Abstract
Uracil DNA Glycosylase (UNG2) is the primary enzyme in humans that prevents the stable incorporation of deoxyuridine monophosphate into DNA in the form of U/A basepairs. During S-phase, UNG2 remains associated with the replication fork through its interactions with two proteins, Proliferating Cell Nuclear Antigen (PCNA) and Replication Protein A (RPA), which are critical for DNA replication and repair. In this work, we used protein semisynthesis and fluorescence anisotropy assays to explore the interactions of UNG2 with PCNA and RPA and to determine the effects of two UNG2 phosphorylation sites (Thr6 and Tyr8) located within its PCNA-interacting motif (PIP-box). In binding assays, we found that phosphorylation of Thr6 or Tyr8 on UNG2 can impede PCNA binding without affecting UNG2 catalytic activity or its RPA interaction. Our data also suggests that unmodified UNG2, PCNA, and RPA can form a ternary protein complex. We propose that the UNG2 N-terminus may serve as a flexible scaffold to tether PCNA and RPA at the replication fork, and that post-translational modifications on the UNG2 N-terminus disrupt formation of the PCNA-UNG2-RPA protein complex.
Introduction
The nuclear uracil DNA glycosylase (UNG2) initiates the base excision repair pathways that remove uracil from genomic DNA. The human enzyme consists of a globular catalytic domain that is preceded by a disordered N-terminus. The catalytic domain itself can efficiently detect and remove uracil bases from DNA, whereas the N-terminus regulates UNG2 abundance and localization inside the nucleus (1, 2). During the cell cycle, UNG2 expression peaks during late G1/early S-phase, in part to ensure removal of genomic deoxyuridine monophosphate that is misincorporated by polymerases during replication (3, 4, 5). To support this function, UNG2 has been shown to localize to the replication fork by interacting with either Proliferating Cell Nuclear Antigen (PCNA), the dsDNA sliding clamp, or Replication Protein A (RPA)—the major ssDNA binding protein in the nucleus (2, 3, 6).
The ∼90-residue N-terminus of UNG2 contains 14 sites that can be acetylated, ubiquitinated, or phosphorylated (7). Some post-translational modifications (PTMs) are cell-cycle dependent and mediate UNG2 degradation after DNA replication (1), and other residues are modified in response to cellular stress (8). However, the functional roles of most UNG2 modifications identified in proteomic screens have not been elucidated. We focused our work here on two specific N-terminal phosphorylation sites on UNG2 at residues Thr6 and Tyr8. Thr6 phosphorylation occurs after cellular UV exposure (8), whereas Tyr8 phosphorylation has been identified in proteomic screens using multiple cancer cell lines (7, 9, 10, 11, 12). We used protein semisynthesis (13, 14) and in vitro assays to investigate whether phosphorylation of the UNG2 sites affects enzyme activity or protein-protein interactions.
Materials and Methods
Peptide synthesis
A standard fluorenylmethyloxycarbonyl (Fmoc) solid-phase peptide synthesis strategy with Wang resin was used to prepare N-terminal UNG2(aa 1–19) peptides with and without phosphorylation at position Thr6 or Tyr8. Twenty-percent piperidine in dimethyl formamide (DMF) was used for deprotection, and residues were coupled in DMF using five molar equivalents of protected amino acid, 4.75 molar equivalents of HATU, and 10 molar equivalents of diisopropylethylamine (DIPEA). The progress of the couplings was monitored using the Kaiser test.
The peptides PogoLigase (PL) (15) and SMARCAL1(aa 5–30) (SMARCAL) were synthesized in a similar manner, and Fmoc-6-aminohexanoic acid was coupled to the N-termini of these peptides. To label these with fluorescein, the final Fmoc group was removed with 20% piperidine. The peptides were washed with DMF followed by 5% N-methylmorpholine (NMM) in DMF, then 0.21 molar equivalents of 5/6-carboxyfluorescein succinimidyl ester (NHS-fluorescein; Thermo Fisher Scientific, Waltham, MA) dissolved in 5% NMM were added for 16 h. After synthesis, the peptides were washed extensively with DMF and dichloromethane (DCM) before being cleaved from the resin with Reagent K (82.5% trifluoroacetic acid, 5% phenol, 5% water, 5% thioanisole, and 2.5% 1,2-ethanedithiol).
The N-terminal UNG2 11-mer and 29-mer thioester peptides used for native chemical ligation were synthesized on Dawson Dbz NovaSynTGR resin (EMD Millipore, Billerica, MA) (16). For the 11-mer used in pThr6 ligations, 20% piperidine in DMF was used for deprotection, and residues were coupled using five molar equivalents of protected amino acid, 4.75 molar equivalents of HBTU, 4.75 molar equivalents of hydroxybenzotriazole, and 10 molar equivalents of DIPEA. For the 29-mer used in pTyr8 ligations, 20% piperidine in DMF was used for deprotection, and residues were coupled in 0.4 M NMM/DMF using four molar equivalents of protected amino acid and 3.75 molar equivalents of HATU. For both the 11-mer and 29-mer, a Boc-protected methionine was added to the N-terminus. After elongation, these peptides were washed with DCM then reacted for 1 h with five molar equivalents of 4-nitrophenyl chloroformate dissolved in DCM. After washing with DCM, 0.5 M DIPEA in DMF was added for 30 min. The peptides were washed with DMF and DCM, then cleaved from the resin with a mixture containing 90% trifluoroacetic acid, 5% DCM, 2.5% water, and 2.5% triisopropylsilane. The resulting peptides contain a C-terminal N-acyl-benzimidazolinone group (Nbz), and were used in ligations as described below.
After cleavage from the resin, all peptides were ether-precipitated then purified (>95%) by reversed-phase high-pressure liquid chromatography on a C18 column using an acetonitrile/water gradient (0.1% trifluoroacetic acid). The identities of the final peptides were confirmed with mass spectrometry (Figs. S1–S7).
Expression and purification of UNG2(aa 12–313, S12C)
The truncated human UNG2 gene encoding residues 12-313 and containing a S12C mutation was inserted into an empty pET21a vector with an 8×His-tag, a Gly-Ser-Gly-Ser-Gly linker, then the SUMO protein (SMT3 from Saccharomyces cerevisiae) fused in-frame to its N-terminus. The plasmid was transformed into BL21(DE3)pLysS cells, which were plated on Luria Broth (LB)-Agar containing ampicillin and chloramphenicol. Colonies were picked for overnight starter cultures grown at 37°C in Miller’s LB broth containing 50 mg/L carbenicillin and 25 mg/L chloramphenicol. For protein expression, 10 mL of starter culture was added to 1 L of Miller’s LB broth (also containing 50 mg/L carbenicillin and 25 mg/L chloramphenicol), and the Escherichia coli were grown at 35°C while shaking at 180 rpm. When O.D. ∼0.6, the flask was cooled on ice to room temperature, and isopropylthiogalactoside was added to a final concentration of 0.25 mM. The flask then continued to shake at 21°C and 180 rpm for 16 h. The cells were then pelleted, and the pellet was stored frozen at −80°C.
