Abstract
Promoter hypermethylation of tumor suppressor genes has been confirmed to serve a pivotal role in tumorigenesis. Protocadherin 8 (PCDH8), a novel tumor suppressor gene, has been reported to be inactivated by promoter hypermethylation a number of cancer types, including bladder cancer and renal cell carcinoma. The aim of the present study was to investigate the occurrence of PCDH8 hypermethylation in prostate cancer and its potential as a novel biomarker of prostate cancer. The transcriptional levels of PCDH8 were examined by quantitative polymerase chain reaction (PCR) in 82 prostate cancer tissues as well as 30 prostate hyperplasia tissues, and verified the protein level by western blot analysis of representative samples. PCDH8 expression levels were found to be reduced to 0.30±0.10 in 70.7% (58/82) of prostate cancer tissues. To identify the possible reason for mRNA downregulation, the methylation status of the PCDH8 promoter was assessed in prostate cancer tissues and prostate hyperplasia tissues by methylation-specific PCR (MSP). A total of 47 prostate cancer patients who exhibited reduced PCDH8 expression (57.3%; 47/82) also showed promoter hypermethylation (47/58). None of the samples (0/30) in the benign prostate hyperplasia group were positive on MSP. Furthermore, the associations between the methylation status of the PCDH8 promoter and various clinicopathological features of prostate cancer were analyzed, revealing that the methylation status of PCDH8 was closely associated with tumor size, tumor shape (papillary/non-papillary), tumor stage and tumor grade (all P<0.05), while there were no correlations with the age of the patients or the number of tumors (P>0.05). Additionally, patients with hypermethylation of the PCDH8 gene promoter had a relapse rate of 36.17% and a mortality rate of 29.79%, which were significantly higher than the hypermethylation-negative patients (P<0.05), indicating a poorer prognosis. Therefore, the methylation status of the PCDH8 gene in prostate cancer may be an important marker for use in the early diagnosis and prediction of prognosis in prostate cancer.
Keywords: prostate cancer, protocadherin 8, promoter hypermethylation, prognosis
Introduction
Prostate cancer is the second leading cause of cancer-associated mortality in men worldwide, and the incidence of the disease is increasing year by year (1). Currently, the level of prostate-specific antigen, the clinical stage and the grade of the tumor (Gleason score) are the predominant methods for predicting the prognosis of prostate cancer (2). However, the epigenetic silencing of key tumor suppressors genes during prostate cancer tumorigenesis and progression may be translated into a promising clinical diagnostic marker (2). CpG islands at transcription initiation sites are generally poorly methylated. However, in certain cancer tissues, hypermethylation of CpG may silence gene transcription (3). In prostate cancer tissues, a number of large-scale genome-wide analyses have suggested that >30 key genes may be regulated through promoter hypermethylation (3). At present, there are still relatively few biomarkers for the prediction of prostate cancer (2).
Protocadherin 8 (PCDH8) is a member of the cadherin family and has been identified to be a novel tumor suppressor gene that is silenced in a number of cancer types, including gastric cancer, breast cancer, bladder cancer and renal cell carcinoma (4–7). Transcriptional silencing may occur via genetic mutation (5,6) or epigenetic promoter hypermethylation (4–7).
The present study analyzed the methylation status of the promoter of the PCDH8 gene in prostate cancer tissues, with the aim of identifying a potential association between the methylation status and the diagnosis and prognosis of prostate cancer.
Materials and methods
Patients and specimens
According to the prostate cancer diagnostic criteria of the World Health Organization and National Cancer Institute for prostate cancer (8), 82 patients with prostate cancer and 30 hospitalized patients with benign prostatic hyperplasia at Yantai Yuhuangding Hospital (Yantai, China) were enrolled between May 2010 and May 2012. The inclusion criteria were as follows: i) no radiation therapy, chemotherapy or hormone therapy administered prior to surgery; ii) general information, including clinical stage and histological grade, is available; iii) diagnosis of primary prostate cancer; iv) confirmed prostate cancer by pathological report. The exclusion criteria were as follows: i) history of other malignant tumors; ii) incomplete clinical data; iii) presence of remote metastatic or recurrent prostate cancer. The prostate cancer patients were aged between 37 and 77 years, with a mean age of 58.1±3.9 years. There were 34 cases with a single tumor, and 48 cases with multiple tumors. With regard to tumor size, 48 cases had a tumor diameter of ≤3 cm, and 34 cases had a diameter of >3 cm. A total of 57 cases were papillary and 25 cases were non-papillary. There were 55 cases of grade (G)1-G2, and 27 cases of G3. Regarding tumor stage, a total of 49 cases were Ta-T1 and 33 cases were T2-T4 (9,10).
DNA extraction
All surgical specimens were placed immediately in liquid nitrogen following the surgery. The specimens (~25 mg) were ground in liquid nitrogen and DNA was extracted using QIAamp DNA Mini Kit (Qiagen GmbH, Hilden, Germany; lot 130217) according to the manufacturer's instructions. The purity and concentration of DNA was measured using a DU800 ultraviolet (UV) spectrophotometer (Beckman-Coulter, Inc., Brea, CA, USA).
