c; d, right; e, right) are 50 µm. For (b and d, left) the scale bars are 1 mm. All panels are confocal images of native XFP fluorescence. The efficiency (Eff) and specificity (Spec) values for transduction by FEV/Ple67, mTH, mDlx vectors are giving in the DRN, VTA, SNc, cortex (CTX), and hippocampus (HPC).
Similarly, to achieve cell type-restricted expression within the PNS, we used AAV-PHP.S to co-administer the 3XFP cocktail under the control of the hSyn1 promoter (ssAAV-PHP.S:hSyn-3XFP) or by expressing the 3XFP cocktail from the CAG promoter in a double-floxed inverted open reading frame (ssAAV-PHP.S:CAG-DIO-3XFP) for expression in cholinergic neurons when used with ChAT-IRES-Cre transgenic mice (Supplementary Fig. 8). Using this strategy together with the two-component TRE-tTA system described above, we were able to sparsely label cholinergic neurons within the myenteric plexus in the proximal colon and trace the processes of multiple neurons for several millimeters (Supplementary Fig. 8).
In Supplementary Table 3, we summarize the available constructs presented here, along with their applications within this paper.
DISCUSSION
Using the previously described CREATE21 selection method, we have developed and characterized two novel capsids, AAV-PHP.eB and AAV-PHP.S, that enable efficient and noninvasive gene delivery throughout the CNS or PNS and visceral organs, respectively (Fig. 1 and Supplementary Fig. 4). As in our prior study21, these variants were identified after only two rounds of in vivo selection, further demonstrating how CREATE can be used to identify AAV capsids with enhanced transduction properties.
Previously, we selected for AAV capsids that more efficiently crossed the BBB and transduced GFAP-expressing astrocytes. Several of the identified variants, most notably AAV-PHP.B, also transduced neurons, although in general not with the same efficiency. In the current work, we sought to improve neuronal transduction while maintaining strong astrocytic transduction by performing parallel selections for capsids that transduced two widely distributed glutamatergic and GABAergic neuronal populations as well as astrocytes. This led to the development of the AAV-PHP.eB capsid, a variant that was consistently enriched through selections in each of the three Cre lines. Remarkably, AAV-PHP.eB differs from AAV-PHP.B at only two AAs adjacent to the original 7-mer insertion, yet at the relatively low systemic dose of 1 × 1011 vg/mouse it transduces the majority of neurons in the cortex and striatum, as well as over 75% of cerebellar Purkinje cells. The success of this approach suggests that the continued evolution of the AAV-PHP.B-based capsid using CREATE in multiple Cre lines is a viable strategy to improve transduction of other cell types, such as oligodendrocytes, while maintaining the overall CNS tropism.
We also describe the AAV-PHP.S capsid, which we identified by selecting for capsids that transduce GFAP-expressing cells. Although the mechanism underlying the selection of this variant is not clear, its discovery is nevertheless important; to date, there have been no reports of recombinant vectors capable of efficient, widespread transduction of peripheral neurons via systemic delivery in adult animals. Consequently, studies of PNS circuitry are arguably lagging behind those in the CNS. Many peripheral neuronal populations are challenging to access surgically, especially in small rodents, owing to their location or broad distribution. For example, the DRG are a key target for gene therapy or genetic manipulation in the study of pain13,33. Our finding that AAV-PHP.S transduces at least 80% of DRG neurons should enable high coverage delivery of transgenes to multiple DRG. More broadly, AAV-PHP.S also transduces neurons in the enteric nervous system and in other peripheral ganglia, such as the cardiac ganglia, and could provide system-wide gene delivery to these populations, or to subsets of these populations when used with Cre transgenic lines or cell type-specific promoters. Encouragingly, several recent studies have demonstrated that the tools developed for studying the brain can also be applied to probe the function and projections of specific cell types within the PNS12,14,15. The importance of anatomically and functionally mapping the PNS has been recognized by the recent NIH initiative for Stimulating Peripheral Activity to Relieve Conditions (SPARC), and the novel tropism of AAV-PHP.S may bring about new strategies to modulate PNS circuits through genetic tools such as optogenetics4 or chemogenetics5. For example, systemic Flp- and Cre-dependent effectors delivered via AAV-PHP.S to Cre transgenic animals or used in combination with circuit-specific retrograde Cre viruses should allow for circuit-based interrogation of PNS function and enable the types of experiments now routinely conducted in the CNS4,34.
The improved transduction efficiency we observed from both AAV-PHP.eB and AAV-PHP.S relative to their parental capsids was apparent from both the increased number of transduced cells and the increased expression level per cell (Fig 2, 3 and Supplementary Fig. 1). The higher expression in individual cells likely results from an increased mean number of vector genomes per cell, as supported by the high frequency of cells expressing multiple XFPs (Fig. 4a). This suggests that for applications that do not require high expression levels per cell, the viral doses could be lowered from those used in this study. Lowering the viral load used for systemic gene delivery will reduce cost and production burden1 and minimize the risk for adverse reactions to the viral components35.