For purification, the pellet was thawed and resuspended in ice-cold lysis buffer (20 mM sodium phosphate, pH 7.5, 300 mM NaCl, 20 mM imidazole, and 1 mM triscarboxyethylphosphine (TCEP)) containing Roche complete protease inhibitor cocktail (1 tablet/300 mL buffer). All subsequent steps were performed at 4°C. The cells were lysed with a French press before centrifugation at 20,000× g. The supernatant was gravity flowed through a column containing Ni2+ Sepharose 6 Fast Flow resin (GE Healthcare, Little Chalfont, UK). The resin was washed twice with 10 mL lysis buffer, followed by consecutive treatment (8 mL each) with lysis buffer containing 50, 100, 150, or 250 mM imidazole. The fusion protein (8×His-SUMO-UNG2(aa 12–313, S12C)) elutes with the 150 and 250 mM imidazole treatments. These fractions were pooled and dialyzed against 1.5 L of 50 mM Tris, pH 8.0, 140 mM NaCl, 1 mM TCEP, and 2.5% glycerol for ∼4 h before transferal to 1.5 L of fresh, identical buffer for overnight dialysis. The protein was then concentrated to ∼2–3 mg/mL with an Amicon 10-kDa molecular-mass cutoff centrifugal filter (EMD Millipore) before being aliquoted, snap frozen in liquid N2, and stored at −80°C. Approximate yield of the fusion protein was 16 mg/L of bacterial cell culture.
To remove the 8×His-SUMO tag, the protein was thawed then digested with 300 μg SUMO protease (ubiquitin-like-specific protease, ULP1, which contained a 6×His-tag) for 5 h at 4°C. The protein mixture was then reapplied to the Ni2+ column, and UNG2(aa 12–313, S12C) was eluted with lysis buffer containing 50 mM imidazole. The SUMO-free UNG2 protein was dialyzed overnight against 20 mM sodium phosphate, pH 7.5, 100 mM NaCl, 1 mM TCEP, and 10% glycerol, then the protein was concentrated to ∼1.5 mg/mL with an Amicon 10-kDa molecular-mass cutoff centrifugal filter (EMD Millipore). UNG2(aa 12–313, S12C) was ∼95% pure based on SDS-PAGE.
Semisynthesis of pThr6-UNG2
The 11-mer peptide MIGQKpTLYSFF-Nbz was suspended in 6 M guanidinium-HCl, 10 mM sodium phosphate, pH 7.0, at a concentration of 12 mg/mL. To install a C-terminal thioester, this was slowly diluted 5.7-fold with 312 mM mercaptoethylsulfonate (MESNA), 10 mM sodium phosphate, pH 6.8, and the pH of the reaction was adjusted to 7.0 with NaOH. After 20 min at room temperature, the peptide was slowly added to a gently stirring solution that contained 25 μM UNG2(aa 12–313, S12C) in 15 mM sodium phosphate, pH 7.5, 60 mM NaCl, 150 mM MESNA, 4 mM TCEP, and 5.8% glycerol; the final concentration of UNG2(aa 12–313, S12C) after peptide addition was 15 μM. The solution pH was adjusted to 7.5 with NaOH, and the mixture maintained at room temperature for 24 h.
The reaction mixture was subsequently dialyzed against 10 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM dithiothreitol (DTT), and 6% glycerol at 4°C. The protein solution was then diluted 5.3-fold with 25 mM Tris-acetate, pH 8.0, 10 mM NaCl, and 0.5 mM DTT. This was immediately injected onto an equilibrated Mono S 5/50 GL cation exchange column (GE Healthcare). For fast protein liquid chromatography, Buffer A was 50 mM Tris-acetate, pH 8.0, 20 mM NaCl, and 1 mM DTT, and Buffer B was 50 mM Tris-acetate, pH 8.0, 300 mM NaCl, and 1 mM DTT. The protein was eluted from the Mono S column with a linear gradient of 0–100% Buffer B over 198 column volumes, and elution fractions were analyzed by SDS-PAGE. Glycerol was added to fractions containing purified pThr6-UNG2 such that the final concentration was 15%, then the protein was concentrated to >1 mg/mL, aliquoted, and snap frozen in liquid N2 before storing at −80°C.
Semisynthesis of pTyr8-UNG2
The 29-mer peptide MHHHHHHKRKGENLYFQGMIGQKTLpYSFF-Nbz was dissolved in 6 M guanidinium-HCl, 10 mM sodium phosphate, pH 7.0, at a concentration of 13.5 mg/mL. The peptide was then diluted 5.7-fold with 310 mM MESNA, 10 mM sodium phosphate, pH 6.8, and the solution pH was adjusted to 7.0 with NaOH. After 30 min at room temperature, the peptide was slowly added to a gently stirring solution that contained 29 μM UNG2(aa 12–313, S12C) in 15 mM sodium phosphate, pH 7.5, 70 mM NaCl, 150 mM MESNA, 4 mM TCEP, and 6.8% glycerol; the final concentration of UNG2(aa 12–313, S12C) after peptide addition was 15 μM. The solution pH was adjusted to 7.5 with NaOH, and the mixture maintained at room temperature for 26 h.
The reaction mixture was subsequently diluted fourfold with 20 mM sodium phosphate, pH 7.5, 150 mM NaCl, and 1 mM DTT and then applied to an equilibrated Ni2+ column. The column was washed with lysis buffer (as above) that contained 20, 50, or 100 mM imidazole, then the desired product was eluted with lysis buffer containing 250 mM imidazole. This protein was dialyzed into 20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM DTT, and 5% glycerol, then digested until completion with tobacco etch virus (TEV) protease at 4°C. After digestion, the mixture was reapplied to the Ni2+ column and was subsequently eluted with lysis buffer containing 50 mM imidazole. The desired semisynthetic protein (pTyr8-UNG2) was dialyzed against 20 mM sodium phosphate, pH 7.5, 200 mM NaCl, 1 mM DTT, and 15% glycerol, then was concentrated to >1 mg/mL before being snap frozen in liquid N2.
Electrospray ionization mass spectrometry
UNG2 proteins were desalted with C4 ZipTips (EMD Millipore), eluted with 70% acetonitrile in water with 0.1% formic acid, then diluted twofold with water containing 0.1% formic acid. Proteins were injected onto a BioBasic C4 column (Thermo Fisher Scientific) using a nanoAcquity ultra-performance liquid chromatography system (Waters, Milford, MA), then eluted using a 20–72% gradient of acetonitrile in water with 0.1% formic acid. The samples were electrosprayed online into a Thermo TSQ Vantage mass spectrometer, and data was collected using the software Xcalibur (Thermo Fisher Scientific).
Protein elution over ∼20 s was an intense peak on the total ion current chromatogram, and the spectra within that peak were averaged in the Xcalibur Qual Browser (Thermo Fisher Scientific) to yield a characteristic protein spectrum for each sample. On the spectrum for each protein, the m/z for each peak was extracted, the protein charge state (z) that corresponded to that m/z was deduced, and the experimentally observed protein mass (massobs) for each peak was calculated using the equation
| (1) |
where H+ is the mass of a proton. The observed masses from the peaks for each spectrum were then averaged.
We assessed the accuracy and precision of our mass spectrometer for measuring intact protein masses by using the UNG catalytic domain as a standard control. This protein has been extensively characterized both structurally and biochemically (17, 18) and has a predicted monoisotopic mass of 25,489.0 Da. The observed mass from the +22 to +34 charge state ions of catalytic UNG was 25,494.6 ± 4.0 Da (mean ± SD), indicating an experimental error of this measurement of ∼ ±10 Da (Fig. S8).