Methylation-specific polymerase chain reaction (MSP)
The methylation status of PCDH8 was determined by MSP (11). A total of 1 µg genomic DNA was modified by methylation using an EZ DNA Methylation-Gold Kit (UZymo Research Corporation, Irvine, CA, USA; lot 130522) according to the manufacturer's instructions. In this method, following the genomic DNA methylation reaction, unmethylated cytosine is converted to uracil, whereas methylated cytosine remains unchanged. Therefore, specific primers may be used to differentiate the methylation status. The primers were designed as previously described (12). All primers were synthesized by BayGene Biotech Company Limited (Beijing, China). The primers for the methylated reaction were as follows: Sense, 5′-CGGTTATTGGTTATTCGGTTCC-3′; and antisense, 5′-ACGAACTCTAAAAACGCGCG-3′; product size, 94 bp. The primers for the unmethylated reaction were as follows: Sense, 5′-GGTGGTTATTGGTTATTTGGTTT-3′; antisense, 5′-CCAACAAACTCTAAAAACACACA-3′; product size, 97 bp. Water was used as a blank control in each assay. PCR products (10 µl) were separated on a 2% agarose gel, stained with ethidium bromide and visualized under UV illumination. Specimens were scored as methylation-positive when a specific band was amplified by methylated reaction-specific primers, and negative when amplified only by unmethylated reaction-specific primers. The thermocycling conditions were as follows for a total of 40 cycles: Denaturing at 95°C for 30 sec, annealing at 60°C for 5 sec and extension at 72°C for 45 sec. The 2−ΔΔCq method (13) was used for the quantification of the PCR results.
Western blotting
Western blotting was performed to study the protein expression levels of PCDH8 in the clinical samples. A total of 1 mg tissue was solubilized in 100 µl lysis buffer (30 mM Tris, 2 M Thiourea, 4% CHAPS and 7 M urea; pH 8.5) on ice for 30 min. The lysis buffer was subsequently centrifuged at 1,000 × g for 15 min and the supernatant was collected for further testing. Primary antibodies were used for incubation at 4°C overnight and secondary antibodies at room temperature for 1 h. Primary antibodies targeting PCDH8 (1:1,000 dilution; catalog no. ab85561; Abcam, Cambridge, UK) and actin (1:1,000; rabbit; polyclonal; cat. no. ab8227; Abcam, Cambridge, UK) were used. The following secondary antibodies were used: Anti-rabbit IgG-biotin (cat. no. BA1020, Boster Biological Technology, Wuhan, China). Bands were detected using the DAB Chromogenic Reagent kit (cat. no. AR1021; Boster, Biological Technology).
Postoperative follow-up
Following surgery, patients were followed up for 6–24 months for the analysis of recurrence and survival.
Statistical analysis
All data were statistically analyzed using SPSS 18.0 (SPSS, Inc., Chicago, IL, USA). The different mRNA and protein expression between cancer tissues and controls were analyzed with the Student's t-test. For survival data, differences between two groups were assessed with the log-rank test. Kaplan-Meier curves of ovarall survival were constructed. The differences between clinicopathological features and the status of methylation were analyzed using the χ2 test. The data were presented as the mean ± standard deviation. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of PCDH8 is reduced in prostate cancer tissues
As a tumor suppressor, the expression of PCDH8 is reduced in various cancer types (4–7). Therefore, the present study investigated the expression levels of PCDH8 in prostate cancer tissues by quantitative polymerase chain reaction (qPCR). The results revealed that the mRNA level of PCDH8 was reduced to 0.30±0.10 in 70.7% (58/82) of prostate cancer tissues (Table I). Cases 22–25 are shown as representative samples for the mRNA results in Fig. 1. This result was also verified at the protein level by western blot analysis (Fig. 2). The mRNA and protein levels in cases 22 and 24, but not in cases 23 and 25, were reduced compared with the control (Table I; Figs. 1 and 2).
Table I.
Relative PCDH8 expression and methylation summary for patients with prostate cancer (n=82) or benign prostatic hyperplasia.