As AAV vector engineering efforts continue to improve the efficiency of transduction of specific organs or cell populations36, widespread gene transfer will become a more viable alternative to the development of germline transgenics. To facilitate studies in nontransgenic animals, it will be necessary to develop additional vehicles with improved transduction efficiency and specificity and to identify gene regulatory elements that can be used to restrict expression to defined populations. Finding short enhancers that provide the expected restriction in expression has proven challenging37, but more recent efforts31 are expanding the collection of enhancer-promoters available. Here, we evaluated several gene regulatory elements and found that it was possible to achieve expression that was largely restricted to the expected cell types. If the separate vector components are translatable across species, vector-based genetic manipulations of specific cell types may become more feasible in genetically intractable organisms. Notably, intravenously administered AAV-PHP.B provided improved transduction of the rat CNS, relative to AAV938. Recently, a Dlx5/6 enhancer was found to drive expression that was largely restricted to GABAergic neurons in the telencephalon of a panel of vertebrate species32. Here, we further validate the Dlx5/6 enhancer vector for forebrain-wide GABAergic neuron expression when delivered using intravenous AAV-PHP.eB (Fig. 5e).
Cell morphology both informs and determines cell function39,40, and numerous tools have been developed for morphology and connectivity applications41,42, especially in the brain43,44. To enable tracing of cellular processes, sparse labeling of cells has been a key enabler – whether this comes from restricted gene or dye delivery (e.g. single-cell electroporation45,46) or from stochastic multicolor labeling and color-channel separation24,43,44. Both viral and transgenic approaches have been employed to address this problem. Although transgenic mice with sparse gene expression47,48 are useful, AAVs24 provide a versatile platform that can be quickly customized with up-to-date tools, promoters, and recombinase-dependent expression control systems. Furthermore, vectors can be delivered through different routes or at a specific dose to control the location and fraction of cells labeled. For these reasons, we developed an AAV-based expression system that allows for sparse multicolor labeling. This system uses two AAV components: a cocktail of XFP encoding vectors that exist in the off-state until induced by a second AAV vector that can be delivered at an empirically determined dose to provide expression in the desired fraction of co-transduced cells. We demonstrated that this system is well suited to the use of systemically delivered AAV-PHP.eB and AAV-PHP.S to achieve relatively uniform and sparse expression in the CNS and PNS. In addition, decoupling the relationship between the labeling density and the expression level may have additional applications. For example, sparse, high-level expression could be beneficial for in vivo monitoring of neural activity with genetically encoded activity indicators by lowering the background fluorescence during wide-field imaging49 or large area 2-photon imaging50. Furthermore, a similar framework could be applied to the understanding of how neuronal circuitry and morphology change during normal and diseased states. Future applications of this two-component approach could also be used to achieve intersectional gene expression in defined populations through circuit-specific delivery of one or both components or through the use of unique capsid-promoter or capsid-capsid combinations that serve to restrict expression to more specific populations.
Overall, the vector resources we present here expand the AAV toolbox for the genetic modification of neurons with the central and peripheral nervous systems.
METHODS
Animals
All procedures were approved by the California Institute of Technology Institutional Animal Care and Use Committee (IACUC). Mice were purchased from Jackson Laboratory (JAX) - C57Bl/6J mice (000664), GFAP-Cre (012886)51, Vgat-IRES-Cre (016962)52, Vglut2-IRES-Cre (016963)52, and Tbx21-Cre (024507)53. Male or female mice were used. Mice were randomly assigned to groups of predetermined sample size. All experiments with direct comparisons were performed in parallel to minimize variability.
In vivo selection
The AAV-PHP.eB capsid was identified by selecting for AAV-PHP.B variants that transduced cells in Vglut2-IRES-Cre, Vgat-IRES-Cre, and GFAP-Cre mice through two rounds of selection as previously described21. Intravenous administration of AAV vectors was performed by injection into the retro-orbital sinus of adult mice (6–8 weeks of age). The AAV-PHP.B capsid was modified by partial randomization of the original AAV-PHP.B 7-mer insertion and 4 flanking AAs (AAV-PHP.B AA positions 587 – 597). The AAV-PHP.B-based library was generated by PCR using primer XF21 and a mixture of 5 primers identical to the AAV-PHP.B template sequence with the exception of mixed bases at 3 consecutive randomized codons (NNK). In each primer mix, the position of the 3 randomized NNK codons was shifted by 6 bp (2 codons) such that each position in the 7-mer and the 2 AAs on each side of the 7-mer AAs were randomized, 3 AA at a time, in an overlapping manner. An equal amount of each of the 5 PCR products was assembled into the AAV-(delta)Cap-in-cis lox acceptor vector using the NEB Hi-Fi DNA Assembly Master Mix (New England Biolabs E2621) and used to generate the AAV-PHP.B-based AAV library. AAV-PHP.S was generated from an insertion of a random 7-mer between the 588-589 site of AAV9 and was identified from two rounds of in vivo selection in GFAP-Cre mice21.