Expression and purification of UNG2 and UNG2(S12C)
The full-length human UNG2 gene or the gene containing an S12C mutation was inserted into an empty pET21a vector with an 8×His-tag, a Gly-Ser-Gly-Ser-Gly linker, and the SUMO protein fused in-frame to the N-terminus. Plasmid transformation into BL21(DE3)pLysS cells and subsequent growth and induction was identical to that described above for the truncated protein UNG2(aa 12–313, S12C). The cells were pelleted after growth and stored at −80°C.
The fusion protein 8×His-SUMO-UNG2 was isolated using Ni2+ chromatography as described above, and after dialysis, ULP1 removed the 8×His-SUMO. The protein mixture was then reapplied to the Ni2+ column, which was subsequently eluted with lysis buffer containing 50 mM imidazole. The protein was dialyzed against 10 mM sodium phosphate, pH 7.4, 300 mM NaCl, 20% glycerol, and 1 mM DTT, then concentrated to ∼5 mg/mL with an Amicon 10-kDa molecular-mass cutoff centrifugal filter (EMD Millipore). The protein was then further purified with size exclusion chromatography (Superdex 200 10/300 GL column; mobile phase: 10 mM sodium phosphate, pH 7.4, 150 mM NaCl, and 1 mM DTT; GE Healthcare). Glycerol was then added to the peak fractions containing UNG2 such that the final concentration was 20%, and the protein was snap frozen in liquid N2 before storing at −80°C. Approximate yield of the purified protein was 3 mg/L of bacterial cell culture.
UNG2 uracil excision assay
The 19-mer oligos 5′-GCGGCCAAAUAAAAAGCGC-3′ and 5′-GCGCTTTTTPTTTGGCCGC-3′ (U, uridine deoxyribonucleotide; P, 2-aminopurine deoxyribonucleotide) were purchased from Integrated DNA Technologies (Coralville, IA). These were dissolved in 10 mM Tris-HCl, pH 8.0, 40 mM NaCl, and 0.1 mM EDTA, and mixed in equal concentrations (240 μM each). The oligos were heated to 95°C for 4 min then cooled to room temperature. The oligos were diluted 10-fold, the NaCl concentration was adjusted to 60 mM, and MgCl2 was added to a final concentration of 2.5 mM. Annealing of the oligos was confirmed with native gel electrophoresis (20% polyacrylamide-TBE) followed by ethidium bromide staining.
Steady-state kinetic measurements of uracil excision activity were performed essentially as described in Stivers (19) using 158.5 pM UNG2 and a buffer of 10 mM Tris-HCl, pH 8.0, 60 mM NaCl, 2.5 mM MgCl2, and 1 mM DTT. Substrate concentrations of 0.11, 0.15, 0.25, 0.5, 1, 3, and 5 μM were used, and assays for each protein were performed in triplicate. Temperature was controlled at 23°C.
PCNA expression and purification
The human PCNA gene was inserted into an empty pET21a vector with an N-terminal 6×His-tag that was followed by a Gly-Gly-Gly linker. The plasmid was transformed into BL21(DE3)pLysS cells, and growth and induction conditions were identical to that of the UNG2 proteins. The E. coli were lysed and the protein was purified on a Ni2+ column as described above, with PCNA eluting in the 150 and 250 mM imidazole fractions.
PCNA-PL peptide binding assays
PCNA binding to the PL peptide was determined by fluorescence anisotropy measurements based on a published protocol (20). To maintain constant buffer conditions, proteins used in all anisotropy assays were first dialyzed into the fluorescence anisotropy assay buffer (FA Buffer: 10% glycerol, 0.01% Triton X-100, 100 mM NaCl, 1 mM MgCl2, 1 mM DTT, and 25 mM HEPES-NaOH, pH 7.4). Peptides were also dissolved directly in FA Buffer. Assays were performed at 23°C, and the concentration of PL peptide was 32 nM in all assays. To determine the affinity of PL peptide for PCNA, anisotropy measurements were taken after equilibrating the peptide with increasing concentrations of protein (concentrations were based on amount of PCNA monomer). The measurements were plotted versus PCNA concentration, and the data was fit using the quadratic binding equation
| (2) |
where A(min) and A(max) are the minimum and maximum anisotropy values, [PL] is the total PL peptide concentration, [PCNA] is the PCNA concentration, and Kd is the PCNA concentration at half-maximal saturation (21).
Competition experiments during which the PL peptide was displaced from PCNA were performed using UNG2, its protein variants, and 19-mer UNG2 peptides. PL peptide (32 nM) was equilibrated with 0.23 μM PCNA and the corresponding competitor, then anisotropy measurements were taken. The anisotropy values were plotted versus the log10 of the competing UNG2 protein or peptide concentration. When required, the data were fit with a sigmoidal curve using the equation
| (3) |
with variable Ymin, Ymax, n (Hill coefficient), and IC50 values. The IC50 derived from this curve was used to calculate the affinity (Ki) of the UNG2(aa 1–19) peptides for PCNA using the equation
| (4) |
where L50 is the concentration of free PL peptide at 50% inhibition, P0 is the concentration of free PCNA monomer at 0% inhibition, and Kd is the dissociation constant of the PCNA-PL peptide complex (22). Note that, for competition experiments with pThr6-UNG2(aa 1–19), we were unable to fully saturate the PCNA sites at the highest peptide concentrations. In this case, the IC50 was estimated to be the concentration of pThr6-UNG2(aa 1–19) that caused the anisotropy to decrease to the same level observed at the IC50 for the nonphosphorylated UNG2(aa 1–19) curve.
Alkaline phosphatase dephosphorylation of pTyr8-UNG2
Here, 32 nM PL peptide was equilibrated with 24 μM pTyr8-UNG2 at 23°C in FA Buffer. A quantity of 0.1 μM Calf Intestinal Alkaline Phosphatase (New England Biolabs, Ipswich, MA) was added, and after briefly mixing, anisotropy measurements were recorded every minute. The data was fit to the equation for a one-phase decay:
| (5) |
where Y0 is the initial anisotropy, Ymin is the minimum plateau anisotropy derived from the fitted curve, K is the variable rate constant, and X is the time in minutes. As a control, an identical experiment was performed with 24 μM unmodified full-length UNG2.
RPA purification
Human RPA was expressed in BL21-CodonPlus (DE3) cells using the p11d-tRPA plasmid (23), which was a generous gift from Dr. Marc Wold. The protein was purified essentially as described in Henricksen et al. (23) and Binz et al. (24) by three consecutive chromatography columns: Affi-Gel Blue (Bio-Rad, Hercules, CA), hydroxyapatite (Bio-Rad), and Mono Q (GE Healthcare).
RPA-SMARCAL peptide binding assays
All assays were performed at 23°C in FA Buffer, and the concentration of the fluorescent peptide SMARCAL was 20 nM. To determine the affinity of SMARCAL peptide for RPA protein, anisotropy measurements were taken after equilibrating the peptide with increasing concentrations of protein. The methodology used to determine the Kd of the interaction using the quadratic binding fit was identical to that described above for PCNA and PL peptide (i.e., Eq. 2). In competition experiments where SMARCAL peptide was displaced from RPA with UNG2, the concentration of RPA was 9 μM, and where applicable, the concentration of PCNA was 42 μM; as a control, we determined that PCNA itself had no effect on the anisotropy or fluorescence intensity of free SMARCAL peptide. The anisotropy values were plotted versus the log10 of the competing UNG2 concentration. Also as described above, a sigmoidal curve was fit to this data using Eq. 3 with variable Ymin, Ymax, n, and IC50 values, and the Ki of the UNG2-RPA interaction was calculated using Eq. 4.