| Relative mRNA expression | |||||
|---|---|---|---|---|---|
| Group | Effect | Mean | SEM | P-value | MSP-positive |
| Control (C1-C3) | – | 1.00 | 0.11 | – | – |
| Cases with no PCDH8 reduction (n=24/82; 29.3%) | |||||
| P3 | N | 1.03 | 0.21 | N.S. | n |
| P4 | N | 1.21 | 0.14 | N.S. | n |
| P10 | N | 1.17 | 0.13 | N.S. | n |
| P14 | N | 1.08 | 0.22 | N.S. | n |
| P15 | N | 1.22 | 0.32 | N.S. | n |
| P23 | N | 1.12 | 0.21 | N.S. | n |
| P25 | N | 0.91 | 0.09 | N.S. | n |
| P31 | N | 1.08 | 0.28 | N.S. | n |
| P38 | N | 0.86 | 0.14 | N.S. | n |
| P39 | N | 0.85 | 0.23 | N.S. | n |
| P40 | N | 1.08 | 0.27 | N.S. | n |
| P45 | N | 0.88 | 0.14 | N.S. | n |
| P46 | N | 0.93 | 0.13 | N.S. | n |
| P47 | N | 1.21 | 0.37 | N.S. | n |
| P50 | N | 0.79 | 0.32 | N.S. | n |
| P55 | N | 0.86 | 0.09 | N.S. | n |
| P58 | N | 0.81 | 0.16 | N.S. | n |
| P59 | N | 1.08 | 0.11 | N.S. | n |
| P64 | N | 1.32 | 0.44 | N.S. | n |
| P73 | N | 1.21 | 0.13 | N.S. | n |
| P76 | N | 1.24 | 0.21 | N.S. | n |
| P78 | N | 0.86 | 0.16 | N.S. | n |
| P80 | N | 1.21 | 0.28 | N.S. | n |
| P81 | N | 0.94 | 0.08 | N.S. | n |
| Cases with PCDH8 reduction (n=58/82; 70.7%) | |||||
| P1 | R | 0.27 | 0.03 | <0.005 | y |
| P2 | R | 0.33 | 0.18 | <0.050 | y |
| P5 | R | 0.41 | 0.13 | <0.050 | n |
| P6 | R | 0.39 | 0.18 | <0.050 | y |
| P7 | R | 0.35 | 0.11 | <0.005 | y |
| P8 | R | 0.31 | 0.13 | <0.005 | y |
| P9 | R | 0.25 | 0.18 | <0.050 | n |
| P11 | R | 0.32 | 0.09 | <0.010 | y |
| P12 | R | 0.28 | 0.07 | <0.050 | y |
| P13 | R | 0.39 | 0.13 | <0.050 | n |
| P16 | R | 0.33 | 0.13 | <0.050 | y |
| P17 | R | 0.37 | 0.18 | <0.005 | y |
| P18 | R | 0.17 | 0.04 | <0.010 | y |
| P19 | R | 0.42 | 0.03 | <0.050 | n |
| P20 | R | 0.40 | 0.18 | <0.050 | y |
| P21 | R | 0.38 | 0.11 | <0.005 | y |
| P22 | R | 0.18 | 0.08 | <0.010 | y |
| P24 | R | 0.31 | 0.07 | <0.010 | y |
| P26 | R | 0.24 | 0.18 | <0.005 | y |
| P27 | R | 0.22 | 0.07 | <0.010 | n |
| P28 | R | 0.39 | 0.07 | <0.050 | y |
| Cases with PCDH8 reduction (n=58/82; 70.7%) | |||||
| P29 | R | 0.27 | 0.13 | <0.005 | y |
| P30 | R | 0.40 | 0.02 | <0.010 | n |
| P32 | R | 0.33 | 0.13 | <0.005 | y |
| P33 | R | 0.30 | 0.05 | <0.005 | y |
| P34 | R | 0.32 | 0.07 | <0.010 | y |
| P35 | R | 0.39 | 0.08 | <0.050 | y |
| P36 | R | 0.30 | 0.13 | <0.005 | y |
| P37 | R | 0.17 | 0.04 | <0.005 | n |
| P41 | R | 0.30 | 0.13 | <0.010 | y |
| P42 | R | 0.21 | 0.13 | <0.010 | y |
| P43 | R | 0.19 | 0.13 | <0.010 | y |
| P44 | R | 0.24 | 0.02 | <0.005 | y |
| P48 | R | 0.27 | 0.09 | <0.005 | y |
| P49 | R | 0.36 | 0.18 | <0.010 | n |
| P51 | R | 0.27 | 0.09 | <0.010 | y |
| P52 | R | 0.39 | 0.04 | <0.050 | y |
| P53 | R | 0.30 | 0.05 | <0.005 | y |
| P54 | R | 0.33 | 0.11 | <0.010 | y |
| P56 | R | 0.34 | 0.08 | <0.050 | y |
| P57 | R | 0.24 | 0.04 | <0.010 | y |
| P60 | R | 0.24 | 0.13 | <0.010 | n |
| P61 | R | 0.40 | 0.05 | <0.005 | y |
| P62 | R | 0.37 | 0.08 | <0.010 | y |
| P63 | R | 0.22 | 0.09 | <0.050 | y |
| P65 | R | 0.15 | 0.03 | <0.010 | y |
| P66 | R | 0.34 | 0.14 | <0.050 | y |
| P67 | R | 0.31 | 0.05 | <0.050 | y |
| P68 | R | 0.21 | 0.05 | <0.010 | n |
| P69 | R | 0.33 | 0.08 | <0.010 | y |
| P70 | R | 0.17 | 0.07 | <0.005 | y |
| P71 | R | 0.32 | 0.08 | <0.005 | y |
| P72 | R | 0.24 | 0.18 | <0.010 | y |
| P74 | R | 0.27 | 0.04 | <0.010 | n |
| P75 | R | 0.15 | 0.03 | <0.050 | y |
| P77 | R | 0.32 | 0.09 | <0.050 | y |
| P79 | R | 0.24 | 0.13 | <0.005 | y |
| P82 | R | 0.33 | 0.08 | <0.010 | y |
| Mean | – | 0.30 | 0.10 | – | – |
Samples P1-P82 were from 82 prostate cancer patients, and C1-C3 were from controls with benign prostatic hyperplasia. PCDH8, protocadherin 8; SEM, standard error of the mean; MSP, methylation-specific polymerase chain reaction; N, not affected; R, reduced; N.S., not significant; n, MSP-negative; y, MSP-positive.