Plasmids
The backbones of all AAV plasmids that are double inverted orientation (DIO) were based on pAAV-Ef1a-DIO EYFP (Addgene, #27056), and all non-DIO plasmids were based on pAAV-EF1a-Cre (Addgene, #55636). The backbone of pAAV-CAG-fDIO-mNeonGreen was based on pAAV-Ef1a-fDIO EYFP (Addgene, #55641). The fluorescent protein cDNAs for mTurquoise254, mNeonGreen55, mRuby256, or mKate2.5 were synthesized as gBlocks (Integrated DNA Technologies). The MBP-eYFPf and GFAP-mkate2.5f vectors have a farnesylation sequence attached by overhang PCR. The pAAV-CAG-NLS-GFP vector was modified by PCR to add N- and C-terminal NLS sequences and assembled using NEB Hi-Fi DNA Assembly Master Mix (New England Biolabs E2621). The mouse tyrosine hydroxylase (TH) promoter (mTH) was based on the 2.5 kb region of a published rat TH promoter57 and myelin basic promoter (MBP)58. Both were directly cloned from mouse genomic DNA. The hSyn1 promoter59 was cloned from pAAV-hSyn Con/Foff EYFP (Addgene, #55651). The GFAP (GfABC1D) promoter was previously described60 and was cloned upstream of a synthetic intron. The GFAP-mKate2.5 plasmid also contained 3 tandem copies of sequences complementary to 6 miRNAs: miR-1, miR-122, miR-375, miR-196a, miR-743 and miR-10b inserted between the WPRE and an SV40 pA sequence. The TRE was PCR amplified from a SG-TRE containing plasmid28. The inducible hSyn1 (ihSyn1) promoter was cloned by overlap PCR with the hSyn1 promoter, and oligonucleotides designed with a synthetic intron and 3 tetO binding sites. The AAV vectors expressing GFP from the Ple67 and Ple155 promoters were obtained from (Addgene, #49138 and #49140, respectively). The AAV-mDlx-NLS-mRuby vector was cloned by replacing the GFP reporter in pAAV-mDlx-GFP-Fishell-1 (Addgene, #83900) with a mRuby reporter fused with an N-terminal SV40 NLS.
Virus production and purification
AAVs were generated in HEK 293T cells (ATCC) using Polyethylenimine (PEI)61. 72 hours post transfection, viral particles were harvested from the media and after 120 hours from cells and the media. Viral particles from the media were precipitated with 40% polyethylene glycol (Sigma, 89510-1KG-F) in 500 mM NaCl and combined with cell pellets for processing. The cell pellets were suspended in 500 mM NaCl, 40 mM Tris, 2.5 mM MgCl2, pH 8, and 100 U/mL of salt-activate nuclease (Arcticzymes) at 37°C for 30 minutes. Afterwards, the cells were clarified by centrifugation at 2,000 × g and then purified over iodixanol (Optiprep, Sigma; D1556) step gradients (15%, 25%, 40%, and 60%)62. Viruses were concentrated using Amicon filters (EMD, UFC910024), and formulated in sterile phosphate buffer saline (PBS). Virus titers were measured by determining the number of DNAse I-resistant vg using qPCR using a linearized genome plasmid as a standard61.
Tissue preparation, immunohistochemistry and imaging
Intravenous administration of AAV vectors was performed by injection into the retro-orbital sinus of adult mice (6–8 weeks of age). After allowing time for expression (3 weeks unless otherwise noted in Results), mice were anesthetized with EUTHASOL™ (Virbac) and transcardially perfused first with 0.1M phosphate buffer (PB, pH 7.4) at room temperature (RT) and then with freshly prepared, ice-cold 4% paraformaldehyde (PFA) in 0.1M PB. The tissues were post-fixed in 4% PFA overnight and afterwards transferred to phosphate buffered saline (PBS) with 0.05% sodium azide. The brains, lungs, and liver were sectioned to 100 µm, and the spinal cord to 300 µm, using a Leica VT1200 vibratome. Immunohistochemistry (IHC) was performed using 10% normal donkey serum (NDS), 0.1% Triton X-100 (no detergent was used with GAD67 staining) and 0.05% sodium azide. Primary antibodies used were rabbit Calbindin (1:500; Abcam, ab11426), mouse NeuN (1:500; EMD, MAB377), rabbit S100 (1:500; Abcam ab868), rabbit tyrosine hydroxylase (1:500; Abcam, ab112), goat 5-HT (1:200; Immunostar, 20079), rabbit PGP9.5 (1:500; Abcam, ab10404), and mouse GAD67 (1:250; EMD, MAB5406). Sections were incubated with primary antibodies in blocking solution (PBS with 10% NDS and 0.05% sodium azide) for 16–24 hrs at RT and then washed in PBS overnight and followed by secondary antibody incubation (Jackson Immuno Research, 711-166-152, 711-606-152, 715-166-150, or 715-606-150) for 16–24 hrs at RT.
