Stool consistency and frequency patterns are complex traits that are often altered in GI disease, and recent studies published in Gut highlight the importance of stool frequency in relation to gut microbiota composition and the efficacy of pharmacological and dietary treatments in IBS.1–3
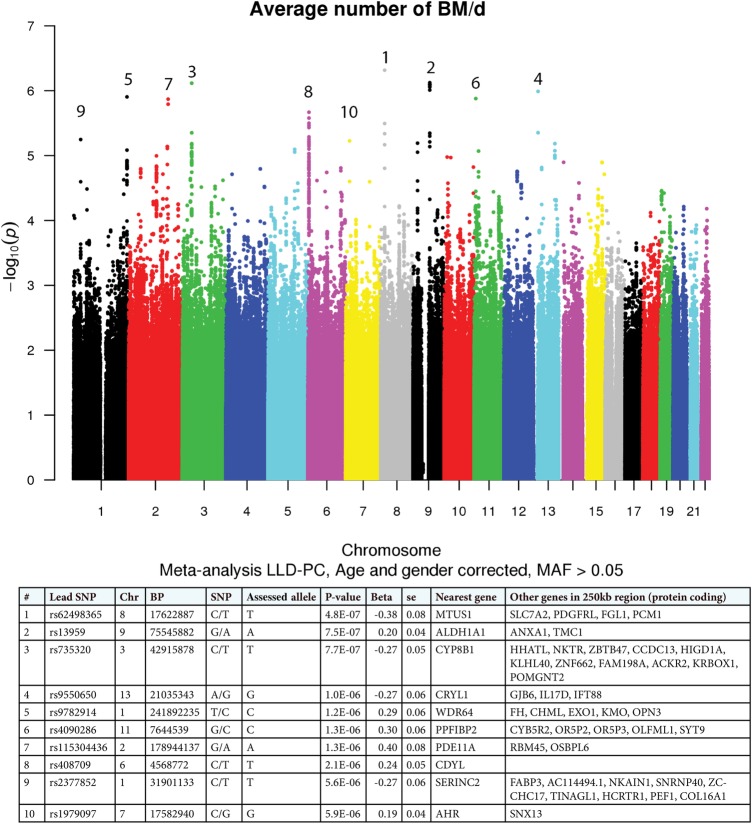
Despite reported heritability in invertebrates4 and similar evidence from open-field defaecation models in rats,5 the genetics of stool frequency has not been explored in humans. We undertook a genome-wide association study (GWAS) in two well-characterised population-based cohorts with genotype and defaecation data available: LifeLines-Deep (LLD) from the Netherlands (N=1546; 58% females; mean age 44 years (range 18–86)) and PopCol (PC) from Sweden (N=284; 60% females; mean age 54 years (range 22–71)).6 7 The average number of bowel movements per day (BM/d) was extracted from daily records kept by both populations and did not differ between cohorts (LLD=1.39±0.64SD; PC=1.42±0.74SD). Available CytoChip+Immunochip (LLD) and HumanOmniExpressExome (PC) Illumina single-nucleotide polymorphism (SNP) genotype data were imputed using IMPUTE2 (https://mathgen.stats.ox.ac.uk/impute/impute_v2.html) with the Genome of the Netherlands (http://www.nlgenome.nl/) as reference. SNPs were filtered on minor allele frequency >0.05 and Hardy–Weinberg equilibrium p>1E-04, samples were filtered on infoscore ≥0.8 and population outliers were excluded using principal component analysis. In total, high-quality genotype data for 5 390 800 common SNPs and BM/d information were obtained for 1022 LLD and 259 PC individuals. Genotype–BM/d association tests were performed in SNPTEST (https://mathgen.stats.ox.ac.uk/genetics_software/snptest/snptest.html) using logistic regression under an additive model correcting for age and sex, followed by a fixed-effect model meta-analysis with META (https://mathgen.stats.ox.ac.uk/genetics_software/meta/meta.html). Summary statistics for the top-10 loci from the meta-analysis and the corresponding effect of associated alleles on the frequency (increased/decreased) of defaecation are given in figure 1.