Figure 1.
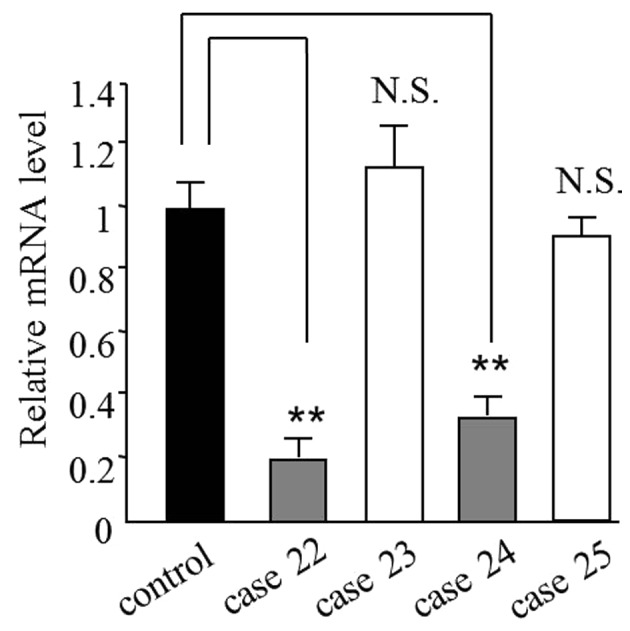
Representative mRNA levels of PCDH8 from 4 patients with prostate cancer and the control. The mRNA level of PCDH8 was reduced in cases 22 and 24. The controls were samples from patients with benign prostatic hyperplasia. **P<0.01 compared with control; N.S., not significant compared with control. PCDH8, protocadherin 8.
Figure 2.
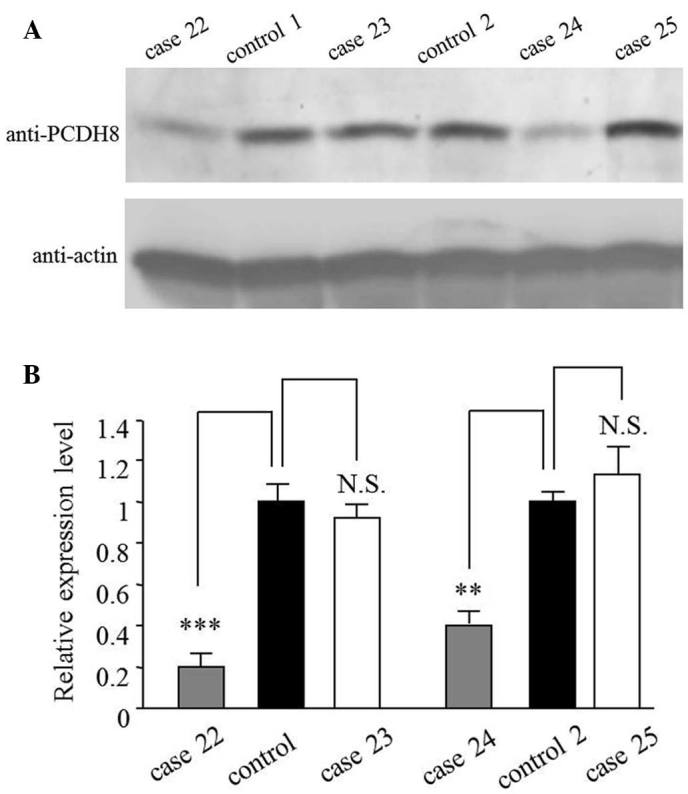
Protein levels of PCDH8 from 4 representative patients with prostate cancer and controls. (A) Western blot and (B) quantification of western blot. The protein level of PCDH8 was reduced in cases 22 and 24. **P<0.01 and ***P<0.005 compared with control; N.S., not significant compared with control. PCDH8, protocadherin 8.
