Abstract
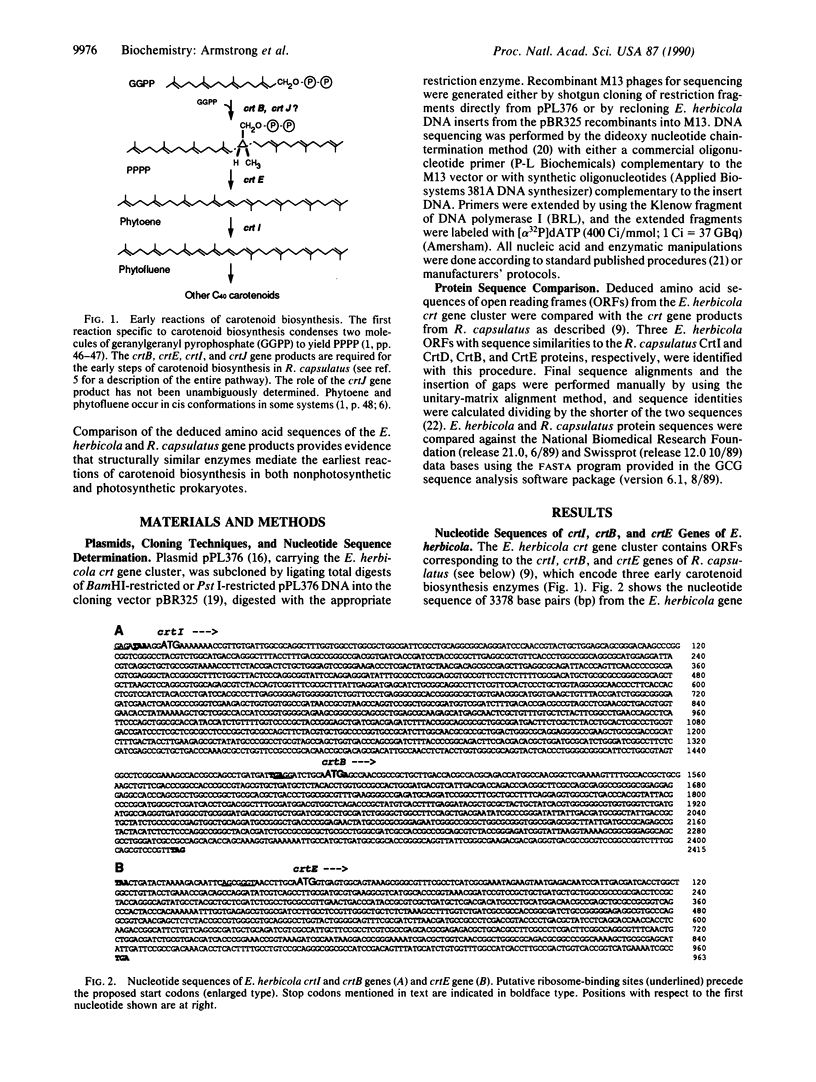
Carotenoids comprise one of the most widespread classes of pigments found in nature. The first reactions of C40 carotenoid biosynthesis proceed through common intermediates in all organisms, suggesting the evolutionary conservation of early enzymes from this pathway. We report here the nucleotide sequence of three genes from the carotenoid biosynthesis gene cluster of Erwinia herbicola, a nonphotosynthetic epiphytic bacterium, which encode homologs of the CrtB, CrtE, and CrtI proteins of Rhodobacter capsulatus, a purple nonsulfur photosynthetic bacterium. CrtB (prephytoene pyrophosphate synthase), CrtE (phytoene synthase), and CrtI (phytoene dehydrogenase) are required for the first three reactions specific to the carotenoid branch of general isoprenoid metabolism. The homologous proteins from E. herbicola and R. capsulatus show sequence identities of 41.7% for CrtI, 33.7% for CrtB, and 30.8% for CrtE. E. herbicola and R. capsulatus CrtI also display 27.2% and 27.9% sequence identity, respectively, with R. capsulatus CrtD (methoxyneurosporene dehydrogenase). All three dehydrogenases possess a hydrophobic N-terminal domain containing a putative ADP-binding beta alpha beta fold characteristic of enzymes known to bind FAD or NAD(P) cofactors. In addition, E. herbicola and R. capsulatus CrtB show 25.2% and 23.3% respective sequence identities with the protein product of pTOM5, a tomato cDNA of unknown function that is differentially expressed during fruit ripening. These data indicate the structural conservation of early carotenoid biosynthesis enzymes in evolutionarily diverse organisms.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armstrong G. A., Alberti M., Leach F., Hearst J. E. Nucleotide sequence, organization, and nature of the protein products of the carotenoid biosynthesis gene cluster of Rhodobacter capsulatus. Mol Gen Genet. 1989 Apr;216(2-3):254–268. doi: 10.1007/BF00334364. [DOI] [PubMed] [Google Scholar]
- Armstrong G. A., Schmidt A., Sandmann G., Hearst J. E. Genetic and biochemical characterization of carotenoid biosynthesis mutants of Rhodobacter capsulatus. J Biol Chem. 1990 May 15;265(14):8329–8338. [PubMed] [Google Scholar]
- Bartley G. E., Schmidhauser T. J., Yanofsky C., Scolnik P. A. Carotenoid desaturases from Rhodobacter capsulatus and Neurospora crassa are structurally and functionally conserved and contain domains homologous to flavoprotein disulfide oxidoreductases. J Biol Chem. 1990 Sep 15;265(26):16020–16024. [PubMed] [Google Scholar]
- Bartley G. E., Scolnik P. A. Carotenoid biosynthesis in photosynthetic bacteria. Genetic characterization of the Rhodobacter capsulatus CrtI protein. J Biol Chem. 1989 Aug 5;264(22):13109–13113. [PubMed] [Google Scholar]
- Bendich A. Carotenoids and the immune response. J Nutr. 1989 Jan;119(1):112–115. doi: 10.1093/jn/119.1.112. [DOI] [PubMed] [Google Scholar]
- Beyer P., Mayer M., Kleinig H. Molecular oxygen and the state of geometric isomerism of intermediates are essential in the carotene desaturation and cyclization reactions in daffodil chromoplasts. Eur J Biochem. 1989 Sep 1;184(1):141–150. doi: 10.1111/j.1432-1033.1989.tb15000.x. [DOI] [PubMed] [Google Scholar]