Similar assays were performed using RPA that was prebound to a 31-nt ssDNA (dT(31), purchased from Integrated DNA Technologies). Before the binding experiments, RPA in FA Buffer was equilibrated with an equimolar concentration of dT(31) for 25 min at room temperature. In these conditions, the Kd of the RPA-dT(31) interaction is 0.22 nM (25). During the titration experiment to determine the affinity between SMARCAL peptide and RPA-dT(31), >95% of RPA should be bound to dT(31) at even the lowest concentration tested (105 nM). In the competition experiment where SMARCAL peptide was displaced from RPA-dT(31) with UNG2, the concentration of RPA-dT(31) was 14.6 μM.
Generation of UNG2(Fluor)
Mutations were inserted into the pET21a plasmid encoding 8×His-SUMO-UNG2 (from above) to generate a plasmid that contained a cysteine residue before Met1 of UNG2 (i.e., immediately downstream from the SUMO protease cleavage site) and also cysteine to alanine mutations at three UNG2 sites (C141A, C166A, and C290A). This protein was expressed, purified with Ni2+ column chromatography, cleaved with ULP1, then reapplied to a Ni2+ column and eluted as described above for UNG2 and UNG2(aa 12–313, S12C). Subsequently, the UNG2 mutant was dialyzed into a buffer containing 10% glycerol, 200 mM NaCl, 1 mM TCEP, and 50 mM sodium phosphate, pH 7.2. After dialysis, the protein was concentrated to 53 μM with an Amicon 10-kDa molecular-mass cutoff centrifugal filter (EMD Millipore). Fluorescein-5-maleimide (Thermo Fisher Scientific), originally dissolved in DMSO at 20 mg/mL, was added dropwise to the protein to a final concentration of 500 μM. The protein reacted with fluorescein-5-maleimide for 20 h at 4°C in the dark, then the reaction was quenched by adding 5 mM DTT and incubating for 1 h. The protein solution was dialyzed extensively over 36 h against 10% glycerol, 200 mM NaCl, 1 mM DTT, and 30 mM sodium phosphate, pH 7.2, with several changes of dialysis buffer to begin removing unconjugated fluorescein-5-maleimide. The fluorescein-labeled protein UNG2(Fluor) was then further purified with size exclusion chromatography.
UNG2(Fluor) binding assays
50 nanomolar of UNG2(Fluor) was used in all assays with FA Buffer. Anisotropy measurements were taken with increasing PCNA concentrations, and the data was fit using Eq. 2 as in PL peptide binding assays. When PCNA and RPA were added simultaneously, 25 μM PCNA and 16 μM RPA were used.
Data statistics and figure preparation
For binding assays, each data point represents the mean measurement with standard error from two to four independent experiments. In many cases, the error bars are smaller than the plotted point. The error associated with IC50 and Kd values also represents SE. All curves were fit using the software GraphPad Prism 6 (https://www.graphpad.com/scientific-software/prism/), which was also used for figure preparation.
Results
Production of semisynthetic phospho-UNG2s
We used the chemoselective N-Cys/C-thioester “native chemical ligation” reaction to generate UNG2 proteins that contained a phosphorylation at either Thr6 or Tyr8 (Fig. 1 A) (26). Initially, we synthesized peptides that corresponded to UNG2 residues 1-11 and contained a phosphorylation at either Thr6 or Tyr8, then installed a C-terminal MESNA thioester on the peptides under aqueous conditions (16). These peptides reacted with truncated UNG2(aa 12–313, S12C), which was generated as a SUMO-fusion protein cleaved by the protease ULP1. Typically, the ligation reaction converted 60–80% of the truncated UNG2 into full-length, post-translationally modified protein that contained a single point mutation (S12C) at the site of ligation.
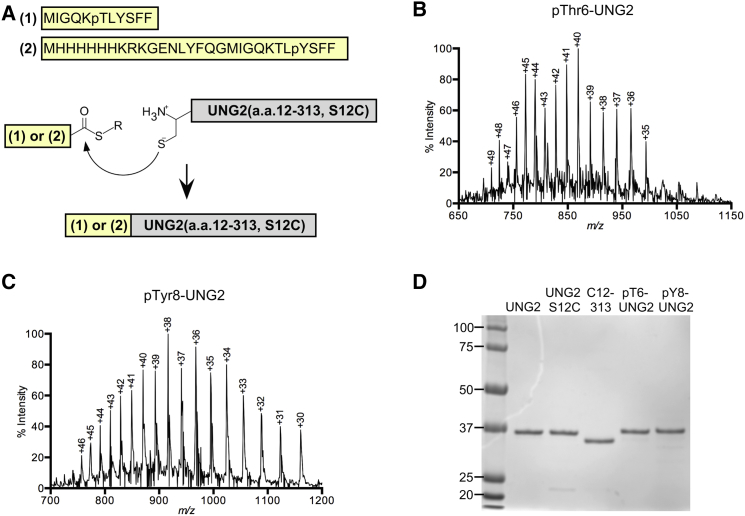
Figure 1.
(A) Synthetic peptides (1) and (2), which contained C-terminal MESNA thioesters, were reacted with a truncated UNG2 containing an N-terminal cysteine at residue 12 of the protein. The native chemical ligation reaction combined the polypeptides to form full-length, post-translationally modified UNG2 proteins. (B) Shown here is a mass spectrum for intact pThr6-UNG2 made with native chemical ligation. This data was obtained using ESI-MS, and the charge states of the peaks are indicated. Eq. 1 was used to calculate an observed mass of 34,728.8 ± 7.3 Da (mean ± SD), and the predicted monoisotopic mass is 34,719.8 Da. Using a standard control, the accuracy of our instrument was determined to be ±10 Da (see Materials and Methods). (C) Shown here is a mass spectrum for intact pTyr8-UNG2. The observed mass was 34,784.1 ± 7.1 Da, and the predicted monoisotopic mass is 34,776.8 Da. (D) Shown here is a Coomassie-stained SDS-PAGE gel showing purity of UNG2 enzymes used for native chemical ligation, ESI-MS, and/or enzyme assays. To see this figure in color, go online.
Semisynthetic pThr6-UNG2 could be purified from remaining UNG2(aa 12–313, S12C) using cation exchange chromatography (Fig. S9); however, semisynthetic pTyr8-UNG2 that was generated with this method coeluted with unligated UNG2(aa 12–313, S12C) in a variety of chromatography conditions including cation exchange, gel filtration, and hydroxyapatite (data not shown). To circumvent this purification challenge, a separate strategy to obtain purified pTyr8-UNG2 was developed. In this alternative approach, we prepared a 29-mer thioester peptide that contained a 6×His-tag, charged residues to enhance peptide solubility, and a TEV protease cleavage site that preceded UNG2 residues 1-11 with a phosphorylation at Tyr8 (Fig. 1 A). After reacting with truncated UNG2, the semisynthetic His-TEV-pTyr8-UNG2 construct was purified using Ni2+ chromatography. The 6×His-tag was removed by cleavage with TEV protease and the mixture was further purified chromatographically to yield full-length pTyr8-UNG2 in high purity with the S12C mutation and a residual glycine on the N-terminus (Fig. S10).