The dorsal root ganglia were post-fixed in 4% PFA at 4° C overnight and then cryopreserved in 30% sucrose at 4° C overnight. Tissues were then embedded in OCT (Tissue-Tek, Torrance, CA, USA), frozen in 2-methybutane on dry ice and stored at −80° C until sectioning. For cryosectioning, the DRGs were cut at a thickness of 16 µm using a Leica CM3050 and adhered to a Superfrost Plus microscope slide (Fisher Scientific, Tustin, CA, USA). Slides were stored at −80° C until staining. For DRG IHC, the slides were incubated in rabbit anti-PGP9.5 antibody (1:250, Abcam, ab10404) in blocking solution (10% horse serum, 0.1% triton X-100, and 0.02% sodium azide in PBS) overnight at RT. The next day, sections were incubated with fluorescence-conjugated secondary antibody for 1 hr at RT (Jackson Immuno Research, 711-606-152). Between each step, sections were thoroughly washed with PBS. ProLong gold, anti-fade mounting medium (P36941; Molecular Probe, Life Technologies, Carlsbad, CA, USA) was applied to the slide before coverslip mounting.
The small intestines and colons were post-fixed in 4% PFA at 4° C overnight. Then the duodenum, jejunum, ileum, and proximal colon were sectioned and blocked in PBS with 0.1% triton X-100, 3% bovine serum albumin, and 0.01% sodium azide overnight at RT. The next day, the tissue was incubated with the mouse on mouse blocking reagent (Vector Laboratories, MKB-2213) in PBS with 0.01% sodium azide at RT for 16 hours. Afterwards, the tissue was stained in the initial blocking solution with guinea pig anti-PGP9.5 antibody (1:100, Abcam, ab10410) and rabbit anti-s100 beta antibody (1:200, Abcam, ab52642) for 48 hours and then washed in PBS with 0.01% sodium azide overnight. The secondary antibodies – goat anti-guinea pig Alexa 647 (A-21450) and goat anti-rabbit Alexa 555 (A-21428) Molecular Probes, Life Technologies, Carlsbad, CA, USA) – were incubated in the initial blocking solution for 48 hours. Unbound antibodies were then washed out with PBS with 0.01% sodium azide overnight. Finally, samples were incubated in a refractive matching solution RIMS63 solution for 6 hours before imaging.
For Fig. 4b and g, samples were made transparent and mounted using ScaleS464. For Fig. 4a,d, Fig. 5c, Supplementary Fig. 5a,b, and Fig. 6b,c samples were made transparent and mounted using RIMS. All other samples were mounted with Prolong Diamond Antifade (ThermoFisher, P36965).
For some images, the 16-bit green channel (GFP) gamma was adjusted to enable visualization of low and high expressing cells while avoiding oversaturation. In all cases, changes to contrast or gamma as well as microscope laser settings remained constant across sets of images that were directly compared. Images were acquired on a Zeiss LSM 780 confocal microscope fitted with the following objectives: Fluar 5×/0.25 M27 Plan-Apochromat 10×/0.45 M27 (working distance 2.0 mm), and Plan-Apochromat 25×/0.8 Imm Corr DIC M27 multi-immersion or Zeiss Axiozoom V16 with a 0.7× objective fitted with a Hamamatsu c11440 camera.
Image processing and data analysis
All image processing was performed using Zen Black 2012, Adobe Photoshop and Illustrator CC, ImageJ, neuTube Build 1.0z2016, and Bitplane Imaris 8.3, and custom-made scripts in Matlab. For data analysis, Microsoft Excel 2016, Matlab, Python, and GraphPad Prism 7.01 were used.
To minimize bias, when direct comparisons for quantification (Fig 2d, 2e, 3b, and 3c) or direct qualitative comparisons were made, (Fig. 3b and 3d) (Supplementary Fig. 3a, 3b, 4a, 4b and 4c) matched regions were selected for imaging by first viewing the cell type-specific immunostaining channel, rather than the GFP expression. Manual cell counting in the striatum and cortex was performed by a blinded observer (Fig. 2e). Automated counting was performed of Purkinje cells in the cerebellum (Fig. 2e).
To mitigate any cross-talk between the 3 fluorescent proteins, linear unmixing was used (Fig. 4d). For a given imaging condition, the expected cross-talk between the three fluorophores was evaluated based on the emission and excitation spectra of each fluorophore. Then, for each pixel in the RGB image the corresponding unmixed signals were calculated using a non-negative least square fitting.
Cell profiling was done using Imaris (Bitplane). To detect neuronal cell bodies from the image data, we manually analyzed cells in the cortex and striatum by drawing a region of interest (ROI) around each cell’s nucleus and intensity values were exported into Microsoft Excel and Graphpad Prism for analysis. For analysis of the cerebellum, we developed a computational pipeline to facilitate the automation of the whole procedure, since the highly organized layout of Purkinje cells allowed for high accuracy automated counting (the true positive rate = 88.0% and the positive predictive value = 95.5% relative to manual detection)
To detect neuronal cell bodies from the image data (Supplementary Fig. 2), the custom Matlab code ‘GFPcount_from_cerebellum.m’ was used and is provided in supplementary software. In short, we applied a circular Hough transform to the maximum projection of the image stack. Prior to this process, morphological filtering with a circular structural element was performed with the images to remove thin fibrous processes. Each circular region detected from the images was considered as a single cell body. To measure the ratio of GFP-positive cells and their intensity in the cerebellum, we applied the abovementioned cell-body detection method to the Calbindin channel to detect the soma of Purkinje cells and used this to define ROIs in the GFP channel. We then subtracted the background from each ROI to exclude the expression (or leakage) of GFPs in cytosol. The background was estimated by applying a 2D Gaussian smoothing kernel to each cell body image. Subsequently, the image was converted to binary, and the convex hull of the connected object therein was calculated. The objects whose area were smaller than 10% of the cell body were removed, and we considered the largest object as a nucleus of interest if there were more than one object. Nuclei with a mean intensity higher than 0.1 (the intensity rescaled to the range of 0–1) was counted as GFP-positive. The mean intensity of the nucleus was calculated by averaging the pixel values inside the object (Supplementary Fig. 1).