Figure 1.
Manhattan plot of the results from the meta-analysis of LifeLines-Deep (LLD) and PopCol (PC) genome-wide association studies. Single-nucleotide polymorphisms (SNPs) which are sorted according to their genomic positions are displayed on the X-axis, and the negative logarithm of the association p value for each SNP after meta-analysis is displayed on the Y-axis; each dot represents a SNP with a certain p value. The top-10 loci are indicated by numbers. Per locus, the statistics of the lead SNPs are shown, including the positions in the genome, the nearest genes and the genes in a 250 kb window around the lead SNPs. The effect of the assessed allele at each locus is indicated by beta; negative betas mean negative effect on the average number of bowel movements per day (BM/d) (decreased number of stool passes) and positive betas mean positive effect on the average number of BM/d (increased number of stool passes). Beta, direction of association; BP, base pair position; Chr, chromosome; SE, SE of the beta.
Although none of these associations achieved genome-wide significance (possibly due to limited sample size), we found excellent functional candidates mapping to these regions. For instance, the second strongest signal included the ALDH1A1 gene, which belongs to the family of aldehyde dehydrogenases, and another member of this family (ALDH1AL1) has been shown to affect human gut microbiota composition.8 Moreover, Gene Network coexpression analysis (http://www.genenetwork.nl/genenetwork/) indicated a role for ALDH1A1 in the cytochrome P450 metabolism of drugs and xenobiotics, and other genes in this pathway also map to top BM/d GWAS loci: the rs735320 signal comes from SNPs in the CYP8B1 gene, which belongs to the cytochrome P450 family; the rs4090 286 locus contains CYB5R2 (cytochrome B5 reductase), which is involved in cholesterol biosynthesis, fatty acid desaturation and elongation; and the rs1979097 locus contains AHR (ligand-activated aryl hydrocarbon receptor), which is a transcription factor modulating gene expression along the cytochrome P450 pathway. The genetic implication of xenobiotic and P450 metabolic pathways is not unexpected, given the interactions linking diet, gut microbiome and pharmaceutical compounds to known effects on human defaecation patterns, but was not reported previously.
A broader pathway analysis (http://129.125.135.180:8080/GeneNetwork/pathway.html), including genes from all 53 loci with p<5E-05, identified the sodium channel complex and voltage-gated sodium channel activity as the most enriched pathways in Gene Ontology (GO) terms for cellular component and molecular function, respectively (table 1). This is remarkable, since genetic defects in the voltage-gated channel SCN5A have been found in a subset of patients with IBS, and normal stool frequency was restored in a severely constipated SCN5A mutant carrier treated with mexiletine, a drug able to rescue SCN5A expression.9
Table 1.
Pathway analysis of GWAS meta-analysis results for the average number of BM/d
| Top associated GO term | p Value |
|---|---|
| Cellular component | |
| Sodium channel complex | 6E-07 |
| Sarcolemma | 2E-05 |
| Voltage-gated sodium channel complex | 2E-05 |
| Ion channel complex | 6E-05 |
| Molecular function | |
| Voltage-gated sodium channel activity | 2E-05 |
| Peptidase regulator activity | 3E-05 |
| Substrate-specific channel activity | 4E-05 |
| Ion channel activity | 4E-05 |
| Passive transmembrane transporter activity | 7E-05 |
| Channel activity | 7E-05 |
| Endopeptidase inhibitor activity | 8E-05 |
Only GO pathways with p<1E-04 are reported.
BM/d, bowel movements per day; GO, Gene Ontology; GWAS, genome-wide association study.