For both pThr6-UNG2 and pTyr8-UNG2, we used electrospray ionization mass spectrometry (ESI-MS) on the intact proteins to confirm that the desired constructs were obtained (Fig. 1, B and C). Having made the desired phosphorylated proteins, we tested whether the modifications influenced protein activity. We measured the enzymatic function of the semisynthetic proteins and compared them to recombinant full-length UNG2 and a recombinant UNG2(S12C) mutant (Fig. 1 D). For these assays, we used a 19-mer duplex DNA substrate that contained uracil opposite the fluorescent base 2-aminopurine (19). We found that the phosphorylated and unmodified proteins had identical activity that was comparable to previously published data for UNG2 (27) (Fig. S11; Table 1).
Table 1.
UNG2 Kinetic Parameters
| Protein | kcat (min−1) | Km (μM) |
|---|---|---|
| UNG2 | 636 ± 68 | 0.7 ± 0.2 |
| UNG2(S12C) | 546 ± 36 | 0.6 ± 0.1 |
| pThr6-UNG2 | 585 ± 74 | 0.7 ± 0.3 |
| pTyr8-UNG2 | 687 ± 58 | 0.8 ± 0.2 |
UNG2-PCNA binding
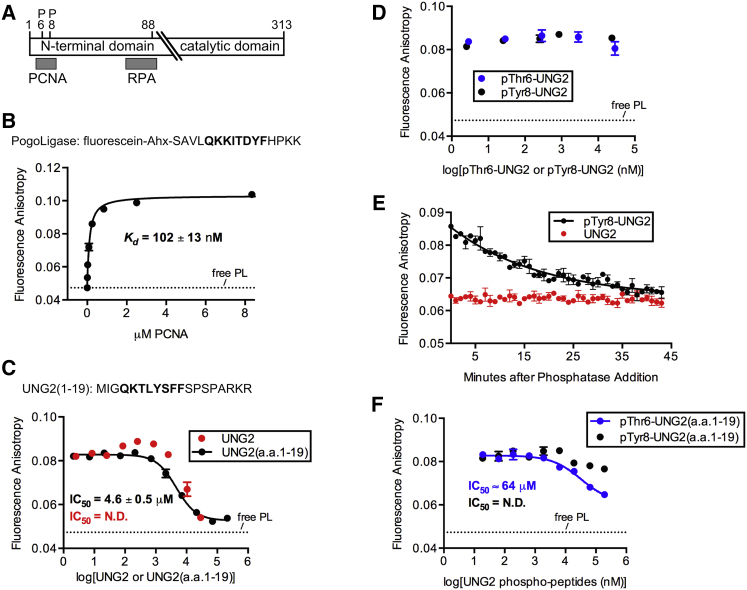
The UNG2 N-terminus contains a canonical PCNA-interacting motif (PIP-box) at residues 4-11 that is thought to bind to the interdomain connecting loop (IDCL) of PCNA (Fig. 2 A) (3). Note that PCNA is a homotrimer, and that each protein contains three IDCL sites. We used an anisotropy assay to measure UNG2 binding to PCNA (20). First, we determined that the fluorescent peptide PL, which is known to interact with the IDCL sites, binds PCNA with a Kd of 102 nM (Fig. 2 B), matching the published Kd of 100 nM determined for PCNA and unlabeled PL peptide (28). Next, after equilibrating PCNA with labeled PL peptide, we titrated full-length UNG2 to saturate the PCNA sites, which manifested as a decrease in fluorescence anisotropy of the labeled peptide (Fig. 2 C).
Figure 2.
(A) Shown here is a schematic of UNG2 showing the relative locations of the PCNA and RPA binding sites on the N-terminal domain, as well as the position of the phosphorylated residues examined in this work. (B) Shown here is fluorescence anisotropy data from titrating increasing concentrations of PCNA into a cuvette with a fixed concentration of PL peptide. The dashed line indicates the anisotropy of free PL peptide in solution (i.e., in the absence of PCNA). The sequence of PL peptide is shown (Ahx, 6-aminohexanoic acid), and its PIP-box is in bold. (C) Given here are anisotropy measurements from competition experiments during which increasing concentrations of full-length UNG2 protein or UNG2(aa 1–19) peptide displaced PL peptide from PCNA. The Hill slope of the curve for UNG2(aa 1–19) was −1.3, and the sequence of UNG2(aa 1–19) is shown with the PIP-box bolded. (D) Given here are anisotropy measurements, similar to (C), but showing that phosphorylated UNG2 proteins do not efficiently displace PL peptide from PCNA. (E) Dephosphorylation of 24 μM pTyr8-UNG2 over time allowed the protein to bind PCNA, thereby competing with PL peptide for binding, resulting in decreased anisotropy measurements. The curve fit to the pTyr8-UNG2 data (R2 = 0.91) determined a Ymin plateau anisotropy = 0.0638. For comparison, over the course of the control experiment using 24 μM unmodified UNG2, the averaged anisotropy was 0.0636. (F) Anisotropy data were determined by displacing PL peptide from PCNA using phospho-UNG2(aa 1–19) peptides. To see this figure in color, go online.
In this competition experiment, we unexpectedly observed a modest increase in anisotropy values at lower UNG2 concentrations (0.08–0.8 μM) that was not compatible with a conventional one-ligand, one-receptor binding isotherm. In comparison, when PL peptide was displaced with the synthetic peptide UNG2(aa 1–19), no increase in the anisotropy signal was observed, and the data fit well to a standard competition curve (Fig. 2 C). We interpret these results as evidence that a ternary PL peptide-PCNA-UNG2 complex formed at low concentrations of UNG2 protein due to the presence of three IDCL sites per PCNA homotrimer, and that the significantly larger size of the PL peptide-PCNA-UNG2 complex compared to that of the PL peptide-PCNA complex or the PL peptide-PCNA-UNG2(aa 1–19) complex was sufficient to increase the anisotropy signal.
In competition experiments with similar protein levels, pThr6-UNG2 and pTyr8-UNG2 were ineffective at displacing PL peptide from PCNA (Fig. 2 D). As a control, we also determined that the protein UNG2(S12C) bound PCNA similarly to wt UNG2 (Fig. S12), demonstrating that inhibition of binding was caused by phosphorylation and not the point mutation. We next tested whether we could generate a PCNA binding partner by dephosphorylating semisynthetic pTyr8-UNG2 to produce an UNG2(S12C) construct that also contains a glycine on its N-terminus. Addition of calf alkaline phosphatase to a mixture of PCNA, PL peptide, and pTyr8-UNG2 caused a time-dependent decrease in anisotropy that was consistent with displacement of PL peptide from PCNA by dephosphorylated semisynthetic protein (Fig. 2 E).
Next, we determined that even at high concentrations (∼200 μM) of synthetic phosphorylated peptides pThr6-UNG2(aa 1–19) and pTyr8-UNG2(aa 1–19), only partial displacement of PL peptide was observed (Fig. 2 F). This experiment did, however, suggest that phosphorylation at Tyr8 had a more disruptive effect on PCNA binding than pThr6. Although we could not completely displace PL peptide from PCNA with pThr6-UNG2(aa 1–19), the estimated IC50 corresponded to a Kd of 19 μM for its interaction with PCNA. In contrast to the relatively weak interactions with the phospho-peptides, we calculated a Kd of 1.3 μM for the unmodified UNG2(aa 1–19) interaction with PCNA (Fig. 2 C). Because complete displacement of PL peptide from PCNA was achieved with similar concentrations of UNG2(aa 1–19) and full-length UNG2 (Fig. 2 C), we reasoned that the UNG2 peptide and protein have similar affinities for PCNA. This would suggest that the PIP-box motif is necessary and sufficient for UNG2 binding to PCNA, that no significant contacts occur between the proteins outside of the PIP-box, and that phosphorylation of the UNG2 PIP-box can impede binding.