The pipeline for automated cell body detection was also applied to other analyses (Supplementary Fig. 7 and Fig. 4f). To measure the density of labeled cells in the sparse-labeling system (Supplementary Fig. 6), the custom Matlab code ‘cell_density.m’ was used and is provided in supplementary software. In short, we applied the cell body detection method to the images and measured the density of detected cells by dividing the cell number by the area of the image. We also examined the distance between the neighboring cells; for each cell, all of the distances to other cells were calculated, and the minimum was selected as the distance to the closest neighbor. The values for all cells were then averaged for each region and condition. We used the same method to determine whether each cell expressed 1, 2, or 3 XPFs (Fig. 4f) in the one- and two-component labeling systems by running a custom Matlab code ‘expressed_channel_count.m’ that can be found in the supplementary software. After detecting each cell body, we collected all the normalized intensity values therein from each channel. If the mean intensity of the channel was above 0.1, we considered that this gene (color) was expressed in the cell. Counting the number of expressed channels in each cell, we calculated the fraction of cells according to the number of expressed gene types.
To determine the mean GFP intensity (Fig. 2d and Fig 3b) the automated cell body detection pipeline above was used for Calbindin+ cells in the cerebellum and PGP9.5+ cells in the DRG along with a custom Python code ‘drg_intensity.ipynb’ that can be found in the supplementary software. Manual ROIs were drawn onto DAPI+ nuclei in the cortex and striatum using Bitplane Imaris 8.3, with the GFP channel turned off to minimize bias. The median fluorescence per ROI was determined and the average across all cells within each replicate were reported as the mean GFP intensity.
Statistics
Microsoft Excel 2016 and GraphPad Prism 7.01 were used for data analysis and graph generation. For the comparison between AAV9 and AAV-PHP.S (n = 5/group) and AAV-PHP.B and AAV-PHP.eB (n = 5/group) the animal group size was chosen based on preliminary data that suggested a large effect size. One animal from each of the AAV-PHP.B and AAV-PHP.eB groups was excluded from data analysis after necropsy due to failed retro-orbital injections. For Figure 1c, 1d, 1e, 1f, and 1g representative images were chosen from AAV9 (n = 5), AAV-PHP.S (n = 5), AAV-PHP.B (n = 4), and AAV-PHP.eB (n = 4). For Figures 2d, 2e, 3b, and 3c each dot is representative of one animal with three technical replicates averaged per animal. For Supplementary Figures 1a, 1b, and 1c, each line represents one animal with three technical replicates per animal. For Supplementary Figures 6c and 6d, each dot represents a technical replicate for each of the three doses. For Figures 2d, 2e, 3b, and 3c, unpaired, two-tailed t-tests were performed. Error bars show SEM. Data distribution was assumed to be normal but this was not formally tested. For Supplementary Figures 1a, 1b, and 1c statistical significance was assessed with a Kolmogorov-Smirnov test. For Supplementary Figure 6c and 6d means were compared by t-tests in which the P value is computed by fewest assumptions and did not assume consistent standard deviation. Statistical significance was determined by the Holm-Sidak method to correct for multiple comparisons with an alpha of 0.05.
Supplementary Material
Acknowledgments
We thank E. Mackey and K. Beadle for assistance with cloning and viral production, P. Anguiano for administrative assistance, and the entire Gradinaru group for discussions. We thank G. Stevens and V. Anand for their efforts in image analysis; P. Rajendran and S. Kalyanam at University of California Los Angeles and C. Fowlkes at the University of Irvine for useful discussions; the University of Pennsylvania vector core for the AAV2/9 Rep-Cap plasmid; M. Brenner at the University of Alabama for the GfABC1D promoter. pAAV-Ef1a-DIO EYFP, pAAV-EF1a-Cre and pAAV-Ef1a-fDIO EYFP were gifts from Karl Deisseroth (Addgene plasmid #27056, #55636 and #55641). pEMS2113 and pEMS2115 were gifts from Elizabeth Simpson (Addgene plasmid # 49138 and #49140).
This work was primarily supported by the National Institutes of Health (NIH) through grants to V.G.: Director’s New Innovator DP2NS087949 and PECASE; SPARC OT2OD023848-01; National Institute on Aging R01AG047664; BRAIN U01NS090577; and National Institute of Mental Health (NIMH) R21MH103824. Additional funding included the Gordon and Betty Moore Foundation through grant GBMF2809 to the Caltech Programmable Molecular Technology Initiative (to V.G.); the Curci Foundation (to V.G.); the Hereditary Disease Foundation (to V.G. and B.E.D.); the Beckman Institute (to V.G. and B.E.D.) and Rosen Center (to C.L. and V.G.) at Caltech; NIH U01 MH109147 02S1 (to C.L. and V.G.); NIH NS085910 (to S.K.M. and V.G.); and the Defense Advanced Research Projects Agency (DARPA) Biological Technologies Office (BTO; to V.G. and B.E.D.) and from the Friedreich’s Ataxia Research Aliance (FARA) and FARA Australasia (to B.E.D). S.K.M and V.G. are Heritage Principal Investigators supported by the Heritage Medical Research Institute.