In conclusion, we report the first GWAS of stool frequency in two harmonised population-based cohorts from the Netherlands and Sweden and highlight plausible candidate genes and biological pathways. Although we are not aware of similar datasets in which our findings may be replicated, the growing interest in this research area warrants larger studies to reach unequivocal conclusions.
Acknowledgments
The authors thank Jackie Senior for editing the text.
Footnotes
Contributors: AZ, MD'A and CW designed the study. AZ, EFT, JF, AA, LA, SW and CW initiated the cohort and collected cohort data. SAJ, AA, LA, SW and EFT generated data. FH, MJB, PD, SAJ, JF and PH analysed data. MD'A, AZ, SAJ, PH and CW wrote the manuscript.
Funding: This work was funded by grants from the Top Institute Food and Nutrition, Wageningen, to CW (GH001), the Netherlands Organization for Scientific Research to JF (NWO-VIDI 864.13.013) and the Swedish Research Council (VR) to MD’A. AZ holds a Rosalind Franklin fellowship (University of Groningen).
Competing interests: None declared.
Patient consent: Obtained.
Ethics approval: The LifeLines-DEEP study was approved by the ethics committee of the University Medical Center Groningen, the Netherlands (document no. METC UMCG LLDEEP: M12.113965). The PopCol study was approved by Karolinska Institutet's ethics committee, Stockholm, Sweden (dnr 394/01). All participants signed an informed consent form prior to study enrolment.
Provenance and peer review: Not commissioned; internally peer reviewed.
References
- 1.Hadizadeh F, Walter S, Belheouane M, et al. . Stool frequency is associated with gut microbiota composition. Gut Published Online First: 28 Apr 2016. 10.1136/gutjnl-2016-311935 [DOI] [PubMed] [Google Scholar]
- 2.Tack J, Schumacher K, Tonini G, et al. . The neurokinin-2 receptor antagonist ibodutant improves overall symptoms, abdominal pain and stool pattern in female patients in a phase II study of diarrhoea-predominant IBS. Gut Published Online First: 15 Apr 2016. 10.1136/gutjnl-2015-310683 [DOI] [PubMed] [Google Scholar]
- 3.McIntosh K, Reed DE, Schneider T, et al. . FODMAPs alter symptoms and the metabolome of patients with IBS: a randomized controlled trial. Gut Published Online First: 14 Mar 2016. 10.1136/gutjnl-2015-311339 [DOI] [PubMed] [Google Scholar]
- 4.Branicky R, Hekimi S. What keeps C. elegans regular: the genetics of defecation. Trends Genet 2006;22:571–9. [DOI] [PubMed] [Google Scholar]
- 5.Blizard DA, Adams N. The Maudsley reactive and nonreactive strains: a new perspective. Behav Genet 2002;32:277–99. 10.1023/A:1020206120248 [DOI] [PubMed] [Google Scholar]
- 6.Tigchelaar EF, Zhernakova A, Dekens JAM, et al. . Cohort profile: LifeLines DEEP, a prospective, general population cohort study in the northern Netherlands: study design and baseline characteristics. BMJ Open 2015;5:e006772 10.1136/bmjopen-2014-006772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walter SA, Kjellström L, Nyhlin H, et al. . Assessment of normal bowel habits in the general adult population: the Popcol study. Scand J Gastroenterol 2010;45:556–66. 10.3109/00365520903551332 [DOI] [PubMed] [Google Scholar]
- 8.Goodrich JK, Davenport ER, Beaumont M, et al. . Genetic determinants of the Gut Microbiome in UK Twins. Cell Host Microbe 2016;19:731–43. 10.1016/j.chom.2016.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beyder A, Mazzone A, Strege PR, et al. . Loss-of-function of the voltage-gated sodium channel NaV1.5 (channelopathies) in patients with irritable bowel syndrome. Gastroenterology 2014;146:1659–68. 10.1053/j.gastro.2014.02.054 [DOI] [PMC free article] [PubMed] [Google Scholar]