UNG2-RPA binding and ternary complex formation
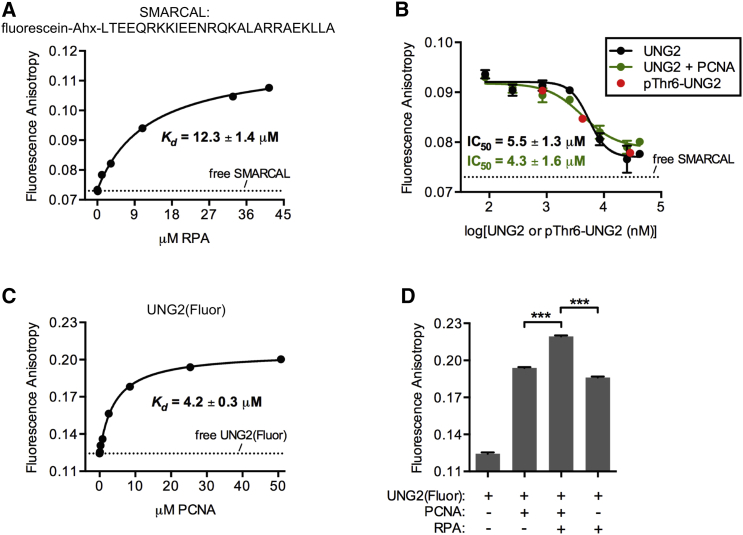
Separate from the PIP-box motif, UNG2 residues 66-91 are thought to mediate binding to RPA through the winged-helix (WH) domain of the RPA32 subunit (Fig. 2 A) (3, 6). The UNG2 binding site on the RPA32 WH domain is shared with several DNA replication/repair proteins (29, 30, 31, 32), and we synthesized a fluorescently labeled peptide called SMARCAL that also binds to this common site (32). With a fluorescence anisotropy assay, we first determined a Kd of 12.3 μM for the SMARCAL peptide interaction with RPA (Fig. 3 A). This is somewhat higher than the measured Kd of 2.9 μM for the interaction of unlabeled SMARCAL peptide with the isolated WH domain (32), although the labeled peptide had sufficient affinity for our competition experiments. SMARCAL peptide was next equilibrated with RPA, then displaced with full-length UNG2 to obtain an IC50, from which we calculated a Kd of 3.2 μM for the UNG2-RPA interaction (Fig. 3 B). This affinity is similar to the measured Kd of 6.6 μM for the peptide UNG2(aa 66–91) interacting with the WH domain (32), which suggested limited UNG2-RPA contacts outside of this binding domain. Consistent with this, phosphorylation of UNG2 in the PIP-box motif had no effect on UNG2 binding to RPA (Fig. 3 B). In a separate set of experiments, we also determined that the affinity of UNG2 for RPA was unchanged when RPA was bound to ssDNA (Fig. S13), which was predicted because the WH domain and the DNA binding core of RPA are separated by a long, flexible linker.
Figure 3.
(A) Shown here are binding data from titrating RPA into a fixed concentration of SMARCAL peptide. The sequence of SMARCAL peptide is shown (Ahx, 6-aminohexanoic acid), and the dashed line indicates its fluorescence anisotropy in solution without RPA. (B) Given here is a competition experiment showing SMARCAL peptide displacement from RPA by UNG2 or pThr6-UNG2. For UNG2 in the absence of PCNA, the Hill slope of the curve was −2.7, and in the presence of PCNA, the Hill slope was −1.4. When added, the PCNA concentration was 42 μM. (C) Shown here are fluorescence anisotropy data obtained by equilibrating 50 nM UNG2(Fluor) with increasing PCNA concentrations. (D) Shown here are fluorescence anisotropy measurements from 50 nM UNG2(Fluor) equilibrated with or without 25 μM PCNA and 16 μM RPA. We used a one-way ANOVA and Bonferroni’s post hoc test to statistically determine that the anisotropy measurements from the three proteins combined were greater than the measurements obtained when UNG2(Fluor) was equilibrated with only PCNA or RPA (∗∗∗p < 0.001). Note that the data for UNG2(Fluor) alone and UNG2(Fluor) + PCNA is taken from (C). To see this figure in color, go online.
We tested whether PCNA could impact the interaction of UNG2 with RPA. In this experiment, we titrated UNG2 to displace SMARCAL peptide from RPA while coincubating with 42 μM PCNA, a concentration that exceeds the Kd for UNG2-PCNA binding by at least 10-fold. If UNG2’s interaction surface with PCNA overlapped with UNG2’s binding surface with RPA, then this high concentration of PCNA would have been expected to reduce the observed binding affinity between UNG2 and RPA. However, the measured IC50 for UNG2 displacement of SMARCAL peptide was unaffected by the presence of PCNA, strongly suggesting that UNG2 interactions with RPA and PCNA are independent and that a ternary complex can form (Fig. 3 B).
To obtain more direct evidence for the formation of a ternary PCNA-UNG2-RPA complex, we used a fluorescein-labeled, full-length UNG2 construct called UNG2(Fluor). To generate UNG2(Fluor), we mutated the three wild-type UNG2 cysteines to alanines and genetically installed a cysteine before Met1. This UNG2 mutant was shown to possess a catalytic efficiency (kcat/Km) within threefold of wild-type UNG2, suggesting that it was folded correctly and functional (Fig. S14). This N-terminal mono-cysteine UNG2 mutant protein reacted with maleimide-fluorescein to furnish UNG2(Fluor) (Fig. S14). We equilibrated UNG2(Fluor) with increasing PCNA concentrations and determined a Kd of 4.2 μM (Fig. 3 C), which is in the same range as the affinity we determined for PCNA and UNG2(aa 1–19) (Kd = 1.3 μM). Next, we equilibrated UNG2(Fluor) with a saturating concentration of PCNA and/or 16 μM RPA. The measured anisotropy from the combination of UNG2(Fluor), PCNA, and RPA was significantly higher than UNG2(Fluor) equilibrated with either PCNA or RPA alone (Fig. 3 D). This relative fluorescence anisotropy increase when the three proteins are combined, compared to the individual protein pairs, is at a level that is entirely consistent with the formation of a PCNA-UNG2-RPA protein complex in the concentration range examined.
Discussion
PCNA, UNG2, and RPA are simultaneously present at the replication fork during normal replication and also times of stress that cause fork stalling and collapse (1, 2, 33). In various contexts, cellular experiments have indicated roles for PCNA and RPA in promoting UNG2 localization to replication sites, which are highly susceptible to DNA damage (2). When expression of UNG2 peaks during S-phase, most of UNG2 is unmodified and therefore fully capable of binding both PCNA and RPA (1), which would support its localization to the replication fork.