All tools and protocols will be available through Addgene or the Beckman Institute for CLARITY, Optogenetics and Vector Engineering Research for technology development and broad dissemination: http://www.beckmaninstitute.caltech.edu/clover.shtml.
Footnotes
Data availability
Cell detection algorithms used in this study are available through https://github.com. The data that support the findings of this study are available from the corresponding authors upon reasonable request.
Accession codes. GenBank: AAV capsid sequences: AAV-PHP.B, KU056473; AAV-PHP.eB: TBD, AAV-PHP.S: TBD.
Contributions
K.Y.C and B.E.D. designed and performed experiments, imaged samples, analyzed data. K.Y.C prepared figures with input from B.E.D and V.G. M.J.J. analyzed data, prepared figures, assisted with experiments and in manuscript preparation. B.B.Y. assisted with tissue processing, imaging, and virus production. A.G. helped with image analysis. N.R. assisted with molecular cloning. W.-L.W. assisted with tissue processing. L.S assisted in tissue processing. C.L and S.K.M assisted in experimental designs. K.Y.C, B.E.D and V.G. wrote the manuscript with support from all authors. B.E.D and V.G. conceived the project. V.G. supervised all aspects of the work.
Competing Financial Interest Statement
The California Institute of Technology has filed patent applications related to this work with B.E.D, K.Y.C. and V.G. listed as inventors. B.E.D. and V.G. receive research support from Voyager Therapeutics, which was not used in preparation of this manuscript.
References
- 1.Hastie E, Samulski RJ. Adeno-associated virus at 50: a golden anniversary of discovery, research, and gene therapy success--a personal perspective. Hum Gene Ther. 2015;26:257–265. doi: 10.1089/hum.2015.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suzuki K, et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016;540:144–149. doi: 10.1038/nature20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Y, et al. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat Biotechnol. 2016;34:334–338. doi: 10.1038/nbt.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rajasethupathy P, Ferenczi E, Deisseroth K. Targeting Neural Circuits. Cell. 2016;165:524–534. doi: 10.1016/j.cell.2016.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Urban DJ, Roth BL. DREADDs (designer receptors exclusively activated by designer drugs): chemogenetic tools with therapeutic utility. Annu Rev Pharmacol Toxicol. 2015;55:399–417. doi: 10.1146/annurev-pharmtox-010814-124803. [DOI] [PubMed] [Google Scholar]
- 6.Marshall JD, et al. Cell-Type-Specific Optical Recording of Membrane Voltage Dynamics in Freely Moving Mice. Cell. 2016;167:1650–1662 e1615. doi: 10.1016/j.cell.2016.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim CK, et al. Simultaneous fast measurement of circuit dynamics at multiple sites across the mammalian brain. Nat Methods. 2016;13:325–328. doi: 10.1038/nmeth.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van der Perren A, Van den Haute C, Baekelandt V. Viral vector-based models of Parkinson's disease. Curr Top Behav Neurosci. 2015;22:271–301. doi: 10.1007/7854_2014_310. [DOI] [PubMed] [Google Scholar]
- 9.Hocquemiller M, Giersch L, Audrain M, Parker S, Cartier N. Adeno-Associated Virus-Based Gene Therapy for CNS Diseases. Hum Gene Ther. 2016;27:478–496. doi: 10.1089/hum.2016.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samulski RJ, Muzyczka N. AAV-Mediated Gene Therapy for Research and Therapeutic Purposes. Annu Rev Virol. 2014;1:427–451. doi: 10.1146/annurev-virology-031413-085355. [DOI] [PubMed] [Google Scholar]
- 11.Bennett J, et al. Safety and durability of effect of contralateral-eye administration of AAV2 gene therapy in patients with childhood-onset blindness caused by RPE65 mutations: a follow-on phase 1 trial. Lancet. 2016;388:661–672. doi: 10.1016/S0140-6736(16)30371-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iyer SM, et al. Virally mediated optogenetic excitation and inhibition of pain in freely moving nontransgenic mice. Nat Biotechnol. 2014;32:274–278. doi: 10.1038/nbt.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samad OA, et al. Virus-mediated shRNA knockdown of Na(v)1.3 in rat dorsal root ganglion attenuates nerve injury-induced neuropathic pain. Mol Ther. 2013;21:49–56. doi: 10.1038/mt.2012.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang RB, Strochlic DE, Williams EK, Umans BD, Liberles SD. Vagal Sensory Neuron Subtypes that Differentially Control Breathing. Cell. 2015;161:622–633. doi: 10.1016/j.cell.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams EK, et al. Sensory Neurons that Detect Stretch and Nutrients in the Digestive System. Cell. 2016;166:209–221. doi: 10.1016/j.cell.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eldridge MA, et al. Chemogenetic disconnection of monkey orbitofrontal and rhinal cortex reversibly disrupts reward value. Nat Neurosci. 2016;19:37–39. doi: 10.1038/nn.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foust KD, et al. Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat Biotechnol. 