- Bolivar F. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique Eco RI sites for selection of Eco RI generated recombinant DNA molecules. Gene. 1978 Oct;4(2):121–136. doi: 10.1016/0378-1119(78)90025-2. [DOI] [PubMed] [Google Scholar]
- Dogbo O., Laferriére A., D'Harlingue A., Camara B. Carotenoid biosynthesis: Isolation and characterization of a bifunctional enzyme catalyzing the synthesis of phytoene. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7054–7058. doi: 10.1073/pnas.85.19.7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano G., Pollock D., Scolnik P. A. The gene crtI mediates the conversion of phytoene into colored carotenoids in Rhodopseudomonas capsulata. J Biol Chem. 1986 Oct 5;261(28):12925–12929. [PubMed] [Google Scholar]
- Krinsky N. I. Carotenoids and cancer in animal models. J Nutr. 1989 Jan;119(1):123–126. doi: 10.1093/jn/119.1.123. [DOI] [PubMed] [Google Scholar]
- Marrs B. Mobilization of the genes for photosynthesis from Rhodopseudomonas capsulata by a promiscuous plasmid. J Bacteriol. 1981 Jun;146(3):1003–1012. doi: 10.1128/jb.146.3.1003-1012.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry K. L., Simonitch T. A., Harrison-Lavoie K. J., Liu S. T. Cloning and regulation of Erwinia herbicola pigment genes. J Bacteriol. 1986 Nov;168(2):607–612. doi: 10.1128/jb.168.2.607-612.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray J., Bird C., Maunders M., Grierson D., Schuch W. Sequence of pTOM5, a ripening related cDNA from tomato. Nucleic Acids Res. 1987 Dec 23;15(24):10587–10587. doi: 10.1093/nar/15.24.10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidhauser T. J., Lauter F. R., Russo V. E., Yanofsky C. Cloning, sequence, and photoregulation of al-1, a carotenoid biosynthetic gene of Neurospora crassa. Mol Cell Biol. 1990 Oct;10(10):5064–5070. doi: 10.1128/mcb.10.10.5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A., Sandmann G., Armstrong G. A., Hearst J. E., Böger P. Immunological detection of phytoene desaturase in algae and higher plants using an antiserum raised against a bacterial fusion-gene construct. Eur J Biochem. 1989 Sep 15;184(2):375–378. doi: 10.1111/j.1432-1033.1989.tb15029.x. [DOI] [PubMed] [Google Scholar]
- Tuveson R. W., Larson R. A., Kagan J. Role of cloned carotenoid genes expressed in Escherichia coli in protecting against inactivation by near-UV light and specific phototoxic molecules. J Bacteriol. 1988 Oct;170(10):4675–4680. doi: 10.1128/jb.170.10.4675-4680.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierenga R. K., Drenth J., Schulz G. E. Comparison of the three-dimensional protein and nucleotide structure of the FAD-binding domain of p-hydroxybenzoate hydroxylase with the FAD- as well as NADPH-binding domains of glutathione reductase. J Mol Biol. 1983 Jul 5;167(3):725–739. doi: 10.1016/s0022-2836(83)80106-5. [DOI] [PubMed] [Google Scholar]
- Wierenga R. K., Terpstra P., Hol W. G. Prediction of the occurrence of the ADP-binding beta alpha beta-fold in proteins, using an amino acid sequence fingerprint. J Mol Biol. 1986 Jan 5;187(1):101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- Zsebo K. M., Hearst J. E. Genetic-physical mapping of a photosynthetic gene cluster from R. capsulata. Cell. 1984 Jul;37(3):937–947. doi: 10.1016/0092-8674(84)90428-8. [DOI] [PubMed] [Google Scholar]