Methylation status of PCDH8 gene promoter
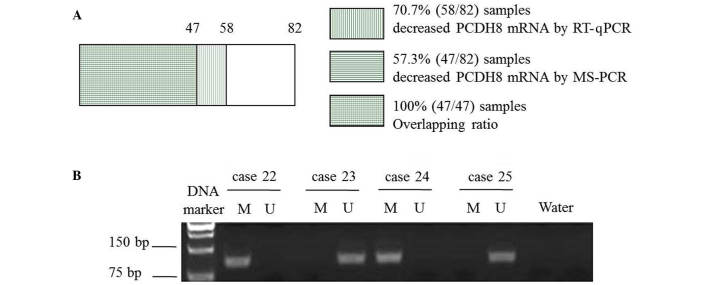
To identify the possible reason for the mRNA downregulation, the methylation status of the PCDH8 promoter was examined. The results revealed that the benign prostatic hyperplasia and prostate cancer samples that did not exhibit PCDH8 reduction were not methylated (Tables I and II). The 47 cases in which methylation was detected were all prostate cancer samples that exhibited PCDH8 reduction (Table I; Fig. 3A). Representative results from cases 22–25 are shown in Fig. 3B.
Table II.
Relative protocadherin 8 expression and methylation summary for controls with benign prostatic hyperplasia (n=30).
| Relative mRNA expression | |||||
|---|---|---|---|---|---|
| Case no. | Effect | Mean | SEM | P-value | MSP-positive |
| Controla | – | 1.00 | 0.11 | – | – |
| C1 | N | 1.07 | 0.13 | N.S. | n |
| C2 | N | 0.96 | 0.08 | N.S. | n |
| C3 | N | 1.17 | 0.12 | N.S. | n |
| C4 | N | 1.24 | 0.08 | N.S. | n |
| C5 | N | 1.22 | 0.22 | N.S. | n |
| C6 | N | 1.05 | 0.13 | N.S. | n |
| C7 | N | 1.21 | 0.21 | N.S. | n |
| C8 | N | 1.20 | 0.09 | N.S. | n |
| C9 | N | 1.06 | 0.16 | N.S. | n |
| C10 | N | 1.08 | 0.14 | N.S. | n |
| C11 | N | 1.17 | 0.14 | N.S. | n |
| C12 | N | 0.84 | 0.22 | N.S. | n |
| C13 | N | 1.04 | 0.28 | N.S. | n |
| C14 | N | 1.21 | 0.08 | N.S. | n |
| C15 | N | 1.24 | 0.21 | N.S. | n |
| C16 | N | 0.86 | 0.23 | N.S. | n |
| C17 | N | 1.21 | 0.19 | N.S. | n |
| C18 | N | 0.94 | 0.16 | N.S. | n |
| C19 | N | 1.10 | 0.11 | N.S. | n |
| C20 | N | 0.91 | 0.08 | N.S. | n |
| C21 | N | 1.23 | 0.20 | N.S. | n |
| C22 | N | 1.11 | 0.07 | N.S. | n |
| C23 | N | 0.89 | 0.19 | N.S. | n |
| C24 | N | 1.24 | 0.28 | N.S. | n |
| C25 | N | 1.20 | 0.10 | N.S. | n |
| C26 | N | 1.02 | 0.19 | N.S. | n |
| C27 | N | 0.98 | 0.05 | N.S. | n |
| C28 | N | 1.26 | 0.23 | N.S. | n |
| C29 | N | 1.14 | 0.16 | N.S. | n |
| C30 | N | 1.24 | 0.19 | N.S. | n |
Average of C1-C3. SEM, standard error of the mean; MSP, methylation-specific polymerase chain reaction; N, not affected; N.S., not significant; n, MSP-negative.
Figure 3.
(A) Summary of RT-PCR and MS-PCR results. 47 samples with PCDH8 methylation all showed decreased PCDH8 expression. The overlapping ratio was 100%. (B) Methylation statuses of PCDH8 from 4 representative patients with prostate cancer. Cases 22 and 24 exhibited PCDH8 promoter methylation. PCDH8, protocadherin 8; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; MS-PCR, methylation-specific polymerase chain reaction; M, methylated; U, unmethylated.
Analysis of the association between methylation status of PCDH8 and various clinicopathological features
The detailed clinicopathological features of patients with prostate cancer are shown in Table III. The results demonstrated that PCDH8 methylation was significantly associated with a larger tumor diameter (>3 vs. ≤3 cm; P=0.016), non-papillary shape (P=0.023), advanced tumor stage (T2-T4 vs. Ta-T1; P=0.016) and advanced pathological grade (G3 vs. G1-2; P=0.009). By contrast, no significant associations were identified between PCDH8 methylation and age (P=0.842) or number of tumors (P=0.500).
Table III.