2009;27:59–65. doi: 10.1038/nbt.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choudhury SR, et al. Widespread Central Nervous System Gene Transfer and Silencing After Systemic Delivery of Novel AAV-AS Vector. Mol Ther. 2016;24:726–735. doi: 10.1038/mt.2015.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pulicherla N, et al. Engineering liver-detargeted AAV9 vectors for cardiac and musculoskeletal gene transfer. Mol Ther. 2011;19:1070–1078. doi: 10.1038/mt.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marchio S, Sidman RL, Arap W, Pasqualini R. Brain endothelial cell-targeted gene therapy of neurovascular disorders. EMBO Mol Med. 2016;8:592–594. doi: 10.15252/emmm.201606407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deverman BE, et al. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat Biotechnol. 2016;34:204–209. doi: 10.1038/nbt.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Betley JN, Sternson SM. Adeno-associated viral vectors for mapping, monitoring, and manipulating neural circuits. Hum Gene Ther. 2011;22:669–677. doi: 10.1089/hum.2010.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beier KT, et al. Circuit Architecture of VTA Dopamine Neurons Revealed by Systematic Input-Output Mapping. Cell. 2015;162:622–634. doi: 10.1016/j.cell.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai D, Cohen KB, Luo T, Lichtman JW, Sanes JR. Improved tools for the Brainbow toolbox. Nat Methods. 2013;10:540–547. [PubMed] [Google Scholar]
- 25.Gombash SE, et al. Intravenous AAV9 efficiently transduces myenteric neurons in neonate and juvenile mice. Front Mol Neurosci. 2014;7:81. doi: 10.3389/fnmol.2014.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inagaki K, et al. Robust systemic transduction with AAV9 vectors in mice: efficient global cardiac gene transfer superior to that of AAV8. Mol Ther. 2006;14:45–53. doi: 10.1016/j.ymthe.2006.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parekh R, Ascoli GA. Neuronal morphology goes digital: a research hub for cellular and system neuroscience. Neuron. 2013;77:1017–1038. doi: 10.1016/j.neuron.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agha-Mohammadi S, et al. Second-generation tetracycline-regulatable promoter: repositioned tet operator elements optimize transactivator synergy while shorter minimal promoter offers tight basal leakiness. J Gene Med. 2004;6:817–828. doi: 10.1002/jgm.566. [DOI] [PubMed] [Google Scholar]
- 29.Raymond CS, Soriano P. High-efficiency FLP and PhiC31 site-specific recombination in mammalian cells. PLoS One. 2007;2:e162. doi: 10.1371/journal.pone.0000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gallia GL, Khalili K. Evaluation of an autoregulatory tetracycline regulated system. Oncogene. 1998;16:1879–1884. doi: 10.1038/sj.onc.1201706. [DOI] [PubMed] [Google Scholar]
- 31.de Leeuw CN, et al. rAAV-compatible MiniPromoters for restricted expression in the brain and eye. Mol Brain. 2016;9:52. doi: 10.1186/s13041-016-0232-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dimidschstein J, et al. A viral strategy for targeting and manipulating interneurons across vertebrate species. Nat Neurosci. 2016 doi: 10.1038/nn.4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fischer G, Pan B, Vilceanu D, Hogan QH, Yu H. Sustained relief of neuropathic pain by AAV-targeted expression of CBD3 peptide in rat dorsal root ganglion. Gene Ther. 2014;21:44–51. doi: 10.1038/gt.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang JC, et al. Cell type-specific manipulation with GFP-dependent Cre recombinase. Nat Neurosci. 2015;18:1334–1341. doi: 10.1038/nn.4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mingozzi F, High KA. Immune responses to AAV vectors: overcoming barriers to successful gene therapy. Blood. 2013;122:23–36. doi: 10.1182/blood-2013-01-306647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tervo DG, et al. A Designer AAV Variant Permits Efficient Retrograde Access to Projection Neurons. Neuron. 2016;92:372–382. doi: 10.1016/j.neuron.2016.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nathanson JL, et al. Short Promoters in Viral Vectors Drive Selective Expression in Mammalian Inhibitory Neurons, but do not Restrict Activity to Specific Inhibitory Cell-Types. Front Neural Circuits. 2009;3:19. doi: 10.3389/neuro.04.019.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson KL, Dayton RD, Deverman BE, Klein RL. Better Targeting, Better Efficiency for Wide-Scale Neuronal Transduction with the Synapsin Promoter and AAV-PHP.B. Front Mol Neurosci. 2016;9:116. doi: 10.3389/fnmol.2016.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Micheva KD, Busse B, Weiler NC, O'Rourke N, Smith SJ. Single-synapse analysis of a diverse synapse population: proteomic imaging methods and markers. Neuron. 2010;68:639–653. doi: 10.1016/j.neuron.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mikuni T, Nishiyama J, Sun Y, Kamasawa N, Yasuda R. High-Throughput, High-Resolution Mapping of Protein Localization in Mammalian Brain by In Vivo Genome Editing. Cell. 2016;165:1803–1817. doi: 10.1016/j.cell.2016.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marshel JH, Mori T, Nielsen KJ, Callaway EM. Targeting single neuronal networks for gene expression and cell labeling in vivo. Neuron. 2010;67:562–574. doi: 10.1016/j.neuron.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wertz A, et al. PRESYNAPTIC NETWORKS. Single-cell-initiated monosynaptic tracing reveals layer-specific cortical network modules. Science. 2015;349:70–74. doi: 10.1126/science.aab1687. [DOI] [PubMed] [Google Scholar]