In addition to phosphorylation sites at Thr6 and Tyr8, UNG2 is known to be phosphorylated and ubiquitinated within its RPA binding domain at positions Thr60, Ser64, and Lys78 (1, 2). Consistent with our findings, these PTMs and PTM-mimicking mutations throughout the N-terminus of UNG2 have no substantial effect on the catalytic activity of the purified enzyme (1, 2, 8, 34). Although we have shown that phosphorylation at Thr6 and Tyr8 impedes binding to PCNA, mutagenesis experiments have suggested that modifications at Thr60, Ser64, and Lys78 can disrupt UNG2 interactions with RPA (1, 2). N-terminal PTMs can therefore promote selective binding to either PCNA or RPA; however, in vitro assays have shown that binding to PCNA or RPA only marginally affects the activity of UNG2 (approximately twofold) (1, 6, 35), suggesting noncatalytic functions for the interactions.
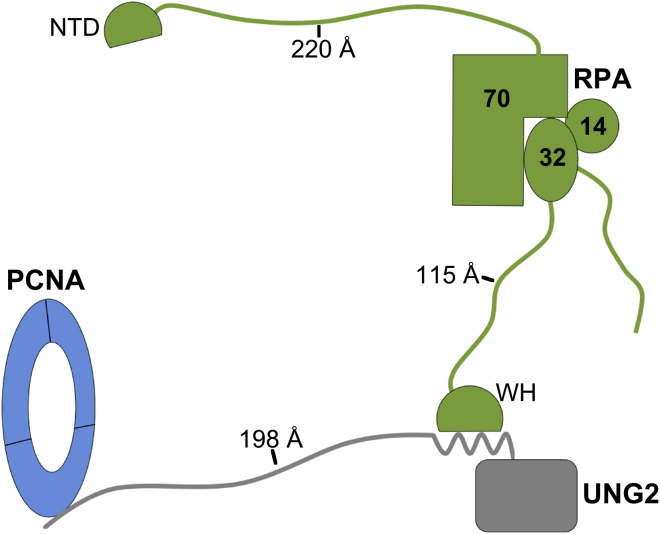
Our findings lead us to hypothesize that PTMs on UNG2 could have a greater effect than simply switching the protein between PCNA- and RPA-bound states at the replication fork. Specifically, we propose that the long flexible N-terminus of UNG2 could act as a scaffold that tethers PCNA and RPA, and that specific PTMs on the N-terminus of UNG2 regulate its ability to form the ternary protein complex (Fig. 4). The length of the UNG2 N-terminus is ideally suited for simultaneous binding to PCNA and RPA. In addition, the WH domain of RPA is also connected to its DNA binding domain by a disordered linker (Fig. 4). These two disordered regions on UNG2 and RPA could accommodate flexible movement of the proteins on DNA at the replication fork, while retaining restricted spatial separation. Because UNG2 does not extensively contact PCNA or RPA outside of the IDCL or WH domains, its presence would not be expected to hinder binding of PCNA or RPA to other proteins. Thus, PCNA could simultaneously interact with more than one of its many binding partners through interactions with vacant IDCL sites (36, 37, 38). Similarly, RPA binding partners would still have unhindered access to the N-terminal domain of the 70-kDa RPA subunit (Fig. 4) (39). We therefore extend the model to speculate that UNG2 could serve as a platform to bring together other PCNA- and RPA-interacting proteins.
Figure 4.
Model of UNG2 tethering together PCNA and RPA with the WH domain of RPA32 and N-terminal domain (NTD) of RPA70 also indicated. The linker lengths were determined by assuming a contour length of 3.6 Å per amino acid (41), and for UNG2, the residues that constitute the PCNA and RPA binding domains were not included in this calculation. If PCNA and RPA maximally extend to ∼300 Å apart, ∼90 DNA bases/bps could fit between the proteins (assuming a contour length of 3.4 Å/bp) (42). To see this figure in color, go online.
Future work will test the relevance of UNG2 as a scaffold in specific contexts, for example, such as at blocked primer-template junctions that arise during translesion DNA synthesis (40). In this case, stalled replication results in a primer-template junction with PCNA on upstream dsDNA and RPA on downstream ssDNA. Recent work has shown that RPA prevents PCNA from advancing past the primer-template junction. Our results suggest that UNG2 could serve as a tether for PCNA and RPA and effectively stabilize PCNA at the junction to prevent its movement in the opposite, upstream direction (40). Should UNG2 serve as a tether in vivo, this could implicate the protein in non-uracil-related DNA damage responses that may be regulated by PTMs of UNG2. Accordingly, our findings suggest expanded functions for UNG2 apart from its role as a DNA glycosylase.
Author Contributions
B.P.W., J.T.S., and P.A.C. designed experiments, analyzed data, and wrote the article. B.P.W. performed experiments.
Acknowledgments
Funding was provided in part by the National Institutes of Health through grants T32-CA9110 and F32-GM119230 (B.P.W.), R01-GM056834 (J.T.S.), and R01-CA74305 (P.A.C.).
Editor: Elizabeth Rhoades.
Footnotes
Fourteen figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30666-5.
Supporting Material
References
- 1.Hagen L., Kavli B., Slupphaug G. Cell cycle-specific UNG2 phosphorylations regulate protein turnover, activity and association with RPA. EMBO J. 2008;27:51–61. doi: 10.1038/sj.emboj.7601958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torseth K., Doseth B., Slupphaug G. The UNG2 Arg88Cys variant abrogates RPA-mediated recruitment of UNG2 to single-stranded DNA. DNA Repair (Amst.) 2012;11:559–569. doi: 10.1016/j.dnarep.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 3.Otterlei M., Warbrick E., Krokan H.E. Post-replicative base excision repair in replication foci. EMBO J. 1999;18:3834–3844. doi: 10.1093/emboj/18.13.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fischer J.A., Muller-Weeks S., Caradonna S. Proteolytic degradation of the nuclear isoform of uracil-DNA glycosylase occurs during the S phase of the cell cycle. DNA Repair (Amst.) 2004;3:505–513. doi: 10.1016/j.dnarep.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 5.Hardeland U., Kunz C., Schär P. Cell cycle regulation as a mechanism for functional separation of the apparently redundant uracil DNA glycosylases TDG and UNG2. Nucleic Acids Res. 2007;35:3859–3867. doi: 10.1093/nar/gkm337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagelhus T.A., Haug T., Krokan H.E. A sequence in the N-terminal region of human uracil-DNA glycosylase with homology to XPA interacts with the C-terminal part of the 34-kDa subunit of replication protein A. J. Biol. Chem. 1997;272:6561–6566. doi: 10.1074/jbc.272.10.6561. [DOI] [PubMed] [Google Scholar]
- 7.Hornbeck P.V., Chabra I., Zhang B. PhosphoSite: a bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics. 2004;4:1551–1561. doi: 10.1002/pmic.200300772. [DOI] [PubMed] [Google Scholar]
- 8.Lu X., Bocangel D., Donehower L.A. The p53-induced oncogenic phosphatase PPM1D interacts with uracil DNA glycosylase and suppresses base excision repair. Mol. Cell. 2004;15:621–634. doi: 10.1016/j.molcel.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 9.Sharma K., D’Souza R.C.J., Mann M. Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Reports. 2014;8:1583–1594. doi: 10.1016/j.celrep.2014.07.036. [DOI] [PubMed] [Google Scholar]