Association between protocadherin 8 methylation and pathological features of patients with prostate cancer.
| Feature | Total, n | Unmethylated, n (%) | Methylated, n (%) | χ2 value | P-value |
|---|---|---|---|---|---|
| Age, years | 0.040 | 0.842 | |||
| ≤65 | 50 | 27 (54.00) | 23 (46.00) | ||
| >65 | 32 | 18 (56.25) | 14 (43.75) | ||
| Tumor number | 0.455 | 0.500 | |||
| Single | 34 | 16 (47.06) | 18 (52.94) | ||
| Multiple | 48 | 19 (39.58) | 29 (60.42) | ||
| Tumor diameter | 5.828 | 0.016 | |||
| ≤3 cm | 46 | 25 (54.35) | 21 (45.65) | ||
| >3 cm | 36 | 10 (27.78) | 26 (72.22) | ||
| Tumor shape | 5.131 | 0.023 | |||
| Papillary | 57 | 29 (50.88) | 28 (49.12) | ||
| Non-papillary | 25 | 6 (24.00) | 19 (76.00) | ||
| Tumor stage | 5.803 | 0.016 | |||
| Ta-T1 | 51 | 27 (52.94) | 24 (47.06) | ||
| T2-T4 | 31 | 8 (25.81) | 23 (74.19) | ||
| Pathological grade | 6.888 | 0.009 | |||
| G1-G2 | 55 | 29 (52.73) | 26 (47.27) | ||
| G3 | 27 | 6 (22.22) | 21 (77.78) |
Postoperative follow-up to assess the association between PCDH8 methylation and prognosis
Survival data were collected for 6–24 months after surgery. The data revealed that the methylation status of PCDH8 is associated with the overall survival time of patients with prostate cancer; those with unmethylated PCDH8 have a better prognosis (recurrence rate, 14.29; and mortality rate, 8.57%) compared with those with methylated PCDH8 (recurrence rate, 36.17%; mortality rate, 29.79%). These differences were statistically significant (recurrence rate, P=0.027; mortality rate, P=0.019) (Table IV; Fig. 4).
Table IV.
Postoperative follow-up analysis (n=82).
| Status | Total, n | Recurrence rate, n (%) | Mortality rate, n (%) |
|---|---|---|---|
| Methylated | 47 | 17 (36.17) | 14 (29.79) |
| Unmethylated | 35 | 5 (14.29) | 3 (8.57) |
| χ2 value | 4.894 | 5.495 | |
| P-value | 0.027 | 0.019 |
Figure 4.
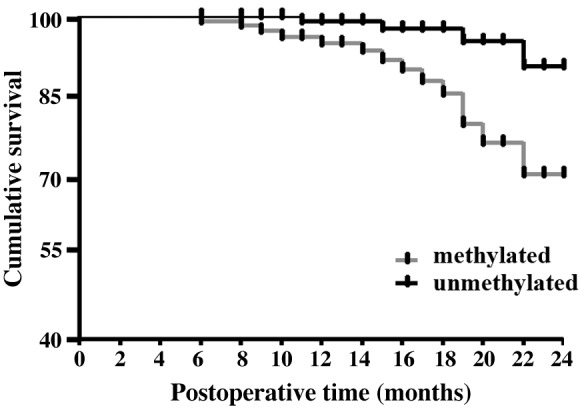
Kaplan-Meier survival curves for patients with different PCDH8 methylation statuses. The curves show the overall survival time.
Discussion
In recent years, the incidence of prostate cancer has been increasing. At present, a comprehensive treatment program consisting of radioactive seed implantation combined with radical prostatectomy has achieved remarkable results (14), particularly in those patients diagnosed early. However, a clinical survey found that distant metastasis occurs in 17–51% of prostate cancer cases, and local recurrence in 6–21% (15). Furthermore, the treatment of patients with metastasis and recurrence is often challenging, and the 5-year survival rate is <77% for such patients (16). Clinical studies have demonstrated that molecular markers, including growth hormone, vascular endothelial growth factor and transforming growth factor β1 may provide information relevant for prostate cancer diagnosis and treatment (10,15). For instance, studies have shown that patients with lymph node metastases had significantly higher expression of VEGF-C than patients without lymph node metastases (17). DNA methylation is a common form of epigenetic modification and changes in its status have been identified in multiple types of cancer. Hypermethylation of a gene inactivates its expression, which is important in the process of tumorigenesis (18). Therefore, the study of DNA methylation is significant for the diagnosis and prognosis of cancer.
PCDH8 is a recently identified member of the cadherin family (19). The protein is composed of an intracellular domain, a transmembrane domain and six repeating extracellular regions. Previous studies have revealed that PCDH8 is important in cytoskeleton formation, intercellular signaling, cell growth and differentiation (20). Létoquart et al found that the methylation of PCDH8 was higher in patients with breast cancer than in those with breast hyperplasia, and that the methylation status of PCDH8 is associated with the stage and metastasis of breast cancer (21). Heichman and Warren found that PCDH8 methylation occurs in pancreatic cancer, whereas there is no occurrence in normal human pancreatic tissues (22). These findings indicate that PCDH8 gene methylation may be closely associated with the occurrence and development of cancer (23). In the present study, the methylation status of the PCDH8 gene promoter was examined and the correlation between methylation status and prostate cancer progression was analyzed. The results revealed that in 82 cases of prostate cancer, PCDH8 gene promoter methylation was present in 57.32%, while no such methylation was present in benign prostatic hyperplasia samples; this is consistent with the results of clinical studies (24). These results demonstrate that the methylation of PCDH8 may be associated with the occurrence of prostate cancer and could potentially be used as a molecular marker for the early diagnosis of prostate cancer.