- 43.Schwarz LA, et al. Viral-genetic tracing of the input-output organization of a central noradrenaline circuit. Nature. 2015;524:88–92. doi: 10.1038/nature14600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loulier K, et al. Multiplex cell and lineage tracking with combinatorial labels. Neuron. 2014;81:505–520. doi: 10.1016/j.neuron.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 45.Kitamura K, Judkewitz B, Kano M, Denk W, Hausser M. Targeted patch-clamp recordings and single-cell electroporation of unlabeled neurons in vivo. Nat Methods. 2008;5:61–67. doi: 10.1038/nmeth1150. [DOI] [PubMed] [Google Scholar]
- 46.Haas K, Sin WC, Javaherian A, Li Z, Cline HT. Single-cell electroporation for gene transfer in vivo. Neuron. 2001;29:583–591. doi: 10.1016/s0896-6273(01)00235-5. [DOI] [PubMed] [Google Scholar]
- 47.Young P, et al. Single-neuron labeling with inducible Cre-mediated knockout in transgenic mice. Nat Neurosci. 2008;11:721–728. doi: 10.1038/nn.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Badea TC, et al. New mouse lines for the analysis of neuronal morphology using CreER(T)/loxP-directed sparse labeling. PLoS One. 2009;4:e7859. doi: 10.1371/journal.pone.0007859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin MZ, Schnitzer MJ. Genetically encoded indicators of neuronal activity. Nat Neurosci. 2016;19:1142–1153. doi: 10.1038/nn.4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sofroniew NJ, Flickinger D, King J, Svoboda K. A large field of view two-photon mesoscope with subcellular resolution for in vivo imaging. Elife. 2016;5 doi: 10.7554/eLife.14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia AD, Doan NB, Imura T, Bush TG, Sofroniew MV. GFAP-expressing progenitors are the principal source of constitutive neurogenesis in adult mouse forebrain. Nat Neurosci. 2004;7:1233–1241. doi: 10.1038/nn1340. [DOI] [PubMed] [Google Scholar]
- 52.Vong L, et al. Leptin action on GABAergic neurons prevents obesity and reduces inhibitory tone to POMC neurons. Neuron. 2011;71:142–154. doi: 10.1016/j.neuron.2011.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haddad R, et al. Olfactory cortical neurons read out a relative time code in the olfactory bulb. Nat Neurosci. 2013;16:949–957. doi: 10.1038/nn.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goedhart J, et al. Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93% Nat Commun. 2012;3:751. doi: 10.1038/ncomms1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shaner NC, et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat Methods. 2013;10:407–409. doi: 10.1038/nmeth.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lam AJ, et al. Improving FRET dynamic range with bright green and red fluorescent proteins. Nat Methods. 2012;9:1005–1012. doi: 10.1038/nmeth.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oh MS, Hong SJ, Huh Y, Kim KS. Expression of transgenes in midbrain dopamine neurons using the tyrosine hydroxylase promoter. Gene Ther. 2009;16:437–440. doi: 10.1038/gt.2008.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gow A, Friedrich VL, Jr, Lazzarini RA. Myelin basic protein gene contains separate enhancers for oligodendrocyte and Schwann cell expression. J Cell Biol. 1992;119:605–616. doi: 10.1083/jcb.119.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kugler S, Kilic E, Bahr M. Human synapsin 1 gene promoter confers highly neuron-specific long-term transgene expression from an adenoviral vector in the adult rat brain depending on the transduced area. Gene Ther. 2003;10:337–347. doi: 10.1038/sj.gt.3301905. [DOI] [PubMed] [Google Scholar]
- 60.Lee Y, Messing A, Su M, Brenner M. GFAP promoter elements required for region-specific and astrocyte-specific expression. Glia. 2008;56:481–493. doi: 10.1002/glia.20622. [DOI] [PubMed] [Google Scholar]
- 61.Gray SJ, et al. Production of recombinant adeno-associated viral vectors and use in in vitro and in vivo administration. Curr Protoc Neurosci. 2011:17. doi: 10.1002/0471142301.ns0417s57. Chapter 4, Unit 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zolotukhin S, et al. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 1999;6:973–985. doi: 10.1038/sj.gt.3300938. [DOI] [PubMed] [Google Scholar]
- 63.Yang B, et al. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell. 2014;158:945–958. doi: 10.1016/j.cell.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hama H, et al. ScaleS: an optical clearing palette for biological imaging. Nat Neurosci. 2015;18:1518–1529. doi: 10.1038/nn.4107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.