- 10.Bai Y., Li J., Haura E.B. Phosphoproteomics identifies driver tyrosine kinases in sarcoma cell lines and tumors. Cancer Res. 2012;72:2501–2511. doi: 10.1158/0008-5472.CAN-11-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bergström Lind S., Artemenko K.A., Pettersson U. Toward a comprehensive characterization of the phosphotyrosine proteome. Cell. Signal. 2011;23:1387–1395. doi: 10.1016/j.cellsig.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 12.Iliuk A.B., Martin V.A., Tao W.A. In-depth analyses of kinase-dependent tyrosine phosphoproteomes based on metal ion-functionalized soluble nanopolymers. Mol. Cell. Proteomics. 2010;9:2162–2172. doi: 10.1074/mcp.M110.000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muir T.W., Sondhi D., Cole P.A. Expressed protein ligation: a general method for protein engineering. Proc. Natl. Acad. Sci. USA. 1998;95:6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Z., Cole P.A. Synthetic approaches to protein phosphorylation. Curr. Opin. Chem. Biol. 2015;28:115–122. doi: 10.1016/j.cbpa.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kontopidis G., Wu S.-Y., Walkinshaw M.D. Structural and biochemical studies of human proliferating cell nuclear antigen complexes provide a rationale for cyclin association and inhibitor design. Proc. Natl. Acad. Sci. USA. 2005;102:1871–1876. doi: 10.1073/pnas.0406540102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blanco-Canosa J.B., Dawson P.E. An efficient Fmoc-SPPS approach for the generation of thioester peptide precursors for use in native chemical ligation. Angew. Chem. Int. Ed. Engl. 2008;47:6851–6855. doi: 10.1002/anie.200705471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parikh S.S., Mol C.D., Tainer J.A. Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J. 1998;17:5214–5226. doi: 10.1093/emboj/17.17.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parker J.B., Bianchet M.A., Stivers J.T. Enzymatic capture of an extrahelical thymine in the search for uracil in DNA. Nature. 2007;449:433–437. doi: 10.1038/nature06131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stivers J.T. 2-Aminopurine fluorescence studies of base stacking interactions at abasic sites in DNA: metal-ion and base sequence effects. Nucleic Acids Res. 1998;26:3837–3844. doi: 10.1093/nar/26.16.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pedley A.M., Lill M.A., Davisson V.J. Flexibility of PCNA-protein interface accommodates differential binding partners. PLoS One. 2014;9:e102481. doi: 10.1371/journal.pone.0102481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seamon K.J., Sun Z., Stivers J.T. SAMHD1 is a single-stranded nucleic acid binding protein with no active site-associated nuclease activity. Nucleic Acids Res. 2015;43:6486–6499. doi: 10.1093/nar/gkv633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nikolovska-Coleska Z., Wang R., Wang S. Development and optimization of a binding assay for the XIAP BIR3 domain using fluorescence polarization. Anal. Biochem. 2004;332:261–273. doi: 10.1016/j.ab.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 23.Henricksen L.A., Umbricht C.B., Wold M.S. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 24.Binz S.K., Dickson A.M., Wold M.S. Functional assays for replication protein A (RPA) Methods Enzymol. 2006;409:11–38. doi: 10.1016/S0076-6879(05)09002-6. [DOI] [PubMed] [Google Scholar]
- 25.Kim C., Paulus B.F., Wold M.S. Interactions of human replication protein A with oligonucleotides. Biochemistry. 1994;33:14197–14206. doi: 10.1021/bi00251a031. [DOI] [PubMed] [Google Scholar]
- 26.Dawson P.E., Muir T.W., Kent S.B. Synthesis of proteins by native chemical ligation. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 27.Grogan B.C., Parker J.B., Stivers J.T. Effect of the thymidylate synthase inhibitors on dUTP and TTP pool levels and the activities of DNA repair glycosylases on uracil and 5-fluorouracil in DNA. Biochemistry. 2011;50:618–627. doi: 10.1021/bi102046h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheleva D.I., Zhelev N.Z., Lane D.P. A quantitative study of the in vitro binding of the C-terminal domain of p21 to PCNA: affinity, stoichiometry, and thermodynamics. Biochemistry. 2000;39:7388–7397. doi: 10.1021/bi992498r. [DOI] [PubMed] [Google Scholar]
- 29.Mer G., Bochkarev A., Chazin W.J. Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell. 2000;103:449–456. doi: 10.1016/s0092-8674(00)00136-7. [DOI] [PubMed] [Google Scholar]
- 30.Feng S., Zhao Y., Xu D. Ewing tumor-associated antigen 1 interacts with replication protein A to promote restart of stalled replication forks. J. Biol. Chem. 2016;291:21956–21962. doi: 10.1074/jbc.C116.747758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kemp M.G., Akan Z., Unsal-Kaçmaz K. Tipin-replication protein A interaction mediates Chk1 phosphorylation by ATR in response to genotoxic stress. J. Biol. Chem. 2010;285:16562–16571. doi: 10.1074/jbc.M110.110304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie S., Lu Y., Qian C. Structure of RPA32 bound to the N-terminus of SMARCAL1 redefines the binding interface between RPA32 and its interacting proteins. FEBS J. 2014;281:3382–3396. doi: 10.1111/febs.12867. [DOI] [PubMed] [Google Scholar]
- 33.Sirbu B.M., McDonald W.H., Cortez D. Identification of proteins at active, stalled, and collapsed replication forks using isolation of proteins on nascent DNA (iPOND) coupled with mass spectrometry. J. Biol. Chem. 2013;288:31458–31467. doi: 10.1074/jbc.M113.511337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baehr C.A., Huntoon C.J., Karnitz L.M. Glycogen synthase kinase 3 (GSK-3)-mediated phosphorylation of uracil N-glycosylase 2 (UNG2) facilitates the repair of floxuridine-induced DNA lesions and promotes cell survival. J. Biol. Chem. 2016;291:26875–26885. doi: 10.1074/jbc.M116.746081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ko R., Bennett S.E. Physical and functional interaction of human nuclear uracil-DNA glycosylase with proliferating cell nuclear antigen. DNA Repair (Amst.) 2005;4:1421–1431. doi: 10.1016/j.dnarep.2005.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakurai S., Kitano K., Hakoshima T. Structural basis for recruitment of human flap endonuclease 1 to PCNA. EMBO J. 2005;24:683–693. doi: 10.1038/sj.emboj.7600519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moldovan G.-L., Pfander B., Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 38.Gilljam K.M., Feyzi E., Otterlei M. Identification of a novel, widespread, and functionally important PCNA-binding motif. J. Cell Biol. 2009;186:645–654. doi: 10.1083/jcb.200903138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fanning E., Klimovich V., Nager A.R. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hedglin M., Benkovic S.J. Replication protein A prohibits diffusion of the PCNA sliding clamp along single-stranded DNA. Biochemistry. 2017;56:1824–1835. doi: 10.1021/acs.biochem.6b01213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ainavarapu S.R.K., Brujić J., Fernandez J.M. Contour length and refolding rate of a small protein controlled by engineered disulfide bonds. Biophys. J. 2007;92:225–233. doi: 10.1529/biophysj.106.091561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rippe K., Mücke N., Langowski J. Superhelix dimensions of a 1868 base pair plasmid determined by scanning force microscopy in air and in aqueous solution. Nucleic Acids Res. 1997;25:1736–1744. doi: 10.1093/nar/25.9.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.