PCDH8 gene is a tumor suppressor gene that is downregulated in a variety of malignant tumors. To date, studies have found that the methylation of the PCDH8 promoter is an important factor for its downregulation (25,26). Inactivation of PCDH8 weakens its ability to suppress tumorigenesis. In the present study, in cases in which PCDH8 promoter hypermethylation was identified, a reduced mRNA level of these gene was also found. However, in certain cases that exhibited a reduced mRNA level, the promoter of PCDH8 was not hypermethylated, indicating that other mechanisms may regulate the expression of PCDH8.
PCDH8 was found to be methylated in prostate cancer patients with a larger tumor diameter (>3 cm) and advanced tumor stage (T2-T4) and pathological grade (G3) (27), indicating that PCDH8 methylation is significantly associated with the degree of differentiation and depth of invasion of the cancer. Furthermore, the data obtained in the present study revealed that the methylation of PCDH8 is highly associated with tumor shape, with the non-papillary form more common than the papillary form in cases showing promoter methylation. This difference may be related to the geographical location, test methods, sample size, individual differences and other factors (28).
In the present study, postoperative follow-up of the 82 patients revealed that patients with methylated PCDH8 had a poorer outcome compared with those with unmethylated PCDH8, indicating that methylation status may be associated with the prognosis of patients with prostate cancer. Considering the limited sample size of this study, a large multi-center study is necessary to confirm the findings.
In summary, the methylation of the PCDH8 gene promoter in prostate cancer is associated with the development and prognosis of prostate cancer and may be used as a molecular marker to determine the early diagnosis and prognosis of prostate cancer.
References
- 1.Javanmard B, Haddad A Hassanzadeh, Yaghoobi M, Lotfi B. Diode laser ablation of prostate and channel transurethral resection of prostate in patients with prostate cancer and bladder outlet obstruction symptoms. Urol J. 2014;11:1788–1792. [PubMed] [Google Scholar]
- 2.Park JY. Promoter hypermethylation in prostate cancer. Cancer Control. 2010;17:245–255. doi: 10.1177/107327481001700405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris MR, Ricketts CJ, Gentle D, McRonald F, Carli N, Khalili H, Brown M, Kishida T, Yao M, Banks RE, et al. Genome-wide methylation analysis identifies epigenetically inactivated candidate tumour suppressor genes in renal cell carcinoma. Oncogene. 2011;30:1390–1401. doi: 10.1038/onc.2010.525. [DOI] [PubMed] [Google Scholar]
- 5.Yu JS, Koujak S, Nagase S, Li CM, Su T, Wang X, Keniry M, Memeo L, Rojtman A, Mansukhani M, et al. PCDH8, the human homolog of PAPC, is a candidate tumor suppressor of breast cancer. Oncogene. 2008;27:4657–4665. doi: 10.1038/onc.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang D, Zhao W, Liao X, Bi T, Li H, Che X. !, !Frequent silencing of protocadherin 8 by promoter methylation, a candidate tumor suppressor for human gastric cancer. Oncol Rep. 2012;28:1785–1791. doi: 10.3892/or.2012.1997. [DOI] [PubMed] [Google Scholar]
- 7.Lin YL, Ma JH, Luo XL, Guan TY, Li ZG. Clinical significance of protocadherin-8 (PCDH8) promoter methylation in bladder cancer. J Int Med Res. 2013;41:48–54. doi: 10.1177/0300060513475571. [DOI] [PubMed] [Google Scholar]
- 8.Welch HG, Albertsen PC. Prostate cancer diagnosis and treatment after the introduction of prostate-specific antigen screening: 1986–2005. J Ntl Cancer Inst. 2009;101:1325–1329. doi: 10.1093/jnci/djp278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Partin AW, Mangold LA, Lamm DM, Walsh PC, Epstein JI, Pearson JD. Contemporary update of prostate cancer staging nomograms (Partin Tables) for the new millennium. Urology. 2001;58:843–848. doi: 10.1016/S0090-4295(01)01441-8. [DOI] [PubMed] [Google Scholar]
- 10.Ugolkov AV, Eisengart LJ, Luan C, Yang XJ. Expression analysis of putative stem cell markers in human benign and malignant prostate. Prostate. 2011;71:18–25. doi: 10.1002/pros.21217. [DOI] [PubMed] [Google Scholar]
- 11.Barfeld SJ, East P, Zuber V, Mills IG. Meta-analysis of prostate cancer gene expression data identifies a novel discriminatory signature enriched for glycosylating enzymes. BMC Med Genomics. 2014;7:513. doi: 10.1186/s12920-014-0074-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tong SJ, Liu J, Wang X, Qu LX. microRNA-181 promotes prostate cancer cell proliferation by regulating DAX-1 expression. Exp Ther Med. 2014;8:1296–1300. doi: 10.3892/etm.2014.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Frank SJ, Levy LB, van Vulpen M, Crook J, Sylvester J, Grimm P, Pugh TJ, Swanson DA. Outcomes after prostate brachytherapy are even better than predicted. Cancer. 2012;118:839–847. doi: 10.1002/cncr.26307. [DOI] [PubMed] [Google Scholar]
- 15.Monma F, Hozumi Y, Ikematsu S, Kawaguchi M, Kadomatsu K, Suzuki T. Expression of midkine in normal human skin, dermatitis and neoplasms: Association with differentiation of keratinocytes. J Dermatol. 2013;40:980–986. doi: 10.1111/1346-8138.12333. [DOI] [PubMed] [Google Scholar]
- 16.Tangen CM, Faulkner JR, Crawford ED, Thompson IM, Hirano D, Eisenberger M, Hussain M. Ten-year survival in patients with metastatic prostate cancer. Clin Prostate Cancer. 2003;2:41–45. doi: 10.3816/CGC.2003.n.011. [DOI] [PubMed] [Google Scholar]
- 17.Jennbacken K, Vallbo C, Wang W, Damber JE. Expression of vascular endothelial growth factor C (VEGF-C) and VEGF receptor-3 in human prostate cancer is associated with regional lymph node metastasis. Prostate. 2005;65:110–116. doi: 10.1002/pros.20276. [DOI] [PubMed] [Google Scholar]
- 18.Sheyhidin I, Hasim A, Zheng F, Ma H. Epigenetic changes within the promoter regions of antigen processing machinery family genes in Kazakh primary esophageal squamous cell carcinoma. Asian Pac J Cancer Prev. 2014;15:10299–10306. doi: 10.7314/APJCP.2014.15.23.10299. [DOI] [PubMed] [Google Scholar]
- 19.Yu JS, Koujak S, Nagase S, Li CM, Su T, Wang X, Keniry M, Memeo L, Rojtman A, Mansukhani M, et al. PCDH8, the human homolog of PAPC, is a candidate tumor suppressor of breast cancer. Oncogene. 2008;27:4657–4665. doi: 10.1038/onc.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bray NJ, Kirov G, Owen RJ, Jacobsen NJ, Georgieva L, Williams HJ, Norton N, Spurlock G, Jones S, Zammit S, et al. Screening the human protocadherin 8 (PCDH8) gene in schizophrenia. Genes Brain Behav. 2002;1:187–191. doi: 10.1034/j.1601-183X.2002.10307.x. [DOI] [PubMed] [Google Scholar]
- 21.Létoquart J, Huvelle E, Wacheul L, Bourgeois G, Zorbas C, Graille M, Heurgué-Hamard V, Lafontaine DL. Structural and functional studies of Bud23-Trm112 reveal 18S rRNA N7-G1575 methylation occurs on late 40S precursor ribosomes; Proc Natl Acad Sci USA; 2014; pp. E5518–E5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heichman KA, Warren JD. DNA methylation biomarkers and their utility for solid cancer diagnostics. Clin Chem Lab Med. 2012;50:1707–1721. doi: 10.1515/cclm-2011-0935. [DOI] [PubMed] [Google Scholar]
- 23.Redshaw N, Huggett JF, Taylor MS, Foy CA, Devonshire AS. Quantification of epigenetic biomarkers: An evaluation of established and emerging methods for DNA methylation analysis. BMC Genomics. 2014;15:1174. doi: 10.1186/1471-2164-15-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LePage DP, Jernigan KK, Bordenstein SR. The relative importance of DNA methylation and Dnmt2-mediated epigenetic regulation on Wolbachia densities and cytoplasmic incompatibility. PeerJ. 2014;2:e678. doi: 10.7717/peerj.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng L, Wei H, Liren L. Promoter methylation assay of SASH1 gene in hepatocellular carcinoma. J BUON. 2014;19:1041–1047. [PubMed] [Google Scholar]
- 26.Jeffries MA, Sawalha AH. Autoimmune disease in the epigenetic era: How has epigenetics changed our understanding of disease and how can we expect the field to evolve? Expert Rev Clin Immunol. 2015;11:45–58. doi: 10.1586/1744666X.2015.994507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sircar K, Huang H, Hu L, Cogdell D, Dhillon J, Tzelepi V, Efstathiou E, Koumakpayi IH, Saad F, Luo D, et al. Integrative molecular profiling reveals asparagine synthetase is a target in castration-resistant prostate cancer. Am J Pathol. 2012;180:895–903. doi: 10.1016/j.ajpath.2011.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yi JM, Dhir M, Guzzetta AA, Iacobuzio-Donahue CA, Heo K, Yang KM, Suzuki H, Toyota M, Kim HM, Ahuja N. DNA methylation biomarker candidates for early detection of colon cancer. Tumour Biol. 2012;33:363–372. doi: 10.1007/s13277-011-0302-2. [DOI] [PMC free article] [PubMed] [Google Scholar]